A Glutamine/Asparagine-Rich Fragment of Gln3, but not the Full-Length Protein, Aggregates in Saccharomyces cerevisiae
K. S. Antonets1,2, H. M. Sargsyan1, and A. A. Nizhnikov1,2,3*
1St. Petersburg State University, Department of Genetics and Biotechnology, 199034 St. Petersburg, Russia; E-mail: ant.nizhnikov@gmail.com2Vavilov Institute of General Genetics, St. Petersburg Branch, Russian Academy of Sciences, 199034 St. Petersburg, Russia
3All-Russia Research Institute for Agricultural Microbiology, 196608 Pushkin, St. Petersburg, Russia
* To whom correspondence should be addressed.
Received October 14, 2015; Revision received December 16, 2015
The amino acid sequence of protein Gln3 in yeast Saccharomyces cerevisiae has a region enriched with Gln (Q) and Asn (N) residues. In this study, we analyzed the effects of overexpression of Gln3 and its Q/N-rich fragment fused with yellow fluorescent protein (YFP). Being overexpressed, full-length Gln3-YFP does not form aggregates, inhibits vegetative growth, and demonstrates nuclear localization, while the Q/N-rich fragment (Gln3QN) fused with YFP forms aggregates that do not colocalize with the nucleus and do not affect growth of the cells. Although detergent-resistant aggregates of Gln3QN are formed in the absence of yeast prions, the aggregation of Gln3QN significantly increases in the presence of [PIN+] prion, while in the presence of two prions, [PSI+] and [PIN+], the percentage of cells with Gln3QN aggregates is significantly lower than in the strain bearing only [PIN+]. Data on colocalization demonstrate that this effect is mediated by interaction between Gln3QN aggregates and [PSI+] and [PIN+] prions.
KEY WORDS: amyloid, prion, Gln3, Rnq1, Sup35, yeast, S. cerevisiaeDOI: 10.1134/S0006297916040118
Abbreviations: CFP, cyan fluorescent protein; DAPI, 4′,6-diamidino-2-phenylindole (a fluorescent dye specific to AT-rich regions of DNA); Gln3QN, asparagine-glutamine-rich fragment of Gln3 protein (a.a. 166-242); [PIN+], prion isoform of Rnq1 protein; [PSI+], prion isoform of Sup35 protein; SDD-AGE, Semi-Denaturing Detergent Agarose Gel Electrophoresis; Sup35NM, prion-forming region of Sup35 protein lacking the C-terminal domain functioning as a translation release factor; YFP, yellow fluorescent protein.
Protein fibrils with the cross-beta structure are called amyloids. The
term “cross-beta” structure implies that protein monomers
form intermolecular beta-sheets that are stabilized by numerous
hydrogen bonds and located near perpendicular to the fibril axis [1]. Such organization of amyloid fibrils makes them
extremely stable and ensures their resistance to solubilization with
ionic detergents such as sodium dodecyl sulfate and sodium lauryl
sarcosinate. Amyloids have been found in all the three domains of the
living world, and they are known to play both pathogenic and functional
roles [1]. Dozens of incurable human and animal
diseases called amyloidoses are associated with amyloid aggregation of
certain proteins [2]. On the other hand, amyloids
are necessary for polymerization of melanin [3] and
storage of hormones in mammals [4], formation of
the long-term memory in mollusks [5] and insects
[6], and production of biofilms in bacteria [1, 7] and archaea [8], as well as for some other functions in various
organisms [1].
Infectious amyloids, or prions, are a special group. The first prion detected, PrPSc [9], is responsible for some lethal neurodegenerative amyloidoses in mammals including humans. The most diverse prions are described for the yeast Saccharomyces cerevisiae. This organism is known to have at least eight prions: [PSI+] [10], [URE3] [11], [PIN+] [12], [SWI+] [13], [OCT+] [14], [MOT3+] [15], [ISP+] [16], and [MOD+] [17], as well as a prion-like determinant [NSI+] [18, 19] whose structural protein has not yet been identified but has been shown to be involved in the regulation of mRNA [20-22]. Amino acid sequences of structural proteins of nearly all yeast prions have a property in common: they are rich in asparagine (N) and glutamine (Q). It is well known that Q/N-rich sequences are prone to produce amyloid-like aggregates. However, various Q/N-rich sequences can either produce or not produce aggregates in vivo [15]. Features determining the capacity of such sequences to aggregate are not yet established, and it is also unclear how amino acid sequences surrounding a Q/N-rich region in a full-size protein molecule can influence aggregation.
