Phylogenomic Analysis Identifies a Sodium-Translocating Decarboxylating Oxidoreductase in Thermotogae
O. I. Klimchuk1, D. V. Dibrova2, and A. Y. Mulkidjanian1,2,3*
1School of Bioengineering and Bioinformatics, Lomonosov Moscow State University, 119991 Moscow, Russia2Belozersky Institute of Physico-Chemical Biology, Lomonosov Moscow State University, 119991 Moscow, Russia
3School of Physics, Osnabrueck University, 49069 Osnabrueck, Germany; E-mail: amulkid@uos.de
* To whom correspondence should be addressed.
Received November 11, 2015; Revision received January 27, 2016
Bacterial sodium-dependent decarboxylases were the first enzymes exemplifying sodium-dependent bioenergetics. These enzyme complexes couple decarboxylation of organic acids with the export of sodium ions via a special membrane subunit. In 711 representative prokaryotic genomes, we have analyzed genomic neighborhoods of the genes that code the membrane subunit of sodium decarboxylases. In representatives of Thermotogae, the operons with the gene of this subunit lack the genes of subunits that perform non-oxidative decarboxylation. Instead, these operons contain the genes of alpha- and delta-subunits of decarboxylating oxidoreductases of alpha-ketoacids. The genes of beta- and gamma-subunits of the decarboxylating oxidoreductases were found within the genomes of respective Thermotogae species as separate, two-gene operons. We suggest that the described two operons code together for sodium-translocating decarboxylating oxidoreductases capable of coupling oxidative decarboxylation of alpha-ketoacids with the export of sodium ions, which is a novel type of bioenergetic coupling.
KEY WORDS: sodium transport, membrane bioenergetics, comparative genomics, phylogenetic analysis, ferredoxin, molecular evolution, anaerobic bacteriaDOI: 10.1134/S0006297916050059
In the early 1980s, Vladimir Skulachev and coauthors proved that a gradient of sodium ions could serve, in several bacteria, as a membrane-associated energetic intermediate, the same as the proton gradient in most organisms [1-3]. In the following years, sodium-dependent bioenergetics was recognized as one of the fundamental mechanisms of biological energy conversion [4, 5]. Initially, sodium-dependent bioenergetics was considered an exotic adaptation of some anaerobic, alkaliphilic, and/or thermophilic prokaryotes to their unusual habitats. Later, however, it was shown that sodium-dependent bioenergetics is common for pathogenic prokaryotes [6] and, therefore, deserves detailed investigation. The reconstruction of the ancestral sequences of Na+- and H+-translocating rotary membrane ATP synthases showed that sodium-dependent bioenergetics was the original (ancestral) form of membrane bioenergetics and should have prevailed in anoxic conditions of the ancient Earth [7-10].
The key elements of sodium-dependent bioenergetics are primary sodium pumps capable of utilizing available energy sources for pumping sodium ions out of cells, which yields a transmembrane difference of electrochemical potentials of Na+ – the sodium potential; the latter can be used to satisfy energy needs of the cell, primarily to drive ATP synthesis. As discussed in detail previously [7-10], the function of exporting Na+ could precede the emergence of sodium-dependent bioenergetics proper. Many key cell systems and, most remarkably, the protein synthesis system require prevalence of K+ over Na+ in the cytoplasm, which is likely due to emergence of the first cells in a specific, potassium-rich environment [11-15]. Since both freshwater and marine reservoirs contain more sodium than potassium, the primordial cells could have inhabited them only after the emergence of Na+-tight cell membranes and systems for exporting Na+. The interaction of several such initially independent systems could eventually cause the emergence of sodium-dependent membrane bioenergetics, as discussed elsewhere [10].
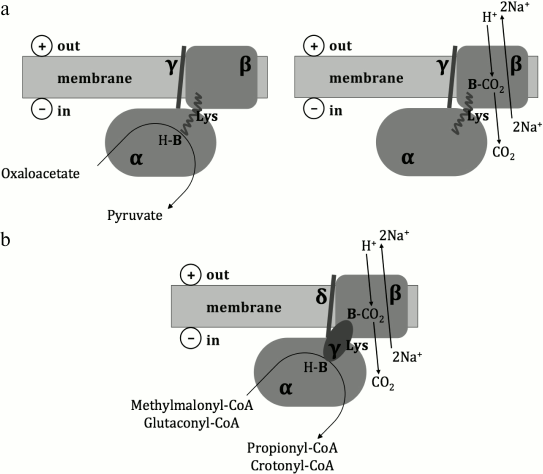
Historically, the first primary sodium pump to be described was the oxaloacetate decarboxylase from Klebsiella pneumoniae [16], which couples decarboxylation of oxaloacetate with the translocation of Na+ across the cell membrane against the electrochemical gradient. Several similar enzymes were later described in different anaerobic microorganisms [17]; some of them, in particular the methylmalonyl-CoA decarboxylase from Veillonella parvula [18], the glutaconyl-CoA decarboxylase from Acidaminococcus fermentans [19], and the malonate decarboxylase system from Malonomonas rubra [20] were experimentally studied. It was shown that the oxaloacetate decarboxylase consists of α-, β-, and γ-subunits; the methylmalonyl-CoA and glutaconyl-CoA decarboxylases consist of α-, β-, γ-, and δ-subunits, wherein the γ-subunit of the oxaloacetate decarboxylase is related to the δ-subunit of the methylmalonyl-CoA and glutaconyl-CoA decarboxylases, and the γ-subunit of the methylmalonyl-CoA and glutaconyl-CoA decarboxylases is functionally equivalent to the C-terminal biotin-binding domain of the α-subunit of the oxaloacetate decarboxylase [17]. As depicted in Fig. 1, the α-subunit of these enzymes is localized in the cytoplasm and catalyzes carboxyl group transfer from a carboxylic acid to biotin. The β-subunit is localized within the cell membrane and couples decarboxylation of carboxybiotin in the cytoplasmic α-subunit with the translocation of Na+ out of the cell. The γ/δ-subunit serves as a membrane anchor for the α-subunit [17]. The malonate decarboxylase system is more complicated, being encoded by an operon of 14 genes [21] whose functions are not fully understood.
