Type 1 Metallothionein (ZjMT) Is Responsible for Heavy Metal Tolerance in Ziziphus jujuba
Lan-Song Li1,2, Yu-Ping Meng2, Qiu-Fen Cao2,3*, Yong-Zhen Yang3, Fan Wang2, Hu-Sheng Jia3, Shu-Biao Wu2,4*, and Xu-Guang Liu1,3*
1College of Chemistry and Chemical Engineering, Taiyuan University of Technology, 030024 Taiyuan, China; fax: +86-351-6014138; E-mail: liuxuguang@tyut.edu.cn2Biotechnology Research Center, Shanxi Academy of Agricultural Sciences, 030031 Taiyuan, China; fax: +86-354-6215341; E-mail: qiufengcao@163.com
3Taiyuan University of Technology, Key Laboratory of Interface Science and Engineering in Advanced Materials, Ministry of Education, 030024 Taiyuan, China
4University of New England, School of Environmental and Rural Science, NSW 2351 Armidale, Australia; fax: +61-2-6773-3922; E-mail: shubiao.wu@une.edu.au
* To whom correspondence should be addressed.
Received November 16, 2015; Revision received February 3, 2016
Metallothioneins (MTs) are a family of low molecular weight, cysteine-rich, metal-binding proteins that are able to make cells to uptake heavy metals from the environment. Molecular and functional characterization of this gene family improves understanding of the mechanisms underlying heavy metal tolerance in higher organisms. In this study, a cDNA clone, encoding 74-a.a. metallothionein type 1 protein (ZjMT), was isolated from the cDNA library of Ziziphus jujuba. At the N- and C-terminals of the deduced amino acid sequence of ZjMT, six cysteine residues were arranged in a CXCXXXCXCXXXCXC and CXCXXXCXCXXCXC structure, respectively, indicating that ZjMT is a type 1 MT. Quantitative PCR analysis of plants subjected to cadmium stress showed enhanced expression of ZjMT gene in Z. jujuba within 24 h upon Cd exposure. Escherichia coli cells expressing ZjMT exhibited enhanced metal tolerance and higher accumulation of metal ions compared with control cells. The results indicate that ZjMT contributes to the detoxification of metal ions and provides marked tolerance against metal stresses. Therefore, ZjMT may be a potential candidate for tolerance enhancement in vulnerable plants to heavy metal stress and E. coli cells containing the ZjMT gene may be applied to adsorb heavy metals in polluted wastewater.
KEY WORDS: Z. jujuba Mill., metallothionein, heavy metal tolerance, heavy metal hyperaccumulationDOI: 10.1134/S000629791606002X
Many higher plants have evolved the ability to tolerate trace elements including metals. These plants are often excluders, which exclude the metals by preventing them from entering the plants or minimizing root-to-shoot translocation of trace metals. On the other hand, a group of rare plants called hyperaccumulators has extremely high tolerance to trace elements, so that high concentration of these metals does not necessarily damage their cells [1]. The overall understanding of such tolerance at genomic level is still limited due to the great complexity of tolerance and limited availability of sequence information from hyperaccumulator species, albeit the physiological and genetic mechanisms underlying heavy metal tolerance have been explored during recent decades [2]. Nevertheless, genes involved in metal tolerance have been identified in the hyperaccumulators, and many of them are functionally associated with metal uptake (ZIP family), metal vacuolar sequestration (MTP family), metal remobilization (NRAMP family), xylem loading/unloading of metal (YSL family), and the synthesis of metal ligands (NAS family) [3-9].
Metallothioneins (MTs) are a family of low molecular weight, cysteine-rich, metal-binding proteins [10, 11]. They are involved in metalloregulatory processes that include cell growth and reproduction [12], and they are present in a vast range of taxonomic groups representing a high-heterogeneity sequence without general homology [13]. In angiosperms, MTs consist of four types, MT1, MT2, MT3, and MT4, with different levels of expressions in varying tissues during plant development and with different metabolic functions [14]. It has been reported that MTs showed high binding capability to heavy metals and are able to remove heavy metals even in low concentrations [15-17]. Although the role of MTs in Cd detoxification was considered to be secondary as compared with phytochelatins and stress proteins [18], the importance of this protein family in Cd detoxification has been demonstrated at least in some of plant species such as wheat and rice [14, 19].
Chinese jujube (Ziziphus jujuba Mill.) is a unique and economically important fruit tree. It is native to, and has a long history of cultivation, in China. The species is well known for its tolerance to drought, salinity, and different environmental conditions in the field. Although limited investigations have been conducted in the drought tolerance of Ziziphus species [20, 21], the molecular mechanisms underlying the tolerance to abiotic stress is still largely unknown in the Ziziphus species such as Chinese jujube. The isolation and identification of potential genes responsible for the tolerance will provide essential insight into the understanding of the molecular mechanisms in these tolerant plants.
We hypothesize that the MT gene family present in Z. jujuba is responsible for its tolerance to abiotic stress, and bacteria such as E. coli expressing the gene will be a potential candidate for the remediation of heavy metal contaminated wastewater. In this study, a novel metallothionein type 1 gene (ZjMT) was isolated and cloned from the cDNA library of Z. jujuba, and the differential expression of ZjMT in cultured Z. jujuba seedlings stressed with different levels of cadmium was analyzed. We found that ZjMT expressed in heterologous E. coli also enhances tolerance to, and the accumulation of, heavy metals, showing potential application perspective in the treatment of heavy metal wastewater.
