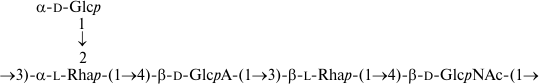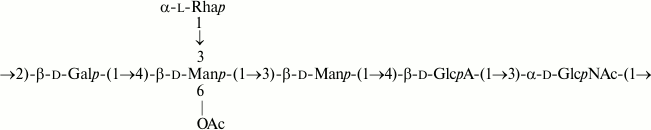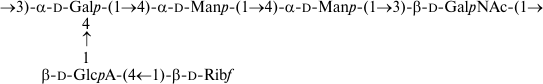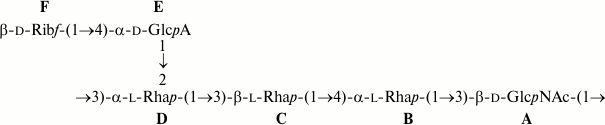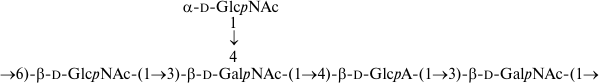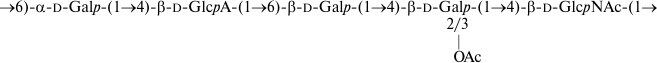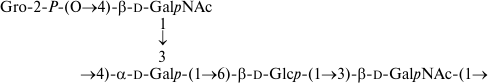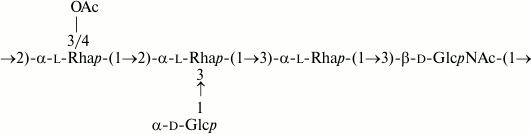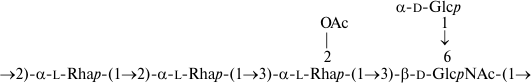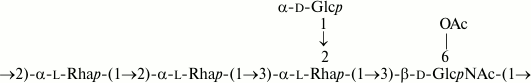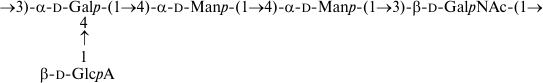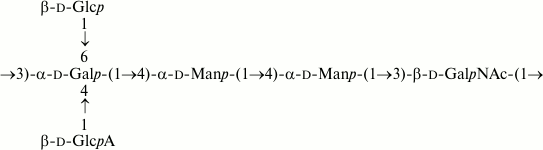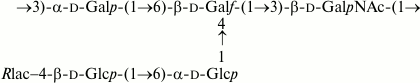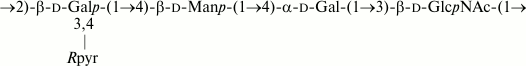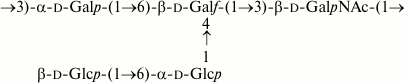Structural Relationships Between Genetically Closely Related O-Antigens of Escherichia coli and Shigella spp.
Y. A. Knirel1*, Chengqian Qian2, A. S. Shashkov1, O. V. Sizova1, E. L. Zdorovenko1, O. I. Naumenko1, S. N. Senchenkova1, A. V. Perepelov1, and Bin Liu2
1Zelinsky Institute of Organic Chemistry, Russian Academy of Sciences, 119991 Moscow, Russia; fax: +7 (499) 135-5328; E-mail: yknirel@gmail.com2Nankai University, TEDA Institute of Biological Sciences and Biotechnology, Key Laboratory of Molecular Microbiology and Technology, Ministry of Education, 300457 Tianjin, China; E-mail: striker198126@aliyun.com
* To whom correspondence should be addressed.