Gln3 is a transcriptional regulator of nitrogen catabolism [23-25] and one of the richest in Q and N proteins in the yeast proteome. It was shown earlier that the Gln3 Q/N-rich fragment fused with YFP (yellow fluorescent protein) (Gln3QN-YFP) can form detergent-resistant aggregates when overexpressed in the presence of the [PIN+] prion. Moreover, being fused with reporter constructs, this fragment has features of a prion [15]; therefore, the full-size protein Gln3 is one of the most probable candidates for novel prion-forming proteins of S. cerevisiae. In the present work, we compared the ability of the full-length Gln3 protein and of its Q/N-rich fragment to aggregate and analyzed the dependence of this aggregation on the yeast prions, [PSI+] and [PIN+].
MATERIALS AND METHODS
Strains of microorganisms, media, and culture conditions. In the present work, routine methods of S. cerevisiae genetic manipulations were used [26]. The yeast was cultured at 30°C on solid and liquid complete medium YAPD or on selective medium MD [27]. To activate expression of genetic constructs controlled by the promoter CUP1, the media were supplemented with 150 µM copper sulfate. Genotypes of the yeast strains used in the work are listed in Table 1. Strain 2-D-701 [PSI+][pin–] was obtained as a result of selection by the corresponding selective markers of haploid segregants in the progeny of the diploid strain resulting on crossing of strains 2-OT56 and 9-10-7A-D832.
Table 1. Strains of S. cerevisiae
The plasmid DNA was amplified using the Escherichia coli DH5α strain. The bacteria were grown on solid medium LB at 37°C [28].
Plasmids. Plasmids used in the work are described in Table 2. The pL-GPD-Sup35NM-CFP and pU-GPD-Sup35NM-YFP plasmids encoding the prion-forming region of protein Sup35 (Sup35NM) fused with cyan (CFP) or yellow (YFP) fluorescent protein were prepared from the plasmid pGPD-PrP23-CFP(LEU2) or from pGPD-PrP23-YFP(URA3) [32, 33], respectively, as by substitution at sites BamHI and SacII of the sequence encoding PrP23-231 with sequence Sup35NM amplified by polymerase chain reaction (PCR) with primers Sup35NFBam and Sup35MRSII (Table 3).
Table 2. Plasmids
* cen – centromeric plasmid.
Table 3. Primers

The plasmids pU-CUP1-YFP and pL-CUP1-YFP carry the sequence encoding YFP under control of promoter CUP1. In these plasmids, between the YFP sequence and promoter, there are restriction sites HindIII and BamHI allowing insertion of the needed gene fragments flanked by these restriction sites without frame shift. The pU-CUP1-YFP plasmid was prepared based on vector pRS316, in which the promoter CUP1 sequence amplified by PCR with primers Cup1SalIF and Cup1Hind3R (Table 3), flanked by SalI and HindIII sites, was initially inserted, and then the YFP sequence cut from the plasmid pGPD-YFP(URA3) [32] by the BamHI and SacI sites was inserted. Plasmid pLCup1-YFP was prepared based on vector pRS315 by inserting the CUP1-YFP sequence from plasmid pU-CUP1-YFP by the SalI and SacI sites.
Plasmid pU-CUP1-GLN3-YFP carrying the chimeric gene GLN3-YFP controlled by an inducible promoter of gene CUP1 was constructed by inserting the PCR-fragment of GLN3 amplified with primers GLN3HindIIIF and GLN3YFPBamHIR (Table 3) and the genomic DNA of strain OT56 as a template into the plasmid pU-CUP1-YFP by the restriction sites HindIII and BamHI.
The plasmids pU-CUP1-GLN3QN-YFP and pL-CUP1-GLN3QN-YFP carrying the chimeric gene encoding the fragment Gln3, a.a. 166-242, fused with YFP were obtained by inserting the PCR-fragment GLN3QN amplified with primers GLN3QNHindIIIF and GLN3QNBamHIR (Table 3) by the sites HindIII and BamHI into the plasmids pU-CUP1-YFP and pL-CUP1-YFP, respectively.
Analysis of protein aggregation. Semi-Denaturing Detergent Agarose Gel Electrophoresis (SDD-AGE) [34, 35] was performed as described in [32] with the following modifications: we used 1% agarose gel, and samples were treated with ionic detergent sodium lauryl sarcosinate (3%) for 10 min at room temperature. The proteins were transferred onto Immobilon-P PVDF-membrane (GE Healthcare, USA). Proteins fused with YFP were detected using monoclonal primary rabbit antibodies against GFP [E385] (ab32146) (Abcam, Great Britain) and an Amersham ECL Prime Western Blotting Detection Reagent kit (GE Healthcare).