MATERIALS AND METHODS
Plant material and treatments. The stress-tolerant Z. jujuba Mill. “Hupingzao”, grown under field conditions at Pomology Institute of Shanxi Academy of Agricultural Sciences (Taiyuan, Shanxi, China), was used in the present study. The seeds were surface-sterilized using 0.1% (w/v) mercuric chloride for 10 min and washed thoroughly with sterile water. They were germinated in Petri dishes containing sterile wet blotting paper. Seedlings were transferred to pots 6 days after germination and grown in a greenhouse under controlled environmental conditions: temperature at 25 ± 1°C, relative humidity of 65-70%, and light density of ~2500 lx at 12 : 12 h dark/light cycle. Thirty-day-old seedlings of Z. jujuba Mill. were subjected to stress of 100 mM CdCl2 in 1/2 MS solution (pH 6.0) for 6, 24, and 48 h. Control seedlings were grown in 1/2 MS solution. Harvested leaves at appropriate time points were quick-frozen in liquid nitrogen and stored at –80°C until required for RNA isolation.
Construction of cDNA library from Chinese jujube. Total RNA was extracted using a modified cetyltrimethylammonium bromide (CTAB) method [22], and the quality was assessed on a denaturing 1.5% agarose gel. mRNA was isolated with the PolyATtract mRNA Isolation System II kit (Promega, USA) following manufacturer’s instruction. The purified mRNA was then reverse-transcribed, and double-stranded cDNA was synthesized using the cDNA Library Construction Kit (Invitrogen, USA). The cDNA was ligated to the SalI adapters, digested with NotI, and size-fractionated by a column, and then the resulting fragments of more than 500 bp were ligated into the NotI-SalI-cut pSPORTl vector according to the manufacturer’s instructions (Invitrogen). Ligated products were transformed into E. coli strain DH5α (Invitrogen) to generate a cDNA library.
cDNA library screening and isolation of ZjMT from Z. jujuba. The cDNA clones were plated onto LB plates. Recombinant plasmids were isolated, NotI-SalI digested, and cDNA inserts sequenced. Protein prediction was performed by the alignment of the sequences obtained with homologous sequences in ExPASy (http://expasy.org) and GenBank by blastx (http://www.ncbi.nlm.nih.gov). The signal peptides were detected using the SignalP 3.0 software (http://www.cbs.dtu.dk/services/SignalP/). Sequence alignment of ZjMT proteins from different species was performed using the ClustalX-2.1 program with the default settings [23] and then processed with the online BOXSHADE 3.21 software (http://www.ch.embnet.org/software/BOX_form.html).
Quantitative PCR. Approximately 1 µg of RNA was reverse-transcribed with PrimeScript® RT Master Mix kit (TaKaRa, China), and SYBR® Premix Ex TaqTMII (TaKaRa) was used to perform quantitative PCR. Amplifications were carried out on an ABI 7300 Real Time PCR System (Applied Biosystems, USA) at a final volume of 20 µl containing 1 µl of diluted cDNA template (100 ng/µl), 8 pmol of each primer, 10 µl SYBR® Premix Ex TaqTMII, 0.4 µl of Rox, and 7.8 µl of ddH2O. The ZjH3 gene was used as the housekeeping gene [24]. The ZjMT primers were designed using the Primer Premier 5.0 software. Both the primers for ZjH3 (forward primer 5′-GAGGAAGCAACTGGCAACTAAGG-3′ and reverse primer 5′-ACCAGCCTCTGGAATGGAAGTTTG-3′) and ZjMT (forward primer 5′-ACGAATTCTGATGTCTGGCTGTGG-3′ and reverse primer 5′-TATCCCGGGATTTGCACGTGCATG-3′) were synthesized by Shanghai Sangon, Ltd. (China). PCR amplification was performed under the following cycle conditions: 30 s at 94°C, followed by 40 cycles of 5 s at 95°C, 31 s at 60°C. PCR amplification was performed with samples representing six seedlings, each in triplicates. The results are expressed as the relative expression of the ZjH3 gene using the 2−ΔΔCT method [25].
ZjMT cDNA cloning and transformation in E. coli. Forward primer 5′-CTGGATCCATGTCTGGCTGTGGTTGT-3′ and reverse primer 5′-AGAAGCTTTCATTTGCACGTGCATG-3′ were designed and synthesized by Shanghai Sangon to amplify the full-length ZjMT cDNA coding sequence using recombinant plasmid from cDNA library as a template.
The PCR was performed in a 50-µl reaction mixture containing 5 µl of 10× buffer, 1.5 U Taq polymerase (TaKaRa), 10 pmol of each primer, 50 ng of DNA template, and 50 µmol of dNTPs. The PCR amplification was performed under the following conditions: 95°C for 5 min, 35 cycles at 94°C for 30 s, 52°C for 30 s, 72°C for 45 s, followed by 1 cycle at 72°C for 5 min. PCR products were purified using the Gel Extraction Kit (Qiagen, Germany), and cloned into pGEX-4T-2 vector containing the GST (glutathione-S-transferase) gene (GE Healthcare, USA). The pGEX-4T-2-ZjMT fusion plasmid was transformed into E. coli strain BL21(DE3), and pGEX-4T-2-plasmid containing transformant was used as a negative control. Putative clones containing the fusion product were screened by digestion with BamHI/HindIII and PCR amplification under the same conditions as described previously.