Fig. 1. Structural schemes and putative catalytic mechanisms of oxaloacetate, methylmalonyl-CoA, and glutaconyl-CoA decarboxylases (from Buckel [17]). Lys – a biotin-binding lysine residue; H-B – biotin; B-CO2 – carboxybiotin. a) Oxaloacetate decarboxylase; b) methylmalonyl-CoA and glutaconyl-CoA decarboxylases.
According to X-ray diffraction data, the α-subunits of the oxaloacetate decarboxylase form a homodimer [22], and the α-subunits of the methylmalonyl-CoA decarboxylase form a homotrimer [23], whereas the α-subunits of the glutaconyl-CoA decarboxylase form either a homodimer [24] or a homotetramer [25]. A monomer of the α-subunit of the methylmalonyl-CoA and glutaconyl-CoA decarboxylases contains one carboxyltransferase domain (the Pfam protein domain database [26] identifier: PF01039), whereas the monomer of the α-subunit of the oxaloacetate decarboxylase usually consists of three other Pfam domains that are not related to the carboxyltransferase domain. The 3D structure of the membrane part of Na+-translocating decarboxylases has not been reported yet.
Experimentally studied sodium-dependent decarboxylases translocate Na+ against the electrochemical gradient by using the free energy that is released upon cleavage of a chemical bond (decarboxylation). In this report, by analyzing the structure of prokaryotic operons, we predict a new class of primary sodium pumps, namely the Na+-translocating, decarboxylating oxidoreductases.
MATERIALS AND METHODS
As initial data, we used the set of 711 complete prokaryotic genomes whose protein-coding genes were attributed to the Clusters of Orthologous Groups (COGs) [27]. Sequences of 180 membrane β-subunits of Na+-translocating decarboxylases (these proteins form COG1883) were found in 141 complete genomes. Sequences of 255 cytoplasmic α-subunits of oxaloacetate decarboxylases that belong to COG5016 and consist of two Pfam domains, HMGL-like (PF00682) and PYC_OADA (PF02436), were found in 216 complete genomes, and these proteins are designated below as α1-subunits. In particular, 170 of the α1-subunits contain an additional biotin-binding Pfam domain (PF00364 or PF13533). Sequences of 876 proteins that belong to COG4799 and contain one carboxyltransferase Pfam domain (PF01039) were found in 413 complete genomes; these proteins include the cytoplasmic α-subunits of methylmalonyl-CoA and glutaconyl-CoA decarboxylases and are designated below as α2-subunits.
Multiple alignment of the amino acid sequences of the β-subunits (180 proteins) was done using the MUSCLE program [28]. Conserved blocks (alignment regions in which all participating sequences are reliably aligned), were manually selected and contained 367 positions in total. A phylogenetic tree of the β-subunits was constructed by the Neighbor-Joining algorithm using the MEGA 5.2 program [29]. The branch support values supporting branches were calculated from 100 iterations. Genomic neighborhoods of the β-subunit genes (180 proteins) were analyzed using the OLESA program [30]. Operon boundaries were determined according to the following criteria: (i) the genes are located on the same DNA strand, (ii) the distance between the genes is small, (iii) the promoter elements (Shine–Dalgarno sequence and Pribnow box) could be detected, and (iv) the termination elements [31] could be detected (Fig. S1 in the Supplement to this paper on the site of this journal http://protein.bio.msu.ru/biokhimiya and Springer site Link.springer.com).
RESULTS AND DISCUSSION
First, we examined the co-occurrence of the genes of subunits α1, α2, and β of Na+-translocating decarboxylases in 711 complete prokaryotic genomes (Table S1 in Supplement). The α-subunit genes were often found in prokaryotic genomes in the absence of the β-subunit genes. In 382 complete genomes from the 711 examined, the α-subunit genes (α1 and/or α2) were not accompanied by β-subunit genes. The genes of both types of subunits (α and β) were found in 141 complete genomes. The β-subunit genes were found in only six archaeal genomes of 83 examined complete archaeal genomes, namely in all representatives of Thermococci (Thermococcus kodakarensis KOD1, Pyrococcus furiosus DSM 3638, Pyrococcus abyssi GE5), as well as in Archaeoglobus fulgidus DSM 4304, Aciduliprofundum boonei T469, and Staphylothermus hellenicus DSM 12710. The β-subunit genes were also found in all representatives of Thermotogae and Synergistetes, all representatives of Bacteroideae, many representatives of Chlorobi, Chloroflexi, Firmicutes, Spirochaetes, and Verrucomicrobia, many representatives of Desulfobacterales and Syntrophobacterales, diverse representatives of γ- and ε-proteobacteria, as well as in some other bacteria (Table S1 in Supplement).