Microscopy. Aggregation and colocalization of proteins fused with CFP and YFP were analyzed using a Leica TCS SP5 laser scanning confocal microscope (Leica Microsystems, Germany). Fluorescence of proteins fused with CFP was analyzed using a 458-nm argon laser (signal detection at 461-510 nm), and proteins fused with YFP were analyzed with a 514-nm argon laser (signal detection at 518-580 nm); the signal of the DNA-specific dye DAPI (4′,6-diamidino-2-phenylindole) was analyzed with a 405-nm argon laser in the UV range (detection at 425-475 nm). The yeast cells were stained with DAPI using a VECTASHIELD Antifade Mounting Medium with DAPI (1.5 µg/ml DAPI) according to the producer’s protocol (Vector Laboratories, USA). For microscopic analysis, yeast cultures were grown for 48 h at 30°C to induce aggregation of the studied proteins.
Statistical analysis. The samples were compared using the non-parametric Mann–Whitney test using the Statistica 6.0 program (StatSoft, USA). To study frequencies of aggregate production by confocal microscopy, five random visual fields were analyzed for five independent transformants. The fraction of cells containing aggregates was determined for each visual field.
To analyze the colocalization of proteins fused with CFP and YFP, a search was performed for cells with aggregates formed by both proteins. The colocalization frequency was determined as the ratio of the number of cells with co-localizing aggregates to the total number of cells containing aggregates of both analyzed proteins. The colocalization frequencies were evaluated for each pair of proteins analyzed in five independent transformants, and for each of them at least fifteen cells were analyzed.
RESULTS
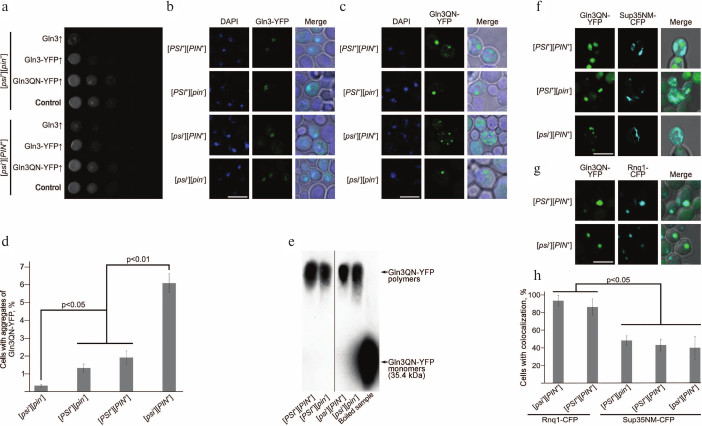
Overexpression of Gln3 and Gln3-YFP is toxic for cells, in contrast to overexpression of Gln3QN-YFP. We found earlier that overexpression of GLN3 had a pleiotropic phenotypic manifestation strongly inhibiting vegetative growth on media containing glucose or galactose as carbon sources and causing nonsense-suppression on the background of mutant variants of SUP35 [30]. We compared effects of overexpression of Gln3, Gln3-YFP, and Gln3QN-YFP in strains 2-OT56 [psi–][pin–] and 1-OT56 [psi–][PIN+]. Inoculums of overnight cultures of strains 1-OT56 and 2-OT56 transformed with constructed plasmids pU-CUP1-GLN3-YFP, pU-CUP1-GLN3, pU-CUP1-GLN3QN-YFP, and pRS316 were matched in optical density and grown for 48 h in liquid selective medium. Then a series of successive tenfold dilutions of the cultures were planted onto solid selective medium supplemented with 150 µM copper sulfate. Results of the experiment (figure, panel (a)) showed that overexpression of Gln3QN-YFP had no influence on vegetative growth, but overexpression of the full-length Gln3 on the genetic background of strain OT56 derivatives led to very strong sublethal inhibition of vegetative growth, which was independent on the status of [PIN+] prion. In the case of Gln3-YFP, the sublethal effect of overexpression was weaker than the same for Gln3, but the inhibition of vegetative growth remained rather strong (figure, panel (a)). Since aggregation of proteins is usually followed by their functional inactivation, these findings suggested that the full-length Gln3 does not aggregate when overproduced and retained functional activity and the ability to inhibit the yeast growth.