Received February 3, 2016; Revision received March 13, 2016
Gene clusters for biosynthesis of 24 of 34 basic O-antigen forms of Shigella spp. are identical or similar to those of the genetically closely related bacterium Escherichia coli. For 18 of these relatedness was confirmed chemically by elucidation of the O-antigen (O-polysaccharide) structures. In this work, structures of the six remaining O-antigens of E. coli O32, O53, O79, O105, O183 (all related to S. boydii serotypes), and O38 (related to S. dysenteriae type 8) were established using 1H and 13C NMR spectroscopy. They were found to be identical to the Shigella counterparts, except for the O32- and O38-polysaccharides, which differ in the presence of O-acetyl groups. The structure of the E. coli O105-related O-polysaccharide of S. boydii type 11 proposed earlier is revised. The contents of the O-antigen gene clusters of the related strains of E. coli and Shigella spp. and different mechanisms of O-antigen diversification in these bacteria are discussed in view of the O-polysaccharide structures established. These data illustrate the value of the O-antigen chemistry and genetics for elucidation of evolutionary relationships of bacteria.
KEY WORDS: Escherichia coli, Shigella dysenteriae, Shigella boydii, O-polysaccharide structure, O-antigen gene clusterDOI: 10.1134/S0006297916060067
Abbreviations: COSY, correlation spectroscopy; Cro-2-P, glycerol 2-phosphate; GalNAc, 2-acetamido-2-deoxygalactose; GlcA, glucuronic acid; GlcNAc, 2-acetamido-2-deoxyglucose; HMBC, heteronuclear multiple-bond correlation; HSQC, heteronuclear single-quantum coherence; Rha, rhamnose; Rlac, (R)-1-carboxyethyl (lactic acid ether); ROESY, rotating-frame nuclear Overhauser effect spectroscopy; Rpyr, (R)-1-carboxyethylidene (pyruvic acid acetal); TOCSY, total correlation spectroscopy.
Enteric bacteria Escherichia coli and Shigella spp.
include both commensal and pathogenic clones. Many Shigella
strains are human pathogens that cause diarrhea and bacillary dysentery
(shigellosis). Specific serotypes of E. coli are associated with
enteritis, hemorrhagic colitis, and hemolytic uremic syndrome. The
O-antigen is a polysaccharide chain (O-polysaccharide) of the
lipopolysaccharide of Gram-negative bacteria, including E. coli
and Shigella spp. It consists of a number of oligosaccharide
repeats (O-units) and is attached to lipid A via a core
oligosaccharide. The O-antigen contributes the major antigenic
variability to the bacterial cell surface and is subject to intense
selection by the host immune system, which may account for the
maintenance of diverse O-antigen forms within species. The current
typing schemes of E. coli and Shigella comprise 181 and
34 basic O-antigen forms, respectively. The O-antigen is also an
important virulence factor that can influence survival and invasiveness
of bacteria.
Escherichia coli and Shigella spp. have long been known to be closely related, but in the 1940s, strains of Shigella spp. were separated from E. coli, put into their own genus, and subgrouped into four species: Shigella boydii, Shigella dysenteriae, Shigella flexneri, and Shigella sonnei. Analysis of housekeeping genes showed that most serotypes of Shigella spp. fall into three clusters within E. coli, which were estimated to have evolved within the last 35,000 to 270,000 years [1]. Clusters 1 and 2 include 19 and 8 serotypes, respectively, due to the presence of different, generally unrelated, O-antigens. Cluster 3 includes all S. flexneri serotypes (except for type 6), whose O-antigens share a common basic structure and differ only in the distribution of side-chain glucose residues, O-acetyl groups, and phosphoethanolamine that are attached by enzymes encoded by prophage or plasmid genes [2]. Shigella boydii type 12 with an unrelated O-antigen also falls in cluster 3. Five Shigella serotypes (S. dysenteriae types 1, 8, and 10, S. boydii type 13, and S. sonnei) constitute independent lineages within E. coli. Therefore, Shigella strains are in effect E. coli with a specific mode of pathogenicity, and the overall picture is of E. coli as a diverse species with a number of pathogenic clones.