Our analysis of genomic neighborhoods of the genes of β-subunits has shown that the genes of β-subunits often, but not always, form putative operons with the α-subunit gene(s) (α1 and/or α2), as well as additional subunits γ and δ. Sometimes such putative operons include genes of other proteins or, alternatively, the β-subunit genes seem not to form operons; further analysis is required to explain such evolutionary events.
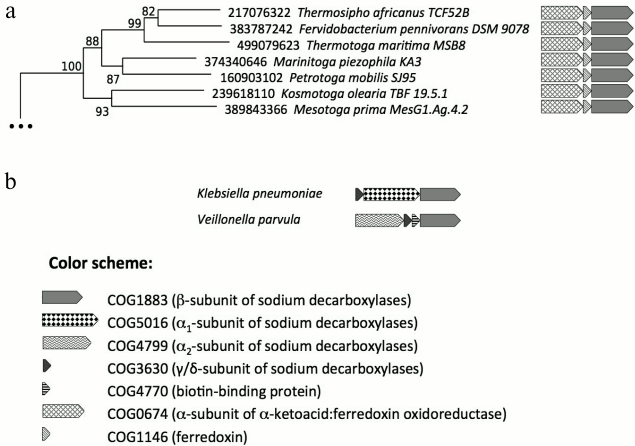
Although the β-subunit genes were not found in prokaryotic genomes in the absence of the genes of α-subunits (α1 and/or α2), analysis of genomic neighborhoods indicates that the β-subunits are not always functionally associated with the α-subunits. In particular, we have noticed that operons with genes of the β-subunit homologs do not include the genes of α-subunits (neither α1, nor α2) in all representatives of Thermotogae; these operons are depicted in Fig. 2. The respective clade, which contains the β-subunit homologs from all representatives of Thermotogae, is separated from the rest of the phylogenetic tree and supported by a reliable bootstrap value (100%). These Thermotogae genomes also contain the genes of α2-subunits; however, these genes are located away from the genes of the β-subunit homologs, being involved in other conservative operon structures (Fig. S2 in Supplement).
Fig. 2. Homologs of membrane β-subunits of Na+-translocating decarboxylases. a) Part of the phylogenetic tree of the membrane β-subunits of Na+-translocating decarboxylases with Thermotogae genes. Genomic neighborhoods of the genes are depicted to the right of the tree. The genes are marked with different shadings in accordance with the COGs they belong to. b) Operon structures typical for experimentally studied oxaloacetate, methylmalonyl-CoA, and glutaconyl-CoA decarboxylases are given for comparison.
This separate Thermotogae clade of β-subunit homologs not associated with the α-subunits is characterized by a special operon structure. In Thermotoga maritima MSB8, the putative operon consists of genes of proteins with GI-identifiers 499079621, 499079622, and 499079623 (corresponding loci: Tmari_0880, Tmari_0881, and Tmari_0882). The protein with GI-identifier 499079623 belongs to the COG1883 that unites the Na+-pumping membrane β-subunits of Na+-translocating decarboxylases (comparison with the β-subunit of the sodium decarboxylase from K. pneumoniae yields an e-value of 3·10–106). The proteins with GI-identifiers 499079621 and 499079622 belong to COG0674 and COG1146, respectively. The functions of these Thermotogae proteins are not experimentally identified, but the database search revealed that the protein with GI-identifier 499079622 belongs to ferredoxins and contains two [4Fe-4S]-clusters (comparison with ferredoxin from Pyrococcus furiosus (PORD_PYRFU) yields an e-value of 3·10–13), whereas the protein with GI-identifier 499079621 can bind α-keto acids and contains thiamine pyrophosphate as a coenzyme (comparison with subunit KorA of the α-ketoglutarate synthase from Methanobacterium thermoautotrophicum (KORA_METTM) yields an e-value of 4·10–117). Corresponding pairwise alignments of amino acid sequences are depicted in Figs. S3-S5 in Supplement.
The two cytoplasmic subunits, whose genes form an operon with homologs of the sodium decarboxylase β-subunit gene in Thermotogae, are otherwise parts of oxidoreductase complexes that perform oxidative decarboxylation of α-keto acids in anaerobic prokaryotes [32-42]. This complex is functionally equivalent, but not homologous, to the mitochondrial α-ketoglutarate dehydrogenase of aerobic organisms that is involved in the Krebs cycle [43]. The functional unit of a bacterial α-ketoacid:ferredoxin oxidoreductase consists of four cytoplasmic subunits (α, β, γ, and δ), which can be represented either by separate polypeptides or by products of fused genes. For example, the enzyme from Desulfovibrio africanus, the first with a deciphered 3D structure, has a fused α + γ + δ + β structure [39, 40]. Another enzyme, whose 3D structure was deciphered recently, belongs to the bacterium Moorella thermoacetica and consists of three subunits: α, β, and an elongated ferredoxin subunit having fused γ + δ structure [41, 42].
The Thermotogae operons shown in Fig. 2, in addition to the homologs of the membrane β-subunit gene, comprise only the genes of subunits α (long) and δ (ferredoxin, short). However, the genes of subunits β and γ could be found in all examined complete Thermotogae genomes, where they form two-gene separate operons (the gene loci of β- and γ-subunits from Thermotoga maritima: Tmari_0402 and Tmari_0403, respectively; Fig. S6 in Supplement). Genomic neighborhoods of the genes of β- and γ-subunits seem to be random in Thermotogae (Fig. S6 in Supplement); therefore, the β- and γ-subunits might form functional complexes with the α- and δ-subunits.