Expression and analysis of ZjMT in E. coli. Escherichia coli cells containing pGEX-4T-2/pGEX-4T-2-ZjMT plasmids or negative control were inoculated in LB medium containing 50 µg/ml of ampicillin and incubated at 37°C overnight with shaking. During the incubation, different concentrations of isopropyl β-D-1-thiogalactopyranoside (IPTG) were added to induce expression of the fusion protein when OD600 of the bacterial culture reached 0.5. The E. coli cells were then harvested by centrifugation at 9000g for 5 min at 4°C. The harvested cells in the loading buffer (20 mM Tris-HCl, pH 8.0, 4% (w/v) SDS, 10% (v/v) β-mercaptoethanol, 20% (v/v) glycerol, 0.04% (v/v) bromophenol blue) were heated in a boiling water bath for 5-10 min and subjected to electrophoresis on a 15% SDS-polyacrylamide gel to determine the expression of the fusion protein.
The harvested cells were also disrupted by sonication, and the supernatant was transferred onto a column of Glutathione Resins (TaKaRa) to purify the fusion protein. The purity and concentration of the protein were assessed by 15% SDS-PAGE.
Metal tolerance and accumulation in the cells containing recombinant ZjMT. Metal tolerance and bioaccumulation of heavy metals (Cd2+, Cr3+, Mn2+, Zn2+, Ni2+, and Cu2+) in E. coli containing pGEX-4T-2/pGEX-4T-2-ZjMT plasmids were assayed. Seed culture (500 µl) was added into 50 ml of MJS medium with 50 µg/ml of ampicillin and incubated at 37°C. IPTG (0.1 mM) and different concentrations of heavy metals were added to the culture when its density (OD600) reached 0.5. The cells were further incubated, and OD600 values were measured to assess the growth of the E. coli cells under the heavy metal addition treatments. When induced for 4 h in the presence of IPTG, the cells were harvested by centrifugation at 9000g for 10 min at 4°C, washed three times with 5 mM HEPES buffer, and vacuum freeze-dried. The cell pellets were digested overnight with 4 ml of 70% HNO3 at 42°C and diluted with 6 ml of deionized water. Heavy metal concentrations were then determined using an AA-6800 atomic absorption spectrophotometer (Shimadzu, Japan). Bioaccumulation capacity was presented as the amount of metals in mg accumulated per kilogram of dry cells. All the experiments were repeated three times under identical conditions.
Statistical analysis. The data were statistically analyzed using the SPSS software package (version 16.0; IBM, USA). Data are expressed by means and corresponding standard errors. One-way ANOVA was performed to compare expression of ZjMT in leaves of Z. jujuba under cadmium stress. Two-way ANOVA was performed to test the effect of the ZjMT gene and metal concentrations on ion accumulation in E. coli cells. Tukey’s post hoc test was used to compare the means. The difference was considered significant when P value was <0.05.
RESULTS
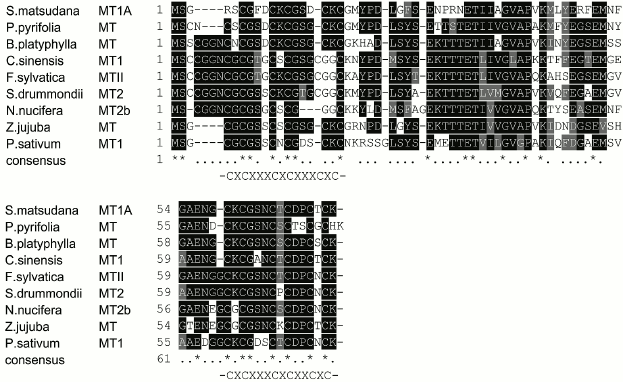
cDNA and putative protein of Z. jujuba metallothionein 1 gene. A cDNA clone with a sequence of 225 bp (GenBank Accession No. AB513130), coding for a polypeptide of 74 a.a. was isolated from the cDNA library of Z. jujuba. Based on blast and alignment analysis, the deduced protein sequence was homologous to the metallothionein proteins of other plant species in the database. Therefore, it was designated as the Z. jujuba metallothionein 1 gene (ZjMT). The deduced amino acid sequence of ZjMT contained six Cys-Xaa-Cys motifs; three of each were identified in the N- (CXCXXXCXCXXXCXC) and C-terminal (CXCXXXCXCXXCXC) domains respectively, which is the common feature of the type 1 MT proteins reported in plants. No cysteine was present in the regions between the cysteine-rich domains, which is also typical of MT proteins (Fig. 1). When the ZjMT protein was subjected to analysis by ExPASy software, it was found to be rich in neutral residues with 17.6% glycine, 16.2% cysteine, and 12.2% serine, and its molecular weight was 7376 Da with theoretical isoelectric point (pI) of 4.83 and mean hydrophobic value of –0.377. However, no signal peptide was detected by the Signal P 3.0 software.
Fig. 1. Alignment of the deduced MT amino acid sequence of Z. jujuba using ClustalX software. MT sequences from eight other species (Betula platyphylla, Fagus sylvatica, Nelumbo nucifera, Sesbania drummondii, Pyrus pyrifolia, Salix matsudana, Pisum sativum, and Camellia sinensis) are available in GenBank. Identical amino acids are marked with asterisks (*), and related amino acids with a single dot (•).
