REVIEW: Diversity of CRISPR-Cas-Mediated Mechanisms of Adaptive Immunity in Prokaryotes and Their Application in Biotechnology
E. E. Savitskaya1,2*, O. S. Musharova1,3, and K. V. Severinov1,2,3
1Skolkovo Institute of Science and Technology, Skolkovo Innovation Center, 143026 Moscow, Russia; E-mail: savitskaya.e@yandex.ru2Institute of Molecular Genetics, Russian Academy of Sciences, 123182 Moscow, Russia
3Institute of Gene Biology, Russian Academy of Sciences, 119334 Moscow, Russia
* To whom correspondence should be addressed.
Received February 16, 2016; Revision received February 19, 2016
CRISPR-Cas systems of adaptive immunity in prokaryotes consist of CRISPR arrays (clusters of short repeated genomic DNA fragments separated by unique spacer sequences) and cas (CRISPR-associated) genes that provide cells with resistance against bacteriophages and plasmids containing protospacers, i.e. sequences complementary to CRISPR array spacers. CRISPR-Cas systems are responsible for two different cellular phenomena: CRISPR adaptation and CRISPR interference. CRISPR adaptation is cell genome modification by integration of new spacers that represents a unique case of Lamarckian inheritance. CRISPR interference involves specific recognition of protospacers in foreign DNA followed by introduction of breaks into this DNA and its destruction. According to the mechanisms of action, CRISPR-Cas systems have been subdivided into two classes, five types, and numerous subtypes. The development of techniques based on CRISPR interference mediated by the Type II system Cas9 protein has revolutionized the field of genome editing because it allows selective, efficient, and relatively simple introduction of directed breaks into target DNA loci. However, practical applications of CRISPR-Cas systems are not limited only to genome editing. In this review, we focus on the variety of CRISPR interference and CRISPR adaptation mechanisms and their prospective use in biotechnology.
KEY WORDS: CRISPR array, cas genes, CRISPR adaptation, CRISPR interferenceDOI: 10.1134/S0006297916070026
Abbreviations: CRISPR, clustered regularly interspaced short palindromic repeats.
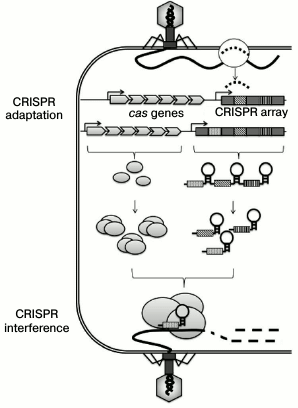
DISCOVERY OF CRISPR-Cas SYSTEMS AND MECHANISMS OF THEIR
ACTION
An unusual Escherichia coli genome locus composed of four short repeats separated by unique spacer sequences was first described in 1987 [1]. With more genomes deciphered, it became evident that similar families of genome repeats are typical for many archaea and eubacteria [2]. Based on their architecture, these families were named CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats). The importance of CRISPR was first predicted in 2005 “at the tip of a pen”, when it was noted that some of the spacers corresponded to nucleotide fragments from plasmids and bacteriophage genomes [3-5]. It was suggested that CRISPR could be involved in cell protection from foreign DNA. At the same time, data has been obtained on the transcription of CRISPR loci [6], and stable association of CRISPR loci with a set of genes (cas, CRISPR-associated) was demonstrated [7]. Three years later, the involvement of CRISPR loci in cell protection against foreign DNA was proven experimentally – it was shown that the presence of cas genes and a CRISPR array spacer complementary to a plasmid sequence fragment (protospacer) prevented plasmid conjugation [8]. The target DNA was destroyed by RNA–protein complexes composed of the cas gene products and short crRNAs formed by the CRISPR array processing, so that each crRNA contained one spacer flanked by fragments of DNA repeats [9] (figure). Thus, one of the most important properties of CRISPR-Cas systems was discovered – the ability to recognize and modify a fragment of a genome complementary to the CRISPR array spacer. This phenomenon was named CRISPR interference. Mechanisms preventing autoimmune response possible in the case of recognition of a cell’s own spacers in the content of a CRISPR array differ in different types of CRISPR-Cas systems, and they will be discussed below.