We tested whether the homologs of the sodium decarboxylase β-subunit, as found in Thermotogae, contain amino acid residues presumably involved in Na+ transport in sodium decarboxylases [44, 45]. As can be seen in Fig. 3, all of the sodium binding residues and most of the other functionally important residues are retained in the Thermotogae subunits. Interestingly, the methionine residue (M386 in the subunit from K. pneumoniae), which is presumably involved in the Na+-translocation [44, 45], is replaced in Thermotogae by aspartate or threonine, which can coordinate Na+ [9, 46] (see also Fig. 4). The conservation of the key residues suggests that these homologs of the sodium decarboxylase β-subunit are likely to be functional and capable of transferring Na+ across the cell membrane.
Fig. 3. Fragments of the multiple sequence alignment of homologs of the membrane β-subunits of Na+-translocating decarboxylases, as found in Thermotogae, with the membrane β-subunit of the oxaloacetate decarboxylase from K. pneumoniae. The topology of helices is given in accordance with Wilde et al. [44]. The amino acid residues presumably responsible for the binding of Na+ are marked with asterisks, and other functionally important residues are marked with “+” [44, 45]. Amino acid residues are shaded in accordance with their physicochemical properties: charged polar residues (D, E, H, K, R) are shaded black, uncharged polar residues (N, Q, S, T, Y) are shaded gray, and nonpolar residues are not shaded.
Fig. 4. Multiple sequence alignment of Thermotogae ATPase rotor c-subunits, which are responsible for ion specificity, with corresponding subunits of experimentally studied Na+-translocating rotary ATPases (F-type ATPases from Ilyobacter tartaricus and Propionigenium modestum, N-type ATPase from Aphanothece halophytica [56, 57]) and H+-translocating rotary ATPases (F-type ATPase from Escherichia coli and a second copy of ATPase from Aphanothece halophytica). Transmembrane regions are shown in accordance with the structure of the ATPL_ILYTA protein from Ilyobacter tartaricus (identifier in PDB database: 2WGM [46]). Amino acid residues forming the Na+-binding site in the Na+-translocating enzymes are marked with asterisks [9, 46]. Amino acid residues are shaded in accordance with their physicochemical properties: charged polar residues (D, E, H, K, R) are shaded black, uncharged polar residues (N, Q, S, T, Y) are shaded gray, and nonpolar residues are not shaded.
The type of bioenergetics of each organism is determined by the ion specificity of its ATP synthase, which in turn entirely depends on the ion specificity of its membrane rotor [8, 9, 47]. Figure 4 shows that the membrane c-subunits of the ATP synthase of the same Thermotogae contain complete sets of known Na+-binding ligands [9, 46], which indicates sodium-dependent bioenergetics in these organisms.
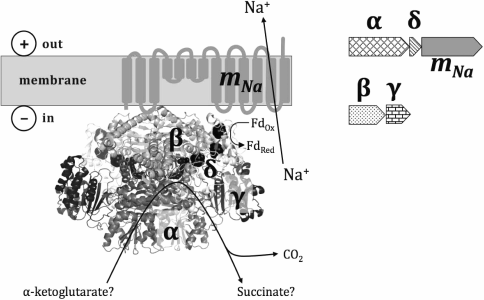
Based on these results, we suggest that Thermotogae possess a Na+-translocating membrane enzyme of a type not previously described, which pumps Na+ out of a bacterial cell on account of oxidative decarboxylation, wherein the transfer of two electrons from an organic α-keto acid to ferredoxin is coupled with removal of a carboxyl group. A hypothetical structure of the enzyme is depicted in Fig. 5, where the oxidoreductase, as formed by four cytoplasmic subunits, performs oxidative decarboxylation of α-keto acids, which is coupled to Na+-translocation by the membrane subunit mNa.
Fig. 5. Tentative structure of a sodium-translocating decarboxylating oxidoreductase. The topology of the membrane mNa-subunit homologous to the membrane β-subunit of sodium decarboxylases is given in accordance with Wild et al. [44] and Schmid et al. [45]. The 3D structure of the pyruvate:ferredoxin oxidoreductase from Desulfovibrio africanus [39], PDB-identifier 4C42, was used for visualization of the cytoplasmic subunits α, β, γ, and δ. The cytoplasmic part of the enzyme presumably forms a homodimer. Three iron–sulfur clusters are shown as black circles; thiamine pyrophosphate is shown as a wireless model. Two operons that together seem to encode the complex are depicted to the right.
From the high sequence similarity with experimentally characterized decarboxylating oxidoreductases of α-keto acids, we suggest that α-ketoglutarate or pyruvate could serve as substrates of the described enzyme complex; to accurately identify the substrate, experimental studies are necessary. The standard redox potential of α-ketoglutarate and pyruvate at pH 7.0 (E07) is approximately –0.7 V [48]. Since the standard redox potential of ferredoxins is usually higher than –0.5 V [49], the released free energy should be sufficient to transfer at least one Na+ out of the cell.
As already mentioned, a plethora of data indicates that bioenergetics based on the transmembrane potential of Na+ is not something specific only for dwellers of unusual habitats, but is the original form of membrane bioenergetics, traces of which are preserved in most modern organisms as various sodium-transferring membrane enzymes [7, 10].