High similarities were observed between the amino acid sequences of ZjMT and its homologs. Respectively, ZjMT showed 72.6% similarity to Betula platyphylla MT, 67.6% to Fagus sylvatica MTII, 66.7% to Nelumbo nucifera MT2b, 65.3% to Sesbania drummondii MT2, 64.4% to Pyrus pyrifolia MT, 61.6% to Salix matsudana MT1A, 60.8% to Pisum sativum MT1, and 60.6% to Camellia sinensis MT1 (Fig. 1).
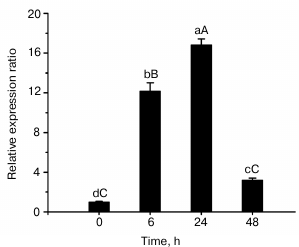
Expression of ZjMT gene in response to cadmium. Quantitative PCR was performed to analyze the expression of ZjMT under cadmium stress, and the results are shown in Fig. 2. A basal level of MT synthesis was seen at the beginning of the experiment. In response to cadmium exposure, ZjMT exhibited higher expression levels in the leaves of Z. jujuba compared with that prior to the stress treatment. Among various exposure times, the strongest response appeared at 24 h with exposure to 100 mM Cd2+, then the expression levels decreased significantly at 48 h.
Fig. 2. Expression level of ZjMT mRNA in Z. jujuba after exposure to 100 mM Cd2+. Expression was measured by SYBR Green I RT-PCR. The relative ZjMT expression level as expressed by 2−ΔΔCT was determined for each group, and vertical bars represented the mean ± S.E. (n = 6).
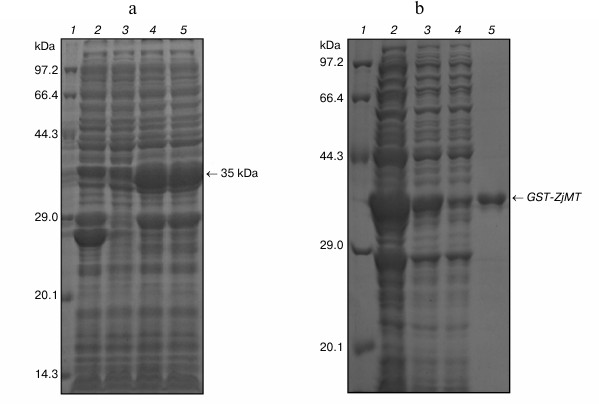
Expression of ZjMT cDNA in E. coli. A 35-kDa protein was expressed in the E. coli transformed by the recombinant plasmid pGEX-4T-2-ZjMT. This is consistent with the calculated molecular weight of the fusion protein, indicating the expression of ZjMT. An increased level of its expression was also seen by IPTG induction compared with that prior to IPTG addition into the culture (Fig. 3).
Fig. 3. SDS-PAGE analysis of GST-ZjMT fusion protein in E. coli cells. a) GST-ZjMT fusion protein in pGEX-4T-2-ZjMT/BL21 before and after IPTG induction. Lanes: 1) protein marker; 2) total protein of pGEX-4T-2/BL21 after IPTG induction; 3) total protein of pGEX-4T-2-ZjMT/BL21 without IPTG induction; 4, 5) total protein of pGEX-4T-2-ZjMT/BL21 after IPTG induction. b) Purification of GST-ZjMT fusion protein by Glutathione Resins. Lanes: 1) marker; 2-4) total protein of pGEX-4T-2-ZjMT/BL21; 5) GST-ZjMT protein purified by Glutathione Resins.
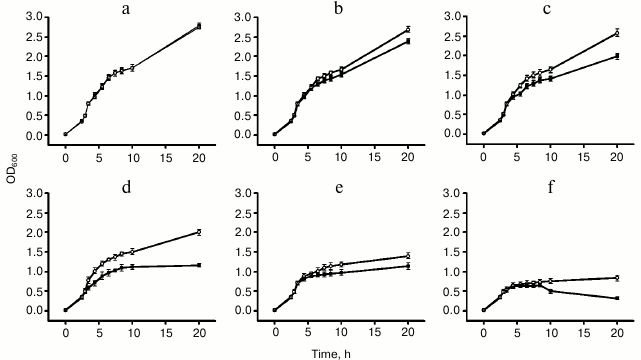
Metal tolerance and ion accumulation in E. coli. To assess the role of ZjMT in metal tolerance, E. coli cells containing pGEX-4T-2-ZjMT or pGEX-4T-2 (as a control) were subjected to different levels of metal stress. Under normal conditions, the growth pattern of cells containing pGEX-4T-2-ZjMT was similar to that of control. However, under heavy metal stress, E. coli cells expressing ZjMT showed greater proliferation than the cells containing only pGEX-4T-2, indicating the ability of ZjMT to tolerate heavy metals, particularly copper and cadmium (Fig. 4).
Fig. 4. Effects of heavy metal concentrations on growth of E. coli BL21 cells containing pGEX-4T-2-ZjMT (open circles) or pGEX-4T-2 (closed squares): a) no metals; b) 400 µM Cu2+; c) 800 µM Cu2+; d) 1600 µM Cu2+; e) 400 µM Cd2+; f) 800 µM Cd2+. The OD600 values with error bars represent the means and S.E. of three independent experiments.