Effects of overexpression of Gln3 and Gln3QN fused with YFP and analysis of colocalization of Gln3QN-YFP aggregates with aggregates of Sup35 and Rnq1 fused with CFP. a) Analysis of the influence of overexpression of different Gln3 variants on the vegetative growth of yeast. Series of tenfold dilutions of cultures of strains 2-OT56 [psi–][pin–] and 1-OT56 [psi–][PIN+] overexpressing Gln3, Gln3-YFP, and Gln3QN-YFP. The strains transformed with pRS316 empty vector were used as a negative control. The cultures were photographed after 48 h of incubation of the cells at 30°C on selective medium supplemented with 150 µM copper sulfate. Laser scanning confocal microscopy of Gln3-YFP (b) and Gln3QN-YFP (c), as well as frequencies of cells with fluorescent aggregates of Gln3QN-YFP (d) in the yeast strains different in [PSI] and [PIN] status. Microscopy and counting of frequencies were performed after 48 h of incubation of the cells at 30°C on selective medium supplemented with 150 µM copper sulfate. The scale bar corresponds to 5 µm. e) Analysis of Gln3QN-YFP aggregation using SDD-AGE with pretreatment of the samples with 3% sodium lauryl sarcosinate. The boiled sample prepared from strain [psi–][PIN+] overexpressing Gln3QN-YFP was used as the negative control. f, g) Analysis of Gln3QN-YFP colocalization with Sup35NM-CFP and Rnq1-CFP, respectively, in the yeast strains different in [PSI] and [PIN] status. h) Frequencies of Gln3QN-YFP colocalization with Sup35NM-CFP and Rnq1-CFP. Laser scanning confocal microscopy and counting of colocalization frequencies were performed after 48 h of incubation of the cells at 30°C on selective medium supplemented with 150 µM copper sulfate. The scale bar corresponds to 5 µm
Frequency of Gln3QN-YFP aggregation depends on prions [PIN+] and [PSI+]. To analyze the ability of Gln3 and Gln3QN fused with YFP to produce aggregates in prion-free yeast strains and in the presence of the prions [PIN+] and/or [PSI+], we used laser scanning confocal microscopy. The strains OT56 [PSI+][PIN+], 2-D-701 [PSI+][pin–], 1-OT56 [psi–][PIN+], and 2-OT56 [psi–][pin–] were transformed with plasmids pU-CUP1-GLN3QN-YFP or pU-CUP1-GLN3-YFP to obtain overexpression of Gln3QN-YFP and Gln3-YFP, respectively. Transformants were chosen on selective medium supplemented with 150 µM copper sulfate, and protein aggregation was analyzed by confocal microscopy 48 h later.
The findings revealed that the full-size Gln3 fused with YFP did not aggregate when overexpressed in all the strains and was colocalized with the nucleus in the majority of the cells (figure, panel (b)). However, its Q/N-rich fragment, a.a. 166-242, fused with the YFP formed clearly distinctive aggregates in the strains carrying [PSI+] and/or [PIN+] and also in the strain [psi–][pin–] (figure, panel (c)). These aggregates had morphology of multiple grains or of a singular large granule; also, the cell could contain both types of aggregates. Note that colocalization of Gln3QN-YFP aggregates with the nucleus was absent (figure, panel (c)).
Thus, Gln3QN-YFP forms aggregates in the presence of prions [PSI+] and [PIN+] and also in their absence. However, quantitative analysis (see “Materials and Methods”) revealed that prions [PSI+] and [PIN+] significantly influenced frequencies of Gln3QN-YFP aggregation (figure, panel (d)). So, these frequencies in strains [psi–][pin–], [PSI+][pin–] and [PSI+][PIN+] were significantly lower than in strain [psi–][PIN+] (figure, panel (d)), i.e. prion [PIN+] acted as an inducer of Gln3QN-YFP aggregation. By contrast, prion [PSI+] suppressed Gln3QN-YFP aggregation in the [PIN+] strains, but this prion itself (in strain [PSI+][pin–] as compared with strain [psi–][pin–]) did not suppress Gln3QN-YFP aggregation (figure, panel (d)). Then we decided to test whether the Gln3QN-YFP aggregates formed in the analyzed strains were resistant to ionic detergents, which characterizes amyloids.
Gln3QN-YFP forms detergent-resistant aggregates in strains with different [PIN] and [PSI] status. The detergent-resistance of Gln3QN-YFP aggregates was analyzed using SDD-AGE. Strains OT56 [PSI+][PIN+], 2-D-701 [PSI+][pin–], 1-OT56 [psi–][PIN+], and 2-OT56 [psi–][pin–] transformed with a plasmid to obtain overproduction of Gln3QN-YFP were grown for 48 h in a liquid selective medium supplemented with 100 µM copper sulfate. Upon isolation of the protein, the lysates were treated at room temperature with 3% sodium lauryl sarcosinate and separated by electrophoresis in agarose gel. A boiled sample of strain 1-OT56 [psi–][PIN+] lysate was used as a negative control.