Most O-antigens of E. coli and Shigella spp. are synthesized by the polymerase Wzy/flippase Wzx-dependent pathway [3]. In the clones that use this pathway, genes for O-antigen synthesis are located in a gene cluster between conserved galF and gnd genes. The cluster typically contains genes for synthesis of nucleotide sugar precursors, genes encoding glycosyltransferases for the O-unit assembly, and O-antigen processing genes wzx and wzy. Gene clusters of 24 of 34 known basic O-antigen forms of Shigella spp. are identical or very similar to those of E. coli (Table 1), and 10 gene clusters are unique to Shigella strains, including eight in S. boydii and one each in S. dysenteriae and S. sonnei [4, 5]. O-antigen structures of all Shigella types are known and summarized in a review [4]. Escherichia coli counterparts of 18 of 24 genetically related serotypes of Shigella spp. have been studied and found to be either identical or very similar also in respect to the O-polysaccharide structure [4, 6, 7]. The O-polysaccharide structures of the six remaining E. coli serogroups O32, O53, O79, O105, O183 (all related to S. boydii strains), and O38 (related to S. dysenteriae type 8), were determined in the present work.
Table 1. Shigella spp. and E.
coli serotypes with related O-antigens
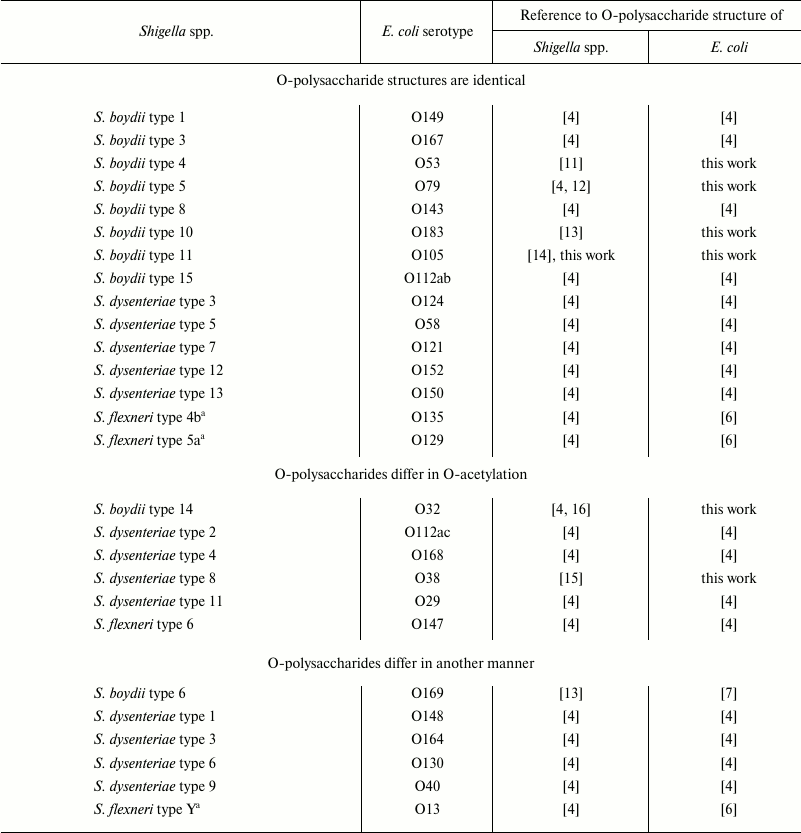
a S. flexneri types 4b, 5a, and Y have the same basic
O-antigen structure.
MATERIALS AND METHODS
Escherichia coli O32, O38, O53, O79, O105, and O183 type strains (laboratory stock numbers G1264, G1648, G1067, G4218, G2806, and G5977, respectively) were obtained from the Institute of Medical and Veterinary Science, Adelaide, Australia. The bacteria were grown to late log phase in 8 liters of Luria–Bertani broth using a 10-liter fermenter (BIOSTAT C-10; B. Braun Biotech International, Germany) under constant aeration at 37°C and pH 7.0. Bacterial cells were washed and dried as described [8].
Lipopolysaccharides were isolated from bacterial cells by the phenol–water method [9], and to remove phenol the crude extract was dialyzed without separation of the layers and then freed from nucleic acids and proteins by treatment with 50% aq. CCl3CO2H to pH 2.0 at 4°C. The supernatant was dialyzed and lyophilized. The lipopolysaccharides were obtained in yields of 7-9%.