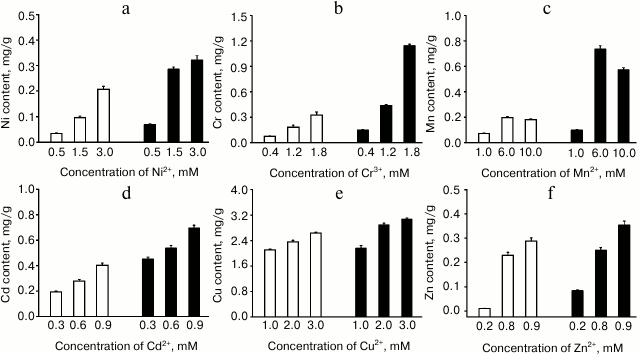
The ability of E. coli cells expressing ZjMT (pGEX-4T-2-ZjMT) to accumulate heavy metals was also investigated using E. coli cell line pGEX-4T-2 as a control. The concentrations of the metal ions in the E. coli cultures were analyzed to assess such bioaccumulations. The overall results showed that recombinant E. coli cells had significantly higher metal bioaccumulation capacity than the control group for all the metals analyzed, as shown in Fig. 5. Meanwhile, the concentrations of the metals contained in the culture significantly contributed to the levels of metal accumulations in both recombinant and control cells. In all the groups except that treated with Mn, higher concentration of metals in the culture resulted in higher levels of metal accumulations in the cells. The increases in the accumulation ability of recombinant E. coli cells expressing ZjMT were different in the groups treated with different metals. The culture of E. coli expressing ZjMT had up to 7.99-fold of Zn2+ compared with that in the control, which was the highest relative accumulation among all the metals. In contrast, the culture of E. coli expressing ZjMT contained only 1.34-fold of Cu2+ compared with that in the control, which was the lowest accumulation among all the metals. Overall, the relative metal adsorption capacity of the E. coli expressing ZjMT was in the order Zn2+ > Mn2+ > Cr3+ > Ni2+ > Cd2+ > Cu2+. When the absolute concentrations of metals in the culture were measured, E. coli cells expressing ZjMT showed the maximum bioaccumulation capacities (mg metal/kg dry weight culture): Ni – 323, Cr – 1145, Mn – 734, Cd – 696, Cu – 3075, and Zn – 354. Accordingly, the absolute metal adsorption capacity of the E. coli expressing ZjMT was in the order: Cu2+ > Cr3+ > Mn2+ > Cd2+ > Zn2+ > Ni2+.
Fig. 5. Bioaccumulation of heavy metals by E. coli containing pGEX-4T-2-ZjMT (solid columns) or pGEX-4T-2 (blank columns) in responses to respective metal concentrations in the culture and ZjMT expression in the E. coli cells: a) Ni(II); b) Cr(III); c) Mn(II); d) Cd(II); e) Cu(II); f) Zn(II). The columns represent means with error bars showing S.E. of three independent experiments. The main effects of ZjMT gene, concentration of heavy metals, and the interaction between the expression of the ZjMT gene and metal concentrations on ion accumulations in E. coli cells were analyzed using the GLM model in SPSS. The main effects were significant at the levels of P < 0.001 on the accumulations of all ions with exception of Cu2+, in which Cu2+ accumulation was significantly affected by Cu2+ concentration in the culture at the level of P < 0.01. Interactions between the expression of ZjMT gene and metal concentrations in the culture were significant at the level of P < 0.001 for Ni2+, Cr3+, and Mn2+ and P < 0.01 for Cu2+ and Zn2+, while no such interaction was detected for Cd2+. The metal contents in the cells are expressed on dry matter basis.
DISCUSSION
Metallothioneins (MTs) are Cys-rich proteins with low molecular weight that are involved in metal tolerance of diverse living organisms. In animals, MTs were found to play a key role in metal detoxification and homeostasis [17], the protection of cells against oxidative stress [26], and the scavenging of free radicals [27]. The affinity and bioaccumulation capacity of MT for heavy metals in aquatic species make these organisms possible candidates to serve as an indicator of environmental exposure to metals and as a biomarker of metal pollution. It has been reported that the exposure of aquatic organisms to metals induced MT expression. Numerous studies have shown the bioaccumulation capacities of aquatic species such as rare minnow [28], mudskipper [29], Mediterranean clam [30], and blue crab [31]. MT genes have also been reported in various plant species, such as Arabidopsis thaliana [32], Olea europaea L. [33], and Helianthus annuus [34]. While mammalian MTs have highly conserved regions [35], plant MTs contain distinctive arrangement(s) of Cys residues although their sequences and their functions may vary [16]. Despite the confirmation of the presence of MT genes in plants, conclusive demonstration of their precise cellular roles in the sequestration/homeostasis of heavy metals has been lacking.
Chinese jujube is highly tolerant to abiotic stresses such as cold, drought, salinity, and alkalinity. However, the mechanisms underlying such stresses have not been extensively investigated. In the present study, we isolated and characterized the ZjMT in the species for the first time. It is highly homologous to the MTs of Betula platyphylla and Fagus sylvatica. The alignment of the deduced amino acid sequence of ZjMT with those from other plants revealed that it belongs to the cysteine-rich protein family that is typical of MT-1 genes. The fact that ZjMT expressed in E. coli significantly enhanced the growth of the cells in comparison with the control under heavy metal stress indicates that the ZjMT gene promotes heavy metal tolerance. This was manifested by the higher heavy metal accumulation in ZjMT-expressing E. coli cells compared with the control. Furthermore, the seedlings of Z. jujuba exposed to Cd2+ showed higher expression of the ZjMT gene. This also confirmed the heavy metal tolerance role of ZjMT in vivo. Similar tolerance nature of the plant MT genes has also been reported elsewhere [32-34, 36]. Therefore, the ability of the ZjMT gene to tolerate heavy metals can at least partially explain the stress tolerance of Z. jujuba under different cultivation conditions.