The samples obtained from all strains under study were shown to contain Gln3QN-YFP polymers resistant to treatment with cold sodium lauryl sarcosinate, although in the [pin–] strains the amount of polymers was lower (figure, panel (e)), which was in agreement with the confocal microscopy data. Thus, notwithstanding a significant influence of [PIN+] and [PSI+] on the frequency of formation of Gln3QN-YFP aggregates, this protein can produce detergent-resistant aggregates also in the absence of the prions.
Gln3QN-YFP aggregates colocalize with aggregates of Sup35NM-CFP and Rnq1-CFP. Both [PIN+] and [PSI+] influenced the frequency of Gln3QN-YFP aggregation; therefore, it was supposed that it could be a result of Gln3QN-YFP interaction with structural proteins of [PIN+] (Rnq1) and [PSI+] (Sup35), and this could be tested by analyzing their colocalization. For the testing, strains OT56 [PSI+][PIN+], 2-D-701 [PSI+][pin–], 1-OT56 [psi–][PIN+], and 2-OT56 [psi–][pin–] were co-transformed with plasmid pU-CUP1-GLN3QN-YFP and also with a plasmid for overexpression of the prion-forming region Sup35 fused with CFP (pGPD-SUP35NM-CFP) or with a plasmid for overexpression of Rnq1 fused with CFP (pCUP1-RNQ1-CFP(LEU2)). The transformants were grown for 48 h on the corresponding selective medium supplemented with 150 µM copper sulfate, and then colocalization was analyzed using laser scanning confocal microscopy. It was found (figure, panels (g) and (h)) that Gln3QN-YFP aggregates were colocalized with the Rnq1-CFP aggregates at the nearly 100% frequency in the [PSI+][PIN+] and [psi–][PIN+] strains (it should be noted that in the [psi–][pin–] and [PSI+][pin–] strains Rnq1-CFP formed microscopically detectable aggregates extremely seldom, and it was impossible to analyze colocalization).
Aggregates of protein Gln3QN-YFP colocalized also with Sup35NM-CFP aggregates (figure, panel (f)), but the colocalization frequency in this case was significantly lower and not higher than 50% (figure, panels (f) and (h)). This was in agreement with data showing that the [PSI+] prion was not an inducer, but it influenced the frequency of Gln3QN-YFP aggregation. In general, although Gln3QN-YFP formed microscopically and biochemically detectable aggregates also in strains deprived of [PSI+] and [PIN+] prions, the results revealed that [PIN+] significantly increased Gln3QN-YFP aggregation, whereas [PSI+], in contrast, suppressed the effect of [PIN+]. Thus, the efficiency of protein Gln3QN-YFP aggregation is mediated by the interaction between two different prions.
DISCUSSION
The comparison of effects of the overexpression of Gln3 and of its Q/N-rich fragment fused with YFP revealed difference in aggregation, localization, and toxicity of these proteins. The full-length protein demonstrated diffuse distribution in the nucleus (figure, panels (a) and (b)), whereas its Q/N-rich fragment formed distinct fluorescent aggregates in the cytoplasm (figure, panels (a) and (c)). The toxicity of the full-length Gln3 overexpression was quite expected, because this protein is a transcriptional activator of genes regulated by nitrogen catabolite repression [23-25]. The aggregation of Gln3 could compensate the toxic effect of the increased number of its molecules, but the full-length Gln3 did not form aggregates when overexpressed. The absence of toxicity of overexpression of the Q/N-rich fragment of Gln3 could be explained either by its inactivation within the aggregates or by localization of the Gln3 domain responsible for its transcriptional activity beyond the Q/N-rich fragment (data from the Saccharomyces Genome Database, http://www.yeastgenome.org/). It should be noted that polyglutamine and polyasparagine tracts have their own transcriptional activity [36, 37]; therefore, the possibility of Gln3QN influence on transcription of some genes cannot be excluded completely.
The aggregation of Gln3QN-YFP depends on prions [PIN+] and [PSI+]: although this protein formed aggregates with low frequency also in strain [psi–][pin–], the frequency of formation of its aggregates increased several-fold on the background of prion [PSI+] (strain [PSI+][pin–]) and enhanced very strongly in the presence of [PIN+] (strain [psi–][PIN+]) (figure, panels (c)-(e)). It is known that pre-existing aggregates of some Q/N-rich proteins can induce aggregation of other proteins [38-42] acting as “seeds”. This seems to explain the increased aggregation of Gln3QN-YFP in the presence of the [PIN+] and [PSI+] prions whose interaction has been demonstrated by data on colocalization (figure, panels (f)-(h)). The frequency of Gln3QN-YFP aggregation in strain [PSI+][PIN+] is lower than in strain [psi–][PIN+], because a part of Gln3QN-YFP is likely to bind with [PSI+] aggregates, which is a weaker inducer of aggregation than the [PIN+].