Mild acid degradation of the lipopolysaccharides was performed with aq. 2% HOAc at 100°C until precipitation of lipid (1-1.5 h). The precipitate was removed by centrifugation (13,000g, 20 min), and an O-polysaccharide (32-52% of the lipopolysaccharide mass) was isolated by gel-permeation chromatography on a column (56 × 2.6 cm) of Sephadex G-50 Superfine (Amersham Biosciences, Sweden) in 0.05 M pyridinium acetate buffer, pH 4.5, monitored using a differential refractometer (Knauer, Germany).
Samples were deuterium-exchanged by freeze-drying from 99.9% D2O and then examined by NMR spectroscopy as solutions in 99.95% D2O. NMR spectra were recorded on an Avance II 600 MHz spectrometer (Bruker, Germany) at 20 (O79), 30 (O32, O38), 40 (O53), or 50°C (O105, O183) using internal sodium 3-trimethylsilylpropanoate-2,2,3,3-d4 (δH 0, δC —1.6) as internal reference for calibration. Two-dimensional NMR spectra were obtained using standard Bruker software, and the Bruker TopSpin 2.1 program was used to acquire and process the NMR data. Spin-lock time of 60 ms and mixing time of 150 ms were used in TOCSY and ROESY experiments, respectively.
RESULTS
Lipopolysaccharides were isolated from E. coli cells by the phenol–water procedure and cleaved with mild acid to release the O-polysaccharides, which were then isolated by Sephadex G-50 gel-permeation chromatography. The O-polysaccharides were studied by 1H and 13C NMR spectroscopy, including two-dimensional 1H,1H COSY, 1H,1H TOCSY, 1H,1H ROESY, 1H,13C HSQC, and 1H,13C HMBC experiments. As a result, the 1H and 13C NMR chemical shifts were assigned, and the spin systems for each monosaccharide residue were identified. The amino sugars (GlcNAc and GalNAc) were distinguished by correlation of the proton at the nitrogen-bearing carbon (H-2) with the corresponding carbon (C-2).
Based on 3JH,H coupling constants estimated from the two-dimensional NMR spectra and C-5 chemical shifts compared with published data [10], relative configurations of the monosaccharides and configurations of the glycosidic linkages were determined. The positions of glycosylation were established by significant downfield displacements of the 13C NMR signals for the linked carbons of the monosaccharide residues in the polysaccharides, as compared with the data of the corresponding non-substituted monosaccharides [10]. The sequences of the monosaccharides were established by 1H,1H ROESY experiments, which revealed correlations between the anomeric protons and the protons at the linkage carbons, and 1H,13C HMBC experiments showing correlations between the anomeric protons and the linkage carbons and vice versa. The positions of the O-acetyl groups, when present, were determined by significant downfield displacements of the proton and carbon signals at the O-acetylation sites due to a deshielding effect.
As a result, the structures of E. coli O53, O79, and O183 were found to be identical to those of related S. boydii types 4 [11], 5 [12], and 10 [13], respectively (Figs. 1-3). The O-polysaccharide of S. boydii type 5 includes an O-acetyl group at position 6 of a mannose residue, and the degree of O-acetylation was reported to vary from 30 to 50% in various batches of bacterial cells [12]. In the related O79-polysaccharide, about half of mannose residues are O-acetylated too.
Fig. 1. O-polysaccharide of E. coli O53 and S. boydii type 4 [11].
Fig. 2. O-polysaccharide of E. coli O79 and S. boydii type 5 [12].
Fig. 3. O-polysaccharide of E. coli O183 and S. boydii type 10 [13].
The O-polysaccharides of related E. coli O105 and S. boydii type 11 [14] include a number of minor O-acetyl groups. The 1H and 13C NMR chemical shifts of the O-deacetylated polysaccharides (Table 2) were essentially identical in the two bacteria, but analysis using two-dimensional NMR spectroscopy showed that an incorrect structure was proposed earlier for S. boydii type 11 O-polysaccharide [14]. Particularly, the revised structure shown below (Fig. 4) differs in the monosaccharide sequence and configuration of one of the rhamnosidic linkages. An attempt to determine the O-acetylation sites failed, as the content of the O-acetyl groups at each position was too low.