Experimental results have shown that the expression of the reported MT-like genes from plants could be induced by various environmental factors other than metals, such as cold, drought, and salt stress [36-38]. The functions of plant MT genes thus appear to be diversified. Nevertheless, the molecular mechanisms underlying the diverse tolerance of MTs to environmental stresses remain largely unknown in plants [16]. It was demonstrated in the present study that IPTG-induced over-expression of the ZjMT gene in ZjMT-containing E. coli enhanced tolerance to not only essential metals such as Mn, Cu, Ni, and Zn, but also to non-essential metals such as Cd and Cr. As this tolerance of MT has also been shown in other species [33, 34, 39, 40], we suggest that the cysteine residue motifs in ZjMT may be responsible for the metal ion binding and sequestration ability [13, 15-17].
Heavy metal contamination is a worldwide environmental concern, and the risk posed to humans is increasing. Although some metals at low levels are essential for metabolisms in organisms, over-accretion of heavy metals in cells is toxic. Industries involving metal plating, mining, leather processing, and photography generate large amounts of heavy metal-containing wastes [41]. Much such waste is discharged into the environment, which inevitably poses threats to ecological systems, plant and animal production, and eventually to human health. Various measures have been applied to minimize such contamination, and removal of heavy metals from contaminated water is one of the most important measures [42, 43]. Biosorption of heavy metals using materials of microbial or plant origin has been considered as an innovative technology for removal of such pollutants [44]. For example, E. coli is biosorptive and has recently been used to adsorb heavy metals [45, 46]. Enhanced uptake and bioaccumulation of mercury by E. coli have been achieved through genetic engineering to produce a recombinant strain [47]. In our study, enhanced expression of the ZjMT gene in E. coli showed increased accumulations of essential and non-essential metal ions, confirming its important role in metal detoxification. The recombinant E. coli cells expressing ZjMT protein also showed high heavy metal tolerance and binding capacity. Therefore, the ZjMT-expressing E. coli can be potentially used in heavy metal sequestration from the environment, especially from aqueous media such as wastewater systems.
In conclusion, a metallothionein gene (ZjMT) was isolated from the cDNA library of Z. jujuba, and its function was characterized using in vitro as well in vivo approaches in the present study. ZjMT is a type 1 metallothionein and is induced in Z. jujuba plants grown under cadmium-stress conditions. Expression of the ZjMT gene in E. coli showed evident heavy metal tolerance and increased accumulations of essential and non-essential metal ions. Therefore, the ZjMT gene can be used as a candidate for transformation to other vulnerable plants to enhance their tolerance to heavy metal stress, and the recombinant E. coli strain can be potentially used in heavy metal sequestration from the environment such as in wastewater systems.
The results obtained in the present study confirmed our hypothesis that the MT gene in Z. jujuba, i.e. ZjMT, is responsible for its tolerance to abiotic stress, at least partially, and the recombinant E. coli expressing the gene will be a potential candidate for the remediation of heavy metal contaminated wastewater. Nevertheless, due to the complexity of the mechanisms underlying heavy metal tolerance and other abiotic stresses, thorough investigation of such mechanisms in plants is warranted in future studies.
This work was supported by Shanxi Provincial Key Innovative Research Team in Science and Technology (2015013002-10), Innovation Initiative Program in Science and Technology, Shanxi Academy of Agricultural Sciences (zzcx22) and Youth Foundation of Taiyuan University of Technology (No. 2012L019).
REFERENCES
1.Verbruggen, N., Hermans, C., and Schat, H. (2009)
Molecular mechanisms of metal hyperaccumulation in plants, New
Phytol., 181, 759-776.
2.Chao, Y.-E., Zhang, M., Feng, Y., Yang, X.-E., and
Islam, E. (2010) cDNA-AFLP analysis of inducible gene expression in
zinc hyperaccumulator Sedum alfredii Hance under zinc induction,
Environ. Exp. Bot., 68, 107-112.
3.Becher, M., Talke, I. N., Krall, L., and Kramer, U.
(2004) Cross-species microarray transcript profiling reveals high
constitutive expression of metal homeostasis genes in shoots of the
zinc hyperaccumulator Arabidopsis halleri, Plant J.,
37, 251-268.
4.Kramer, U., Talke, I. N., and Hanikenne, M. (2007)
Transition metal transport, FEBS Lett., 581,
2263-2272.
5.Talke, I. N., Hanikenne, M., and Kramer, U. (2006)
Zinc-dependent global transcriptional control, transcriptional
deregulation, and higher gene copy number for genes in metal
homeostasis of the hyperaccumulator Arabidopsis halleri,
Plant Physiol., 142, 148-167.
6.Weber, M., Trampczynska, A., and Clemens, S. (2006)
Comparative transcriptome analysis of toxic metal responses in
Arabidopsis thaliana and the Cd2+-hypertolerant
facultative metallophyte Arabidopsis halleri, Plant Cell
Environ., 29, 950-963.
7.Hammond, J. P., Bowen, H. C., White, P. J., Mills,
V., Pyke, K. A., Baker, A. J. M., Whiting, S. N., May, S. T., and
Broadley, M. R. (2006) A comparison of the Thlaspi caerulescens
and Thlaspi arvense shoot transcriptomes, New Phytol.,
170, 239-260.