The difference in the ability of full-length Gln3 and of its Q/N-rich fragment to form aggregates when overexpressed suggests importance of studies on the influence of amyloidogenic regions on the aggregation of full-size proteins. Our data have shown that the ability of the Q/N-rich region to form aggregates does not definitely indicate that the full-size protein can produce aggregates. Unfortunately, at present comparative data on the aggregation of full-size proteins and of their amyloidogenic fragments are almost absent. To understand the principles of interactions of amyloidogenic and non-amyloidogenic regions of the protein molecule, which determine its ability for aggregation and amyloidogenesis, a large bulk of experimental data is needed on various proteins, one of which has been studied in the present work.
In general, the present study has revealed that the aggregation of Q/N-rich proteins is under the control of both cis- (interaction of amyloidogenic and non-amyloidogenic regions within the same protein molecule, the level of protein production) and trans-acting factors (such as prions). Just the balance of these factors determines the ability of the protein to produce aggregates.
We are grateful to A. P. Galkin (SPbSU) for his critical reading the manuscript.
This work was supported by the Grant of the President of the Russian Federation (project No. MK-4854.2015.4, A. A. Nizhnikov and K. S. Antonets), by the Russian Foundation for Basic Research (projects No. 16-34-60153 and 14-04-32213, A. A. Nizhnikov), and also by the St. Petersburg Committee on Science and Higher School (A. A. Nizhnikov and K. S. Antonets). We are grateful to the St. Petersburg State University for the grants 1.50.2543.2013 and 1.37.291.2015 (A. A. Nizhnikov).
The study was performed using equipment of the resource centers “Development of Molecular and Cellular Techniques” and “Chromas” of SPbSU.
REFERENCES
1.Nizhnikov, A. A., Antonets, K. S., and
Inge-Vechtomov, S. G. (2015) Amyloids: from pathogenesis to function,
Biochemistry (Moscow), 80, 1127-1144.
2.Sipe, J. D., Benson, M. D., Buxbaum, J. N., Ikeda,
S., Merlini, G., Saraiva, M. J., and Westermark, P. (2014) Nomenclature
2014: amyloid fibril proteins and clinical classification of
amyloidosis, Amyloid, 21, 221-224.
3.Fowler, D. M., Koulov, A. V., Alory-Jost, C.,
Marks, M. S., Balch, W. E., and Kelly, J. W. (2006) Functional amyloid
formation within mammalian tissue, PLoS Biol., 4, e6.
4.Maji, S. K., Perrin, M. H., Sawaya, M. R.,
Jessberger, S., Vadodaria, K., Rissman, R. A., Singru, P. S., Nilsson,
K. P., Simon, R., Schubert, D., Eisenberg, D., Rivier, J., Sawchenko,
P., Vale, W., and Riek, R. (2009) Functional amyloids as natural
storage of peptide hormones in pituitary secretory granules,
Science, 325, 328-332.
5.Si, K., Giustetto, M., Etkin, A., Hsu, R.,
Janisiewicz, A. M., Miniaci, M. C., Kim, J. H., Zhu, H., and Kandel, E.
R. (2003) A neuronal isoform of CPEB regulates local protein synthesis
and stabilizes synapse-specific long-term facilitation in
aplysia, Cell, 115, 893-904.
6.Majumdar, A., Cesario, W. C., White-Grindley, E.,
Jiang, H., Ren, F., Khan, M. R., Li, L., Choi, E. M., Kannan, K., Guo,
F., Unruh, J., Slaughter, B., and Si, K. (2012) Critical role of
amyloid-like oligomers of Drosophila Orb2 in the persistence of
memory, Cell, 148, 515-529.
7.Chapman, M. R., Robinson, L. S., Pinkner, J. S.,
Roth, R., Heuser, J., Hammar, M., Normark, S., and Hultgren, S. J.
(2002) Role of Escherichia coli curli operons in directing
amyloid fiber formation, Science, 295, 851-855.
8.Chimileski, S., Franklin, M. J., and Papke, R. T.
(2014) Biofilms formed by the archaeon Haloferax volcanii
exhibit cellular differentiation and social motility, and facilitate
horizontal gene transfer, BMC Biol., 12, 65.
9.Bolton, D. C., McKinley, M. P., and Prusiner, S. B.