Table 2. 1H and 13C
NMR chemical shifts (δ, ppm)
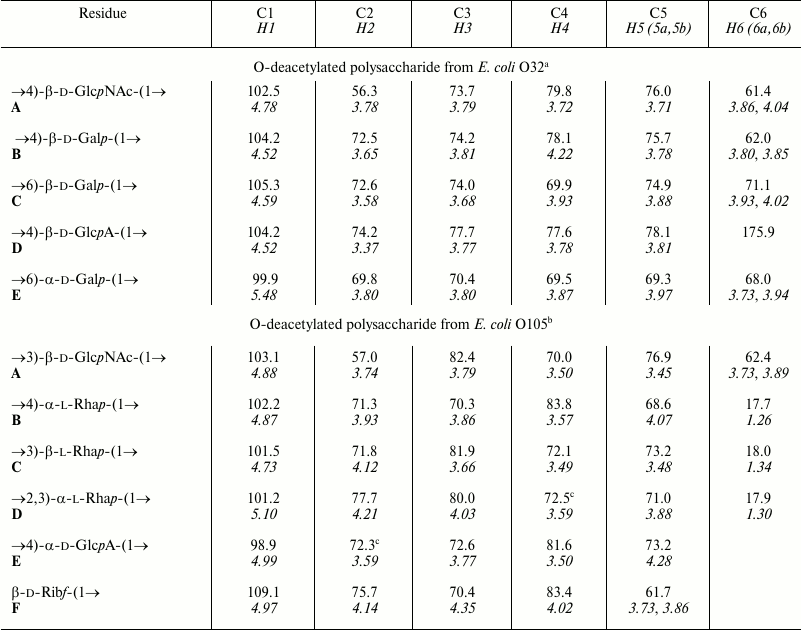
Note: 1H NMR chemical shifts are italicized.
a,b Chemical shifts for the N-acetyl group are: a
δH 2.08, δC 23.5 (Me), and
176.3 (CO); b δH 2.04, δC
23.6 (Me), and 175.5 (CO).
c Assignment could be interchanged.
Fig. 4. O-Deacetylated O-polysaccharide of E. coli O105 and S. boydii type 11 (revised structure).
The O-polysaccharide structures of E. coli O38 and S. dysenteriae type 8 also are identical (Fig. 5), but the former contains minor O-acetyl groups that have not been reported in the latter [15]. The positions of the O-acetyl groups in the O38-polysaccharide were not determined as, again, their content was too low. Different O-polysaccharide structures have been reported for two S. dysenteriae type 8 strains [15], and E. coli O38 shares the structure with S. dysenteriae strain G1221.
Fig. 5. O-Deacetylated O-polysaccharide of E. coli O38 and O-polysaccharide of S. dysenteriae type 8 [15].
Similarly, the O-polysaccharide of E. coli O32 includes O-acetyl groups as opposite to the S. boydii type 14 counterpart [16]. O-Deacetylation by treatment with aqueous ammonia resulted in a modified polysaccharide whose structure was established by two-dimensional NMR spectroscopy (for assigned 1H and 13C NMR chemical shifts see Table 2) and found to be identical to that of S. boydii type 14 (Fig. 6), which was elucidated earlier using other methods [16].
Fig. 6. O-Deacetylated O-polysaccharide of E. coli O32 and O-polysaccharide of S. boydii type 14 [16].
A comparison of the 1H,13C HSQC spectra of the initial and O-deacetylated polysaccharides from E. coli O32 revealed downfield displacement of parts of the H2/C2 and H3/C3 cross-peaks of 4-substituted galactose residue B from δ 3.65/72.5 and 3.81/74.2 to δ 5.02/74.2 and 5.01/75.9, respectively. Therefore, residue B is partially O-acetylated at position either 2 or 3 (Fig. 7). Accordingly, the signals for the neighboring carbons C1 and C3 of Gal2Ac shifted upfield from δ 104.2 and 74.2 to δ 101.9 and 72.9, and those for C2 and C4 of Gal3Ac from δ 72.5 and 78.1 to δ 71.2 and 75.2, respectively (β-effects of O-acetylation). As judged by the ratios of integral intensities of the 1H NMR signals of various forms of residue B, the degree of O-acetylation at position 2 and 3 is ~20 and ~30%, respectively.