The current data on the studied energy converting membrane enzymes are summarized in the table. Many of these enzymes are described as having both Na+- and H+-translocating forms. Redox-dependent proton transporters seem to be more diverse among organisms that rely on proton-dependent bioenergetics, as compared to organisms with sodium-dependent bioenergetics, which may be due to the ability of proton-transporting systems to particularly benefit from high-potential electron acceptors [7, 10]. The dwellers of the “sodium world”, however, are capable of coupling the transfer of Na+ with quite diverse exergonic reactions not described for organisms with the proton-dependent bioenergetics. Thus, in general, the variety of coupling mechanisms is greater in Na+-dependent energy converting systems, which may reflect the evolutionary primacy of sodium-dependent bioenergetics, which could have emerged under the anoxic conditions of the ancient Earth [7, 9].
Classification of energy converting H+ and/or
Na+-translocating enzymes. Indicated are energy sources, the
life domains where particular enzymes have been found, and those
studies in which these enzymes were first described

a Operons of N-ATPases, as discovered in genomes of two
archaea, Methanosarcina acetivorans and Methanosarcina
barkeri, are likely to have originated in bacteria and been
obtained by these archaea as a result of horizontal gene transfer [56].
b This protein family also includes the malonate
decarboxylase system and glutaconyl-CoA decarboxylase [80, 81].
This work was supported by the Russian Science Foundation (projects No. 14-50-00029, O. I. Klimchuk, phylogenomic analysis of redox-dependent decarboxylases; No. 14-14-00592, D. V. Dibrova and A. Y. Mulkidjanian, phylogenetic analysis of sodium-translocating energy converting membrane proteins).
REFERENCES
1.Skulachev, V. P. (1984) Sodium bioenergetics,
Trends Biochem. Sci., 9, 483-485.
2.Skulachev, V. P. (1985) Membrane-linked energy
transductions. Bioenergetic functions of sodium: H+ is not
unique as a coupling ion, Eur. J. Biochem., 151,
199-208.
3.Chernyak, B. V., Dibrov, P. A., Glagolev, A. N.,
Sherman, M. Y., and Skulachev, V. P. (1983) A novel type of energetics
in a marine alkali-tolerant bacterium delta-muna-driven motility and
sodium cycle, FEBS Lett., 164, 38-42.
4.Cramer, W. A., and Knaff, D. B. (1990) Energy
Transduction in Biological Membranes: A Textbook of Bioenergetics,
Springer Verlag.
5.Dimroth, P. (1994) Bacterial sodium ion-coupled
energetics, Antonie Van Leeuwenhoek, 65, 381-395.
6.Hase, C. C., Fedorova, N. D., Galperin, M. Y., and
Dibrov, P. A. (2001) Sodium ion cycle in bacterial pathogens: evidence
from cross-genome comparisons, Microbiol. Mol. Biol. Rev.,
65, 353-370.
7.Mulkidjanian, A. Y., Dibrov, P., and Galperin, M.
Y. (2008) The past and present of sodium energetics: may the
sodium-motive force be with you, Biochim. Biophys. Acta,
1777, 985-992.
8.Mulkidjanian, A. Y., Galperin, M. Y., and Koonin,
E. V. (2009) Co-evolution of primordial membranes and membrane
proteins, Trends Biochem. Sci., 34, 206-215.
9.Mulkidjanian, A. Y., Galperin, M. Y., Makarova, K.
S., Wolf, Y. I., and Koonin, E. V. (2008) Evolutionary primacy of
sodium bioenergetics, Biol. Direct, 3, 13.
10.Dibrova, D. V., Galperin, M. Y., Koonin, E. V.,
and Mulkidjanian, A. Y. (2015) Ancient systems of sodium/potassium
homeostasis as predecessors of membrane bioenergetics, Biochemistry
(Moscow), 80, 495-516.
11.Macallum, A. B. (1926) The paleochemistry of the
body fluids and tissues, Physiol. Rev., 6, 316-357.
12.Mulkidjanian, A. Y., Bychkov, A. Y., Dibrova, D.
V., Galperin, M. Y., and Koonin, E. V. (2012) Origin of first cells at
terrestrial, anoxic geothermal fields, Proc. Natl. Acad. Sci.
USA, 109, 821-830.
13.Mulkidjanian, A. Y., Bychkov, A. Y., Dibrova, D.
V., Galperin, M. Y., and Koonin, E. V. (2012) Open questions on the
origin of life at anoxic geothermal fields, Orig. Life Evol.
Biosph., 42, 507-516.
14.Galimov, E. M., Natochin, Y. V., Ryzhenko, B. N.,
and Cherkasova, E. V. (2012) Chemical composition of the primary
aqueous phase of the Earth and origin of life, Geochem. Int.,
50, 1048-1068.
15.Maruyama, S., Ikoma, M., Genda, H., Hirose, K.,
Yokoyama, T., and Santosh, M. (2013) The naked planet Earth: most
essential pre-requisite for the origin and evolution of life,
Geosci. Front., 4, 141-165.
16.Dimroth, P. (1980) A new sodium-transport system
energized by the decarboxylation of oxaloacetate, FEBS Lett.,
122, 234-236.
17.Buckel, W. (2001) Sodium ion-translocating
decarboxylases, Biochim. Biophys. Acta, 1505, 15-27.
18.Hilpert, W., and Dimroth, P. (1982) Conversion of
the chemical energy of methylmalonyl-CoA decarboxylation into a
Na+ gradient, Nature, 296, 584-585.
19.Buckel, W., and Semmler, R. (1982) A
biotin-dependent sodium pump: glutaconyl-CoA decarboxylase from
Acidaminococcus fermentans, FEBS Lett., 148,
35-38.
20.Hilbi, H., Dehning, I., Schink, B., and Dimroth,
P. (1992) Malonate decarboxylase of Malonomonas rubra, a novel
type of biotin-containing acetyl enzyme, Eur. J. Biochem.,
207, 117-123.