(1982) Identification of a protein that purifies with the scrapie
prion, Science, 218, 1309-1311.
10.Wickner, R., Masison, D. C., and Edskes, H. K.
(1995) [PSI] and [URE3] as yeast prions, Yeast,
11, 1671-1685.
11.Wickner, R. B. (1994) [URE3] as an altered
Ure2 protein: evidence for a prion analog in Saccharomyces
cerevisiae, Science, 264, 566-569.
12.Derkatch, I. L., Bradley, M. E., Hong, J. Y., and
Liebman, S. W. (2001) Prions affect the appiarance of other prions. The
stoty of [PIN], Cell, 106, 171-182.
13.Du, Z., Park, K. W., Yu, H., Fan, Q., and Li, L.
(2008) Newly identified prion linked to the chromatin-remodeling factor
Swi1 in Saccharomyces cerevisiae, Nat. Genet., 40,
460-465.
14.Patel, B. K., Gavin-Smyth, J., and Liebman, S. W.
(2009) The yeast global transcriptional co-repressor protein Cyc8 can
propagate as a prion, Nat. Cell. Biol., 11, 344-349.
15.Alberti, S., Halfmann, R., King, O., Kapila, A.,
and Lindquist, S. (2009) A systematic survey identifies prions and
illuminates sequence features of prionogenic proteins, Cell,
137, 146-158.
16.Rogoza, T., Goginashvili, A., Rodionova, S.,
Ivanov, M., Viktorovskaya, O., Rubel, A., Volkov, K., and Mironova, L.
(2010) Non-Mendelian determinant [ISP+] in yeast is a
nuclear-residing prion form of the global transcriptional regulator
Sfp1, PNAS, 107, 10573-10577.
17.Suzuki, G., Shimazu, N., and Tanaka, M. (2012) A
yeast prion, Mod5, promotes acquired drug resistance and cell survival
under environmental stress, Science, 336, 355-359.
18.Saifitdinova, A. F., Nizhnikov, A. A., Lada, A.
G., Rubel, A. A., Magomedova, Z. M., Ignatova, V. V., Inge-Vechtomov,
S. G., and Galkin, A. P. (2010) [NSI+]: a novel non-Mendelian
nonsense suppressor determinant in Saccharomyces cerevisiae,
Curr. Genet., 56, 467-478.
19.Nizhnikov, A. A., Magomedova, Z. M., Rubel, A.
A., Kondrashkina, A. M., Inge-Vechtomov, S. G., and Galkin, A. P.
(2012) [NSI+] determinant has a pleiotropic phenotypic
manifestation that is modulated by SUP35, SUP45, and
VTS1 genes, Curr. Genet., 58, 35-47.
20.Nizhnikov, A. A., Magomedova, Z. M.,
Saifitdinova, A. F., Inge-Vechtomov, S. G., and Galkin, A. P. (2012)
Identification of genes encoding potentially amyloidogenic proteins
that take part in the regulation of nonsense suppression in yeast
Saccharomyces cerevisiae, Russ. J. Genet. Appl. Res.,
2, 398-404.
21.Nizhnikov, A. A., Kondrashkina, A. M., and
Galkin, A. P. (2013) Interactions of [NSI+] prion-like
determinant with SUP35 and VTS1 genes in Saccharomyces
cerevisiae, Russ. J. Genet., 49, 1004-1012.
22.Kondrashkina, A. M., Antonets, K. S., Galkin, A.
P., and Nizhnikov, A. A. (2014) Prion-like determinant [NSI+] decreases
the expression of the SUP45 gene in Saccharomyces
cerevisiae, Mol. Biol., 48, 688-693.
23.Mitchell, A. P., and Magasanik, B. (1984)
Regulation of glutamine-repressible gene products by the GLN3
function in Saccharomyces cerevisiae, Mol. Cell. Biol.,
4, 2758-2766.
24.Blinder, D., and Magasanik, B. (1995) Recognition
of nitrogen-responsive upstream activation sequences of
Saccharomyces cerevisiae by the product of the GLN3 gene,
J. Bacteriol., 177, 4190-4193.
25.Cox, K. H., Rai, R., Distler, M., Daugherty, J.
R., Coffman, J. A., and Cooper, T. G. (2000) Saccharomyces
cerevisiae GATA sequences function as TATA elements during nitrogen
catabolite repression and when Gln3p is excluded from the nucleus by
overproduction of Ure2p, J. Biol. Chem., 275,
17611-17618.
26.Kaiser, C., Michaelis, S., and Mitchell, A.
(1994) Methods in Yeast Genetics, Cold Spring Harbor Laboratory
Press, N. Y.