Fig. 7. O-polysaccharide of E. coli O32.
DISCUSSION
Elucidation of the O-polysaccharide structures of six remaining E. coli serotypes that are related to Shigella strains enables a complete view on the antigenic relationships between these bacteria. Totally 24 and 27 basic O-antigen forms of Shigella spp. and E. coli, respectively, are involved (Table 1). Of them, 15 pairs possess identical O-polysaccharide structures, and in six pairs, the O-polysaccharides differ only in O-acetylation, which occurs in one of the counterparts. The O-antigen gene clusters of these bacteria have pairwise the same organization and a high level of DNA homology (>97% identity) [4]. This finding indicates that O-acetylation is encoded elsewhere in the genome, most likely by prophage genes as demonstrated for S. flexneri [2].
The O-units of E. coli O130 (Fig. 8) and S. dysenteriae type 6 (Fig. 9) are identical apart from O-acetylation in the latter, but the O-antigen of type 6 comprises only one O-unit. The O-antigen gene clusters of these strains are nearly identical, but in type 6, the polymerase gene wzy and one of the glycosyltransferase genes wffH are fused in one open reading frame due to a single base deletion, which apparently blocks Wzy function [4]. O-Acetylation in type 6 is encoded outside the gene cluster.
Fig. 8. O-polysaccharide of E. coli O130 [4].
Escherichia coli O13, O129, and O135 share the basic O-antigen structure and gene cluster with S. flexneri non-6 serotypes [6]. Whereas O129- and O135-polysaccharides are O-acetylated and glycosylated in the same manner as those of S. flexneri types 5a and 4b, respectively (Figs. 10 and 11), the O13-polysaccharide is distinguished by a unique site of side-chain glycosylation (Fig. 12). No genes for glycosylation are present in the O-antigen gene clusters of O13, O129, and O135, and, most likely, as in S. flexneri [2], this modification is encoded by a gtr gene cluster having phage origin.
Fig. 10. O-polysaccharide of E. coli O129 [6] and S. flexneri type 5a [6].
Fig. 11. O-polysaccharide of E. coli O135 [6] and S. flexneri type 4b [6].
Fig. 12. O-polysaccharide of E. coli O13 [6].
In contrast, in E. coli O169, a glycosyltransferase gene that may be responsible for side-chain glucosylation occurs at the 3′-end of the O-antigen gene cluster [7]. This gene is absent from the related cluster of S. boydii type 6, whose O-polysaccharide lacks any side-chain glucose residue accordingly [13] (compare Figs. 13 and 14). This pair is genetically related to another pair of E. coli O183 and S. boydii type 10 [5], but the O-polysaccharide of the latter pair includes a ribofuranosyl residue terminating a short side chain (see “Results” section). The genetic basis for the lack of ribose from the O-polysaccharides of E. coli O169 and S. boydii type 6 has been elucidated due to different recombination events affecting the ribofuranosyltransferase gene wbaM [7, 13].
Fig. 13. O-polysaccharide of S. boydii type 6 [13].
Fig. 14. O-polysaccharide of E. coli O169 [7].
In two pairs, related O-antigens differ in the presence or absence of a non-sugar substituent: ether-linked (R)-lactic acid (Rlac) or acetal-linked pyruvic acid (Rpyr). They occur in the O-polysaccharides of S. dysenteriae type 3 (identical to E. coli O124) (Fig. 15) and type 9 (Fig. 16) but are absent from E. coli O164 (Fig. 17) and O40 counterparts (Fig. 18), respectively [4]. The loss of the acids is accounted for by inactivation of genes involved in their synthesis in otherwise nearly identical O-antigen gene clusters. The inactivation of the wffR gene responsible for synthesis of the lactic acid ether in O164 is due to an insertion of an IS3 element, and a putative pyruvyltransferase gene lat (wfeP) is inactivated by a frame-shift mutation in O40 [4].
Fig. 15. O-polysaccharide of E. coli O124 [4] and S. dysenteriae type 3 [4].