8.Van De Mortel, J. E., Villanueva, L. A., Schat, H.,
Kwekkeboom, J., Coughlan, S., Moerland, P. D., Van Themaat, E. V. L.,
Koornneef, M., and Aarts, M. G. M. (2006) Large expression differences
in genes for iron and zinc homeostasis, stress response, and lignin
biosynthesis distinguish roots of Arabidopsis thaliana and the
related metal hyperaccumulator Thlaspi caerulescens, Plant
Physiol., 142, 1127-1147.
9.Gendre, D., Czernic, P., Conejero, G., Pianelli,
K., Briat, J. F., Lebrun, M., and Mari, S. (2007) TcYSL3, a
member of the YSL gene family from the hyper-accumulator
Thlaspi caerulescens, encodes a nicotianamine-Ni/Fe transporter,
Plant J., 49, 1-15.
10.Margoshes, M., and Vallee, B. L. (1957) A cadmium
protein from equine kidney cortex, J. Am. Chem. Soc., 79,
4813-4814.
11.Bataineh, Z. M., Heidger, P. M., Jr., Thompson,
S. A., and Timms, B. G. (1986) Immunocytochemical localization of
metallothionein in the rat prostate gland, Prostate, 9,
397-410.
12.Thirumoorthy, N., Kumar, K. M., Sundar, A. S.,
Panayappan, L., and Chatterjee, M. (2007) Metallothionein: an overview,
World J. Gastroenterol., 13, 993-996.
13.Vallee, B. L. (1991) Introduction to
metallothionein, Methods Enzymol., 205, 3-7.
14.Ernst, W. H. O., Krauss, G. J., Verkleij, J. A.
C., and Wesenberg, D. (2008) Interaction of heavy metals with the
sulphur metabolism in angiosperms from an ecological point of view,
Plant Cell Environ., 31, 123-143.
15.Kagi, J. H. R., and Schaffer, A. (1988)
Biochemistry of metallothionein, Biochemistry, 27,
8509-8515.
16.Cobbett, C., and Goldsbrough, P. (2002)
Phytochelatins and metallothioneins: roles in heavy metal
detoxification and homeostasis, Annu. Rev. Plant Biol.,
53, 159-182.
17.Coyle, P., Philcox, J. C., Carey, L. C., and
Rofe, A. M. (2002) Metallothionein: the multipurpose protein, Cell.
Mol. Life Sci., 59, 627-647.
18.Sanita di Toppi, L., and Gabbrielli, R. (1999)
Response to cadmium in higher plants, Environ. Exp. Bot.,
41, 105-130.
19.Shim, D., Hwang, J. U., Lee, J., Lee, S., Choi,
Y., An, G., Martinoia, E., and Lee, Y. (2009) Orthologs of the class A4
heat shock transcription factor HsfA4a confer cadmium tolerance in
wheat and rice, Plant Cell, 21, 4031-4043.
20.Maraghni, M., Gorai, M., Neffati, M., and Labeke,
M. C. V. (2014) Differential responses to drought stress in leaves and
roots of wild jujube, Ziziphus lotus, Acta Physiol.
Plant., 36, 945-953.
21.Kulkarni, M., Schneider, B., Raveh, E., and
Tel-Zur, N. (2010) Leaf anatomical characteristics and physiological
responses to short-term drought in Ziziphus mauritiana (Lamk.),
Sci. Hortic., 124, 316-322.
22.Kotoda, N., Wada, M., Komori, S., Kidou, S., Abe,
K., Masuda, T., and Soejima, J. (2000) Expression pattern of homologues
of floral meristem identity genes LFY and AP1
during flower development in apple, J. Am. Soc. Hortic.
Sci., 125, 398-403.
23.Larkin, M. P., Blackshields, G., Brown, N. P.,
Chenna, R., McGettigan, P. A., McWilliam, H., Valentin, F., Wallace, I.
M., Wilm, A., Lopez, R., Thompson, J. D., Gibson, T. J., and Higgins,
D. G. (2007) Clustal W and Clustal X version 2.0,
Bioinformatics, 23, 2947-2948.
24.Sun, H. F., Meng, Y. P., Cui, G. M., Cao, Q. F.,
Li, J., and Liang, A. H. (2009) Selection of housekeeping genes for
gene expression studies on the development of fruit bearing shoots in
Chinese jujube (Ziziphus jujuba Mill.), Mol. Biol. Rep.,
36, 2183-2190.
25.Livak, K. J., and Schmittgen, T. D. (2001)
Analysis of relative gene expression data using realtime quantitative
PCR and the 2(ΔΔC(T)) method, Methods, 25,
402-408.
26.Hansen, B. H., Romma, S., Garmo, O. A., Olsvik,
P. A., and Andersen, R. A. (2006) Antioxidative stress proteins and
their gene expression in brown trout (Salmo trutta) from three
rivers with different heavy metal levels, Comp. Biochem. Phys.
C, 143, 263-274.
27.Atif, F., Kaur, M., Yousuf, S., and Raisuddin, S.
(2006) In vitro free radical scavenging activity of hepatic
metallothionein induced in an Indian freshwater fish, Channa
punctata Bloch, Chem. Biol. Interact., 162,
172-180.
28.Wang, C., Zhang, F., Cao, W., and Wang, J. (2014)
The identification of metallothionein in rare minnow (Gobiocypris
rarus) and its expression following heavy metal exposure,
Environ. Toxicol. Pharmacol., 37, 1283-1291.