27.Zakharov, I. A., Kozhin, S. A., Kozhina, T. N.,
and Fedorova, I. V. (1984) Collected Methods in Genetics of the
Yeast Saccharomyces [in Russian], Nauka, Leningrad.
28.Sambrook, J., Fritsch, E. F., and Maniatis, T.
(1989) Molecular Cloning. A Laboratory Manual, Cold Spring
Harbor Laboratory Press, N. Y.
29.Derkatch, I. L., Chernoff, Y. O., Kushnirov, V.
V., Inge-Vechtomov, S. G., and Liebman, S. W. (1996) Genesis and
variability of [PSI] prion factors in Saccharomyces cerevisiae,
Genetics, 144, 1375-1386.
30.Nizhnikov, A. A., Kondrashkina, A. M., Antonets,
K. S., and Galkin, A. P. (2014) Overexpression of genes encoding
asparagine-glutamine-rich transcriptional factors causes nonsense
suppression in Saccharomyces cerevisiae, Russ. J. Genet.
Appl. Res., 4, 122-130.
31.Sikorski, R. S., and Hieter, P. (1989) A system
of shuttle vectors and yeast host strains designed for efficient
manipulation of DNA in Saccharomyces cerevisiae,
Genetics, 122, 19-27.
32.Rubel, A. A., Ryzhova, T. A., Antonets, K. S.,
Chernoff, Y. O., and Galkin, A. P. (2013) Identification of PrP
sequences essential for the interaction between the PrP polymers and
Aβ peptide in a yeast-based assay, Prion, 7,
469-476.
33.Rubel, A. A., Saifitdinova, A. F., Lada, A. G.,
Nizhnikov, A. A., Inge-Vechtomov, S. G., and Galkin, A. P. (2008) Yeast
chaperone Hsp104 controls gene expression at the post-transcriptional
level, Mol. Biol., 42, 110-116.
34.Kryndushkin, D. S., Alexandrov, I. M.,
Ter-Avanesyan, M. D., and Kushnirov, V. V. (2003) Yeast
[PSI+] prion aggregates are formed by small Sup35 polymers
fragmented by Hsp104, J. Biol. Chem., 278,
49636-49643.
35.Bagriantsev, S. N., Kushnirov, V. V., and
Liebman, S. W. (2006) Analysis of amyloid aggregates using agarose gel
electrophoresis, Methods Enzymol., 412, 33-48.
36.Benn, C. L., Sun, T., Sadri-Vakili, G.,
McFarland, K. N., DiRocco, D. P., Yohrling, G. J., Clark, T. W.,
Bouzou, B., and Cha, J. H. (2008) Huntingtin modulates transcription,
occupies gene promoters in vivo, and binds directly to DNA in a
polyglutamine-dependent manner, J. Neurosci., 28,
10720-10733.
37.Peters, T. W., and Huang, M. (2007) Protein
aggregation and polyasparagine-mediated cellular toxicity in
Saccharomyces cerevisiae, Prion, 1, 144-153.
38.Derkatch, I. L., Uptain, S. M., Outeiro, T. F.,
Krishnan, R., Lindquist, S. L., and Liebman, S. W. (2004) Effects of
Q/N-rich, polyQ, and non-polyQ amyloids on the de novo formation
of the [PSI+] prion in yeast and aggregation of Sup35 in
vitro, Proc. Natl. Acad. Sci. USA, 101,
12934-12939.
39.Urakov, V. N., Vishnevskaya, A. B., Alexandrov,
I. M., Kushnirov, V. V., Smirnov, V. N., and Ter-Avanesyan, M. D.
(2010) Interdependence of amyloid formation in yeast: implications for
polyglutamine disorders and biological functions, Prion,
4, 45-52.
40.Kochneva-Pervukhova, N. V., Alexandrov, A. I.,
and Ter-Avanesyan, M. D. (2012) Amyloid-mediated sequestration of
essential proteins contributes to mutant huntingtin toxicity in yeast,
PLOS One, 7, e29832.
41.Nizhnikov, A. A., Alexandrov, A. I., Ryzhova, T.
A., Mitkevich, O. V., Dergalev, A. A., Ter-Avanesyan, M. D., and
Galkin, A. P. (2014) Proteomic screening for amyloid proteins, PLoS
One, e116003.
42.Derkatch, I. L., Bradley, M. E., Masse, S. V.,
Zadorsky, S. P., Polozkov, G. V., Inge-Vechtomov, S. G., and Liebman,
S. W. (2000) Dependence and independence of [PSI(+)] and [PIN(+)]: a
two-prion system in yeast? EMBO J., 19, 1942-1952.