Fig. 16. O-polysaccharide of S. dysenteriae type 9 [4].
Fig. 17. O-polysaccharide of E. coli O164 [4].
Fig. 18. O-polysaccharide of E. coli O40 [4].
Escherichia coli O148 (Fig. 19) and S. dysenteriae type 1 (Fig. 20) have similar O-polysaccharides that differ only in replacement of an α-d-glucose residue in the former with an α-d-galactose residue [4]. The O-antigen gene clusters of the two strains have the same organization and a high level of DNA homology (89.8-99.5% identity), except that in S. dysenteriae type 1 the glucosyltransferase gene wbbG is interrupted by a deletion, and a plasmid-borne galactosyltransferase gene wbbP is responsible for the transfer of the galactose residue. It seems evident that the type 1 O-antigen evolved from the O148 antigen by inactivation of wbbG and gain of a wbbP-carrying plasmid [4].
Fig. 19. O-polysaccharide of E. coli O148 [4].
Fig. 20. O-polysaccharide of S. dysenteriae type 1 [4].
Therefore, there are different mechanisms of diversification of the O-antigens of E. coli and Shigella spp. including: (i) acquisition of a gene for O-acetylation or side-chain glucosylation either located in a prophage or incorporated into the O-antigen gene cluster resulting in a complication of the O-polysaccharide structure; (ii) inactivation of a gene for a glycosyltransferase (ribosyltransferase) or for transfer or synthesis of a non-sugar acid constituent (lactic acid or pyruvic acid) resulting in a reduction of the O-antigen structure, and (iii) inactivation of a gene for a glycosyltransferase amended by acquisition of a plasmid carrying another glycosyltransferase gene.
Nine of ten O-antigen structures that are unique for Shigella spp. (those of S. boydii types 2, 7, 9, 12, 13, 16-18 and S. dysenteriae type 10) are rather typical for E. coli [4]. Particularly, as the majority of the O-antigens shared by Shigella spp. and E. coli, they all are acidic. The corresponding parent E. coli strains have been either extinguished from nature or not found yet. In contrast, the O-antigen of S. sonnei is quite atypical for E. coli, and the gene cluster for its biosynthesis is thought to have been transferred on a plasmid from Plesiomonas shigelloides [17].
This work was supported by the Russian Science Foundation (project No. 14-14-01042). Chengqian Qian and Bin Liu were supported by the International Science & Technology Cooperation Program of China (2012DFG31680 and 2013DFR30640), the National Key Program for Infectious Diseases of China (2013ZX10004216-001-001 and 2013ZX10004221-003), National Natural Science Foundation of China (NSFC) Program (31371259 and 81471904), and Research Project of Chinese Ministry of Education (No. 113015A).
REFERENCES
1.Pupo, G. M., Lan, R., and Reeves, P. R. (2000)
Multiple independent origins of Shigella clones of
Escherichia coli and convergent evolution of many of their
characteristics, Proc. Natl. Acad. Sci. USA, 97,
10567-10572.
2.Knirel, Y. A., Sun, Q., Senchenkova, S. N.,
Lan, R., Perepelov, A. V., and Xu, J. (2015) O-Antigen modifications
providing the antigenic diversity of Shigella flexneri and the
underlying genetic mechanisms, Biochemistry (Moscow), 80,
901-914.
3.Raetz, C. R., and Whitfield, C. (2002)
Lipopolysaccharide endotoxins, Annu. Rev. Biochem., 71,
635-700.
4.Liu, B., Knirel, Y. A., Feng, L., Perepelov, A. V.,
Senchenkova, S. N., Wang, Q., Reeves, P. R., and Wang, L. (2008)
Structure and genetics of Shigella O-antigens, FEMS
Microbiol. Rev., 32, 627-653; Corrigendum: (2010) FEMS
Microbiol. Rev., 34, 606.
5.Iguchi, A., Iyoda, S., Kikuchi, T., Ogura, Y.,
Katsura, K., Ohnishi, M., Hayashi, T., and Thomson, N. R. (2014) A
complete view of the genetic diversity of the Escherichia coli
O-antigen biosynthesis gene cluster, DNA Res., 22,
105-107.