29.Han, Y. L., Zhang, S., Liu, G. D., Long, L. L.,
Wang, Y. F., Yang, W. X., and Zhu, J. Q. (2015) Cloning,
characterization and cadmium inducibility of metallothionein in the
testes of the mudskipper Boleophthalmus pectinirostris,
Ecotoxicol. Environ. Safety, 119, 1-8.
30.Santovito, G., Boldrin, F., and Irato, P. (2015)
Metal and metallothionein distribution in different tissues of the
Mediterranean clam Venerupis philippinarum during copper
treatment and detoxification, Comp. Biochem. Phys. C,
174-175, 46-53.
31.Lavradas, R. T., Hauser-Davis, R. A., Lavandier,
R. C., Rocha, R. C. C., Pierre, T. D. S., Seixas, T., Kehrig, H. A.,
and Moreira, I. (2014) Metal, metallothionein and glutathione levels in
blue crab (Callinectes sp.) specimens from southeastern Brazil,
Ecotoxicol. Environ. Safety, 107, 55-60.
32.Murphy, A., and Taiz, L. (1995) Comparison of
metallothionein gene expression and nonprotein thiols in ten
Arabidopsis ecotypes, Plant Physiol., 109,
945-954.
33.Dundar, E., Sonmez, G. D., and Unver, T. (2015)
Isolation, molecular characterization and functional analysis of OeMT2,
an olive metallothionein with a bioremediation potential, Mol.
Genet. Genom., 290, 187-199.
34.Tomas, M., Pagani, M. A., Andreo, C. S.,
Capdevila, M., Atrian, S., and Bofill, R. (2015) Sunflower
metallothionein family characterization. Study of the Zn(II)- and
Cd(II)-binding abilities of the HaMT1 and HaMT2 isoforms, J. Inorg.
Biochem., 148, 35-48.
35.Klaassen, C. D., Liu, J., and Choudhuri, S.
(1999) Metallothionein: an intracellular protein to protect against
cadmium toxicity, Annu. Rev. Pharmacol., 39, 267-294.
36.Yan, L., Yue, Y. C., Shu, G. Y., and Wei, M. T.
(2015) Cloning and characterization of HbMT2a, a metallothionein
gene from Hevea brasiliensis Muell. Arg differently responds to
abiotic stress and heavy metals, Biochem. Biophys. Res. Commun.,
461, 95-101.
37.Sun, H. K., Jeong, J. C., Ahn, Y. O., Lee, H. S.,
and Kwak, S. S. (2014) Differential responses of three sweet potato
metallothionein genes to abiotic stress and heavy metals, Mol. Biol.
Rep., 41, 6957-6966.
38.Akashi, K., Nishimura, N., Ishida, Y., and
Yokota, A. (2004) Potent hydroxyl radical scavenging activity of
drought-induced type-2 metallothionein in wild watermelon, Biochem.
Biophys. Res. Commun., 323, 72-78.
39.Liu, J., Shi, X., Qian, M., Zheng, L., Lian, C.,
Xia, Y., and Shen, Z. (2015) Copper-induced hydrogen peroxide
upregulation of a metallothionein gene, OsMT2c, from Oryza
sativa L. confers copper tolerance in Arabidopsis thaliana,
J. Hazard. Mater., 294, 99-108.
40.Gu, C. S., Liu, L. Q., Deng, Y. M., Zhu, X. D.,
Huang, S. Z., and Lu, X. Q. (2015) The heterologous expression of the
Iris lactea var. chinensis type2 metallothionein
IlMT2b gene enhances copper tolerance in Arabidopsis
thaliana, Bull. Environ. Contam. Toxicol., 94,
247-253.
41.Wu, G., Kang, H., Zhang, X., Shao, H., Chu, L.,
and Ruan, C. (2010) A critical review on the bio-removal of hazardous
heavy metals from contaminated soils: issues, progress,
eco-environmental concerns and opportunities, J. Hazard. Mater.,
174, 1-8.
42.Ai, P. L., and Aris, A. Z. (2014) A review on
economically adsorbents on heavy metals removal in water and
wastewater, Rev. Environ. Sci. Biotechnol., 13,
163-181.
43.Sen, A., Pereira, H., Olivella, M. A., and
Villaescusa, I. (2015) Heavy metals removal in aqueous environments
using bark as a biosorbent, Int. J. Environ. Sci. Technol.,
12, 391-404.
44.Srivastava, S., Agrawal, S. B., and Mondal, M. K.
(2015) A review on progress of heavy metal removal using adsorbents of
microbial and plant origin, Environ. Sci. Pollut. Res. Int.,
22, 15386-15415.
45.Kim, S., Song, M. H., Wei, W., and Yun, Y. S.
(2015) Selective biosorption behavior of Escherichia coli
biomass toward Pd(II) in Pt(IV)-Pd(II) binary solution, J. Hazard.
Mater., 283, 657-662.
46.Khan, Z., Nisar, M. A., Hussain, S. Z., Arshad,
M. N., and Rehman, A. (2015) Cadmium resistance mechanism in
Escherichia coli P4 and its potential use to bioremediate
environmental cadmium, Appl. Microbiol. Biotechnol., 99,
10745-10757.
47.Lin, K. H., Chien, M. F., Hsieh, J. L., and
Huang, C. C. (2010) Mercury resistance and accumulation in
Escherichia coli with cell surface expression of fish
metallothionein, Appl. Microbiol. Biotechnol., 87,
561-569.