21.Berg, M., Hilbi, H., and Dimroth, P. (1997)
Sequence of a gene cluster from Malonomonas rubra encoding
components of the malonate decarboxylase Na+ pump and
evidence for their function, Eur. J. Biochem., 245,
103-115.
22.Studer, R., Dahinden, P., Wang, W. W., Auchli,
Y., Li, X. D., and Dimroth, P. (2007) Crystal structure of the
carboxyltransferase domain of the oxaloacetate decarboxylase
Na+ pump from Vibrio cholerae, J. Mol. Biol.,
367, 547-557.
23.Benning, M. M., Haller, T., Gerlt, J. A., and
Holden, H. M. (2000) New reactions in the crotonase superfamily:
structure of methylmalonyl-CoA decarboxylase from Escherichia
coli, Biochemistry, 39, 4630-4639.
24.Wendt, K. S., Schall, I., Huber, R., Buckel, W.,
and Jacob, U. (2003) Crystal structure of the carboxyltransferase
subunit of the bacterial sodium ion pump glutaconyl-coenzyme A
decarboxylase, EMBO J., 22, 3493-3502.
25.Kress, D., Brugel, D., Schall, I., Linder, D.,
Buckel, W., and Essen, L. O. (2009) An asymmetric model for
Na+-translocating glutaconyl-CoA decarboxylases, J. Biol.
Chem., 284, 28401-28409.
26.Finn, R. D., Bateman, A., Clements, J., Coggill,
P., Eberhardt, R. Y., Eddy, S. R., Heger, A., Hetherington, K., Holm,
L., Mistry, J., Sonnhammer, E. L., Tate, J., and Punta, M. (2014) Pfam:
the protein families database, Nucleic Acids Res., 42
(Database issue), 222-230.
27.Galperin, M. Y., Makarova, K. S., Wolf, Y. I.,
and Koonin, E. V. (2015) Expanded microbial genome coverage and
improved protein family annotation in the COG database, Nucleic
Acids Res., 43 (Database issue), 261-269.
28.Edgar, R. C. (2004) MUSCLE: multiple sequence
alignment with high accuracy and high throughput, Nucleic Acids
Res., 32, 1792-1797.
29.Tamura, K., Peterson, D., Peterson, N., Stecher,
G., Nei, M., and Kumar, S. (2011) MEGA5: molecular evolutionary
genetics analysis using maximum likelihood, evolutionary distance, and
maximum parsimony methods, Mol. Biol. Evol., 28,
2731-2739.
30.Klimchuk, O. I., Dibrova, D. V., and
Mulkidjanian, A. Y. (2015) OLESA: Operon Loci Examination and
Sorting Application, Proceedings of the Moscow Conference
on Computational Molecular Biology (MCCMB), Moscow.
31.Brouwer, R. W., Kuipers, O. P., and Van Hijum, S.
A. (2008) The relative value of operon predictions, Brief.
Bioinform., 9, 367-375.
32.Blamey, J. M., and Adams, M. W. (1994)
Characterization of an ancestral type of pyruvate ferredoxin
oxidoreductase from the hyperthermophilic bacterium, Thermotoga
maritima, Biochemistry, 33, 1000-1007.
33.Eram, M. S., Wong, A., Oduaran, E., and Ma, K.
(2015) Molecular and biochemical characterization of bifunctional
pyruvate decarboxylases and pyruvate ferredoxin oxidoreductases from
Thermotoga maritima and Thermotoga hypogea, J.
Biochem., 158, 459-466.
34.Kletzin, A., and Adams, M. W. (1996) Molecular
and phylogenetic characterization of pyruvate and 2-ketoisovalerate
ferredoxin oxidoreductases from Pyrococcus furiosus and pyruvate
ferredoxin oxidoreductase from Thermotoga maritima, J.
Bacteriol., 178, 248-257.
35.Tersteegen, A., Linder, D., Thauer, R. K., and
Hedderich, R. (1997) Structures and functions of four anabolic
2-oxoacid oxidoreductases in Methanobacterium
thermoautotrophicum, Eur. J. Biochem., 244,
862-868.
36.Yun, N. R., Arai, H., Ishii, M., and Igarashi, Y.
(2001) The genes for anabolic 2-oxoglutarate: ferredoxin oxidoreductase
from Hydrogenobacter thermophilus TK-6, Biochem. Biophys.
Res. Commun., 282, 589-594.
37.Kerscher, L., and Oesterhelt, D. (1981) The
catalytic mechanism of 2-oxoacid:ferredoxin oxidoreductases from
Halobacterium halobium. One-electron transfer at two distinct
steps of the catalytic cycle, Eur. J. Biochem., 116,
595-600.
38.Furdui, C., and Ragsdale, S. W. (2000) The role
of pyruvate ferredoxin oxidoreductase in pyruvate synthesis during
autotrophic growth by the Wood–Ljungdahl pathway, J. Biol.
Chem., 275, 28494-28499.
39.Chabriere, E., Charon, M. H., Volbeda, A.,
Pieulle, L., Hatchikian, E. C., and Fontecilla-Camps, J. C. (1999)
Crystal structures of the key anaerobic enzyme pyruvate:ferredoxin
oxidoreductase, free and in complex with pyruvate, Nat. Struct.
Biol., 6, 182-190.
40.Chabriere, E., Vernede, X., Guigliarelli, B.,
Charon, M. H., Hatchikian, E. C., and Fontecilla-Camps, J. C. (2001)
Crystal structure of the free radical intermediate of
pyruvate:ferredoxin oxidoreductase, Science, 294,
2559-2563.