6.Perepelov, A. V., Shevelev, S. D., Liu, B.,
Senchenkova, S. N., Shashkov, A. S., Feng, L., Knirel, Y. A., and
Wang, L. (2010) Structures of the O-antigens of Escherichia coli
O13, O129 and O135 related to the O-antigens of Shigella
flexneri, Carbohydr. Res., 345, 1594-1599.
7.Perepelov, A. V., Shashkov, A. S., Guo, X.,
Filatov, A. V., Weintraub, A., Widmalm, G., and Knirel, Y. A. (2015)
Structure and genetics of the O-antigen of Escherichia coli O169
related to the O-antigen of Shigella boydii type 6,
Carbohydr. Res., 414, 46-50.
8.Robbins, P. W., and Uchida, T. (1962) Studies on
the chemical basis of the phage conversion of O-antigens in the E-group
Salmonellae, Biochemistry, 1, 323-335.
9.Westphal, O., and Jann, K. (1965) Bacterial
lipopolysaccharides. Extraction with phenol–water and further
applications of the procedure, Methods Carbohydr. Chem.,
5, 83-91.
10.Lipkind, G. M., Shashkov, A. S., Knirel, Y. A.,
Vinogradov, E. V., and Kochetkov, N. K. (1988) A computer-assisted
structural analysis of regular polysaccharides on the basis of
13C-N.M.R. data, Carbohydr. Res., 175,
59-75.
11.Perepelov, A. V., Senchenkova, S. N.,
Shashkov, A. S., Liu, B., Feng, L., Wang, L., and Knirel, Y. A. (2008)
Antigenic polysaccharides of bacteria. 40. Revision of the structures
of the O-specific polysaccharides of Shigella dysenteriae types
3, 9 and Shigella boydii type 4 by NMR spectroscopy, Russ. J.
Bioorg. Chem., 34, 110-117.
12.L’vov, V. L., Shashkov, A. S., Knirel, Y.
A., Arifulina, A. E., Senchenkova, S. N., Yakovlev, A. V., and
Dmitriev, B. A. (1995) Structure of the O-specific polysaccharide chain
of Shigella boydii type 5 lipopolysaccharide: a repeated study,
Carbohydr. Res., 279, 183-192.
13.Senchenkova, S. N., Feng, L., Yang, J.,
Shashkov, A. S., Cheng, J., Liu, D., Knirel, Y. A., Reeves, P., Jin,
Q., Ye, Q., and Wang, L. (2005) Structural and genetic characterization
of the Shigella boydii type 10 and type 6 O-antigens, J.
Bacteriol., 187, 2551-2554.
14.L’vov, V. L., Yakovlev, A. V., Shashkov, A.
S., and Dmitriev, B. A. (1991) Antigenic polysaccharides of bacteria.
The structure of the polysaccharide chain of the lipopolysaccharide of
Shigella boydii type 11, Bioorg. Khim., 17,
111-120.
15.Perepelov, A. V., Senchenkova, S. N.,
Shashkov, A. S., Knirel, Y. A., Liu, B., Feng, L., and Wang, L. (2008)
Antigenic polysaccharides of bacteria. 42. Structures of the
O-polysaccharides from two Shigella dysenteriae type 8 strains,
Russ. J. Bioorg. Chem., 34, 725-729.
16.L’vov, V. L., Yakovlev, A. V., Pluzhnikova,
G. N., Lapina, E. B., Shashkov, A. S., and Dmitriev, B. A. (1987)
Antigenic polysaccharides of bacteria. The structure of the
polysaccharide chain of the Shigella boydii type 14
lipopolysaccharide, Bioorg. Khim., 13, 1256-1265.
17.Shepherd, J. G., Wang, L., and Reeves, P. R.
(2000) Comparison of O-antigen gene clusters of Escherichia coli
(Shigella) sonnei and Plesiomonas shigelloides
O17: sonnei gained its current plasmid-borne O-antigen genes
from P. shigelloides in a recent event, Infect. Immun.,
68, 6056-6061.