41.Gibson, M. I., Brignole, E. J., Pierce, E., Can,
M., Ragsdale, S. W., and Drennan, C. L. (2015) The structure of an
oxalate oxidoreductase provides insight into microbial 2-oxoacid
metabolism, Biochemistry, 54, 4112-4120.
42.Pierce, E., Becker, D. F., and Ragsdale, S. W.
(2010) Identification and characterization of oxalate oxidoreductase, a
novel thiamine pyrophosphate-dependent 2-oxoacid oxidoreductase that
enables anaerobic growth on oxalate, J. Biol. Chem., 285,
40515-40524.
43.Baughn, A. D., Garforth, S. J., Vilcheze, C., and
Jacobs, W. R., Jr. (2009) An anaerobic-type α-ketoglutarate
ferredoxin oxidoreductase completes the oxidative tricarboxylic acid
cycle of Mycobacterium tuberculosis, PLoS Pathog.,
5, e1000662.
44.Wild, M. R., Pos, K. M., and Dimroth, P. (2003)
Site-directed sulfhydryl labeling of the oxaloacetate decarboxylase
Na+ pump of Klebsiella pneumoniae: helix VIII
comprises a portion of the sodium ion channel, Biochemistry,
42, 11615-11624.
45.Schmid, M., Vorburger, T., Pos, K. M., and
Dimroth, P. (2002) Role of conserved residues within helices IV and
VIII of the oxaloacetate decarboxylase β-subunit in the energy
coupling mechanism of the Na+ pump, Eur. J. Biochem.,
269, 2997-3004.
46.Meier, T., Krah, A., Bond, P. J., Pogoryelov, D.,
Diederichs, K., and Faraldo-Gomez, J. D. (2009) Complete
ion-coordination structure in the rotor ring of
Na+-dependent F-ATP synthases, J. Mol. Biol.,
391, 498-507.
47.Skulachev, V. P. (1988) Membrane
Bioenergetics, Springer Verlag, Heidelberg.
48.Skulachev, V. P., Bogachev, A. V., and
Kasparinsky, F. O. (2012) Principles of Bioenergetics, Springer,
Heidelberg.
49.Stephens, P. J., Jollie, D. R., and Warshel, A.
(1996) Protein control of redox potentials of iron-sulfur proteins,
Chem. Rev., 96, 2491-2513.
50.Baltscheffsky, H., Von Stedingk, L. V., Heldt, H.
W., and Klingenberg, M. (1966) Inorganic pyrophosphate: formation in
bacterial photophosphorylation, Science, 153,
1120-1122.
51.Malinen, A. M., Belogurov, G. A., Baykov, A. A.,
and Lahti, R. (2007) Na+-pyrophosphatase: a novel primary
sodium pump, Biochemistry, 46, 8872-8878.
52.Racker, E. (1962) Studies of factors involved in
oxidative phosphorylation, Proc. Natl. Acad. Sci. USA,
48, 1659-1663.
53.Laubinger, W., and Dimroth, P. (1988)
Characterization of the ATP synthase of Propionigenium modestum
as a primary sodium pump, Biochemistry, 27,
7531-7537.
54.Takase, K., Yamato, I., and Kakinuma, Y. (1993)
Cloning and sequencing of the genes coding for the A and B subunits of
vacuolar-type Na+-ATPase from Enterococcus hirae.
Coexistence of vacuolar- and F0F1-type ATPases in
one bacterial cell, J. Biol. Chem., 268, 11610-11616.
55.Solioz, M., and Davies, K. (1994) Operon of
vacuolar-type Na+-ATPase of Enterococcus hirae, J.
Biol. Chem., 269, 9453-9459.
56.Dibrova, D. V., Galperin, M. Y., and
Mulkidjanian, A. Y. (2010) Characterization of the N-ATPase, a
distinct, laterally transferred Na+-translocating form of
the bacterial F-type membrane ATPase, Bioinformatics, 26,
1473-1476.
57.Soontharapirakkul, K., Promden, W., Yamada, N.,
Kageyama, H., Incharoensakdi, A., Iwamoto-Kihara, A., and Takabe, T.
(2011) Halotolerant cyanobacterium Aphanothece halophytica
contains an Na+-dependent F1F0-ATP
synthase with a potential role in salt-stress tolerance, J. Biol.
Chem., 286, 10169-10176.
58.Cheng, J., Guffanti, A. A., and Krulwich, T. A.
(1997) A two-gene ABC-type transport system that extrudes
Na+ in Bacillus subtilis is induced by ethanol or
protonophore, Mol. Microbiol., 23, 1107-1120.
59.Becher, B., Muller, V., and Gottschalk, G. (1992)
N5-methyl-tetrahydromethanopterin:coenzyme M
methyltransferase of Methanosarcina strain Go1 is an
Na+-translocating membrane protein, J. Bacteriol.,
174, 7656-7660.
60.Hatefi, Y., Haavik, A. G., and Griffiths, D. E.
(1961) Reconstitution of the electron transport system. I. Preparation
and properties of the interacting enzyme complexes, Biochem.
Biophys. Res. Commun., 4, 441-446.
61.Roberts, P. G., and Hirst, J. (2012) The deactive
form of respiratory complex I from mammalian mitochondria is a
Na+/H+ antiporter, J. Biol. Chem.,
287, 34743-34751.
62.Castro, P. J., Silva, A. F., Marreiros, B. C.,
Batista, A. P., and Pereira, M. M. (2015) Respiratory complex I: a dual
relation with H+ and Na+? Biochim. Biophys.
Acta, pii: S0005-2728(15)00253-4.
63.Schmehl, M., Jahn, A., Meyer zu Vilsendorf, A.,
Hennecke, S., Masepohl, B., Schuppler, M., Marxer, M., Oelze, J., and
Klipp, W. (1993) Identification of a new class of nitrogen fixation
genes in Rhodobacter capsulatus: a putative membrane complex
involved in electron transport to nitrogenase, Mol. Gen. Genet.,
241, 602-615.
64.Biegel, E., and Muller, V. (2010) Bacterial
Na+-translocating ferredoxin:NAD+ oxidoreductase,
Proc. Natl. Acad. Sci. USA, 107, 18138-18142.
65.Bohm, R., Sauter, M., and Bock, A. (1990)
Nucleotide sequence and expression of an operon in Escherichia
coli coding for formate hydrogenlyase components, Mol.
Microbiol., 4, 231-243.
66.McTernan, P. M., Chandrayan, S. K., Wu, C. H.,
Vaccaro, B. J., Lancaster, W. A., Yang, Q., Fu, D., Hura, G. L.,
Tainer, J. A., and Adams, M. W. (2014) Intact functional
fourteen-subunit respiratory membrane-bound [NiFe]-hydrogenase complex
of the hyperthermophilic archaeon Pyrococcus furiosus, J.
Biol. Chem., 289, 19364-19372.
67.Tokuda, H., and Unemoto, T. (1982)
Characterization of the respiration-dependent Na+ pump in
the marine bacterium Vibrio alginolyticus, J. Biol.
Chem., 257, 10007-10014.
68.Ide, T., Baumer, S., and Deppenmeier, U. (1999)
Energy conservation by the H2:heterodisulfide oxidoreductase from
Methanosarcina mazei Go1: identification of two
proton-translocating segments, J. Bacteriol., 181,
4076-4080.
69.Hatefi, Y., Haavik, A. G., and Griffiths, D. E.
(1962) Studies on the electron transfer system. XLI. Reduced coenzyme Q
(QH2)-cytochrome c reductase, J. Biol. Chem., 237,
1681-1685.
70.Fowler, L. R., Richardson, S. H., and Hatefi, Y.
(1962) A rapid method for the preparation of highly purified cytochrome
oxidase, Biochim. Biophys. Acta, 64, 170-173.
71.Muntyan, M. S., Cherepanov, D. A., Malinen, A.
M., Bloch, D. A., Sorokin, D. Y., Severina, I. I., Ivashina, T. V.,
Lahti, R., Muyzer, G., and Skulachev, V. P. (2015) Cytochrome
cbb3 of Thioalkali vibrio is a
Na+-pumping cytochrome oxidase, Proc. Natl. Acad. Sci.
USA, 112, 7695-7700.
72.Borisov, V. B., Gennis R. B., Hemp, J., and
Verkhovsky, M. I. (2011) The cytochrome bd respiratory oxygen
reductases, Biochim. Biophys. Acta, 1807, 1398-1413.
73.Oesterhelt, D., and Stoeckenius, W. (1973)
Functions of a new photoreceptor membrane, Proc. Natl. Acad. Sci.
USA, 70, 2853-2857.
74.Schobert, B., and Lanyi, J. K. (1982)
Halorhodopsin is a light-driven chloride pump, J. Biol. Chem.,
257, 10306-10313.
75.Yoshizawa, S., Kumagai, Y., Kim, H., Ogura, Y.,
Hayashi, T., Iwasaki, W., DeLong, E. F., and Kogure, K. (2014)
Functional characterization of flavobacteria rhodopsins reveals a
unique class of light-driven chloride pump in bacteria, Proc. Natl.
Acad. Sci. USA, 111, 6732-6737.
76.Inoue, K., Ono, H., Abe-Yoshizumi, R., Yoshizawa,
S., Ito H., Kogure, K., and Kandori, H. (2013) A light-driven sodium
ion pump in marine bacteria, Nat. Commun., 4, 1678.
77.Gushchin, I., Shevchenko, V., Polovinkin, V.,
Kovalev, K., Alekseev, A., Round, E., Borshchevskiy, V., Balandin, T.,
Popov, A., Gensch, T., Fahlke, C., Bamann, C., Willbold, D., Buldt, G.,
Bamberg, E., and Gordeliy, V. (2015) Crystal structure of a
light-driven sodium pump, Nat. Struct. Mol. Biol., 22,
390-395.
78.Bogachev, A. V., Bertsova, Y. V., Verkhovskaya,
M. L., Mamedov, M. D., and Skulachev, V. P. (2016) Real-time kinetics
of electrogenic Na+ transport by rhodopsin from the marine
flavobacterium Dokdonia sp. PRO95, Sci. Rep., 6,
21397.
79.Shalaeva, D. N., Galperin, M. Y., and
Mulkidjanian, A. Y. (2015) Eukaryotic G protein-coupled receptors as
descendants of prokaryotic sodium-translocating rhodopsins, Biol.
Direct., 10, 63.
80.Dimroth, P. (1997) Primary sodium ion
translocating enzymes, Biochim. Biophys. Acta, 1318,
11-51.
81.Dimroth, P., and Von Ballmoos, C. (2008) ATP
synthesis by decarboxylation phosphorylation, Results Probl. Cell
Differ., 45, 153-184.
Supplementary Figures and Table (PDF)