Mechanism of CRISPR-Cas adaptive immunity in prokaryotes. After entering a bacterial cell, fragments of foreign DNA integrate into a CRISPR array in the process of CRISPR adaptation. A CRISPR array is elongated by one new spacer and one repeat. The CRISPR array is then transcribed with the formation of pre-crRNA that is processed into short crRNAs, so that each crRNA contains a spacer flanked by partial repeats. The cas genes code for protein components of the CRISPR interference and CRISPR adaptation complexes. A CRISPR interference complex that includes crRNA and Cas proteins interacts with a protospacer, i.e. a target DNA sequence complementary to the sequence of the crRNA spacer, and this interaction leads to the degradation of the target DNA molecule
Barrangou et al. demonstrated that when Streptococcus thermophilus cells were infected with a bacteriophage, the surviving cells acquired resistance to repeated infection by the same bacteriophage due to the insertion into their CRISPR array of new spacers complementary to fragments of the phage genome [10] – a phenomenon named CRISPR adaptation. CRISPR adaptation is a unique CRISPR-Cas-mediated defensive mechanism that resembles the immunity of higher eukaryotes.
CLASSIFICATION OF CRISPR-Cas SYSTEMS AND MECHANISMS OF CRISPR
INTERFERENCE
CRISPR-Cas systems have been found in more than 40% of eubacteria and 80% of archaea [11, 12]. The availability of high-throughput sequencing and a growing list of completely deciphered genomes have allowed researchers to compare and classify CRISPR-Cas systems. Since proteins responsible for CRISPR adaptation are homologous in all CRISPR-Cas systems, this classification is based mostly on the protein composition of the complexes involved in CRISPR interference. According to the latest data, CRISPR-Cas systems can be subdivided into two classes, five types, and 16 subtypes [13]. The two classes are distinguished based on composition of the interference complexes: CRISPR-Cas systems of Class 1 (Types I, III, IV) are multi-subunit, while systems of Class 2 (Types II and V) contain only one protein (table). The mechanisms of crRNA maturation and the proteins involved in CRISPR interference vary substantially. No evolutionary relation between components of the CRISPR interference systems of the first and second classes has been found [13]. The phylogeny of the CRISPR-Cas system usually does not reflect the phylogeny of the bacteria in whose genomes these systems were found, which indicates the dominating role of horizontal gene transfer in the distribution and evolution of CRISPR-Cas systems.
Classification and variety of interference mechanisms in CRISPR-Cas
systems
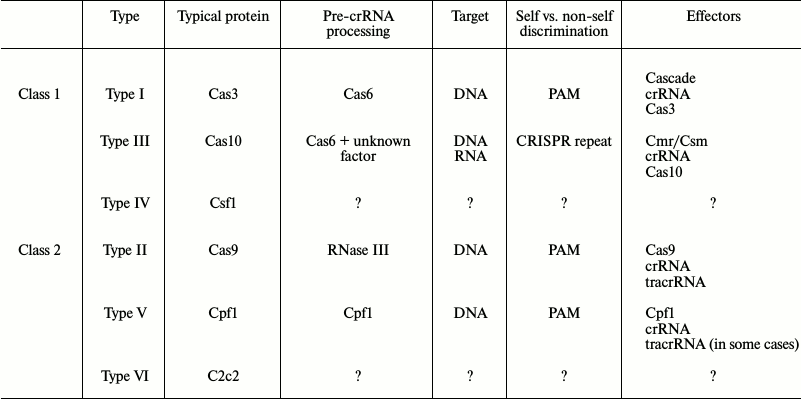
Class 1. Type I. Type I CRISPR-Cas systems are characterized by the presence of a Cas3 protein that displays both nuclease and helicase activities (table). They also contain the multi-subunit crRNA-containing Cascade complex [13] responsible for target recognition in CRISPR interference. Type I includes the well-studied CRISPR-Cas I-E subtype system from E. coli. The 405-kDa Cascade complex from E. coli is composed of the Cse1, Cse2, Cas5, Cas7, and Cas6e proteins in ratio 1 : 2 : 1 : 6 : 1 [9, 14]. The crRNA is formed by processing of the CRISPR array primary transcript (pre-crRNA) by ribonuclease activity of the Cas6e protein [9]. The Cas6e protein is dispensable from the complex if other sources of mature crRNAs exist [15]. In the subtype I-A systems, Cas6 is not a stable constituent of the Cascade complex, which causes considerable variations in the length of crRNA, whose termini remain unprotected [16, 17]. The crRNA-containing Cascade complex recognizes a complementary protospacer in the target DNA; DNA of the protospacer unwinds with the formation of an R-loop, i.e. the heteroduplex between crRNA spacer, target DNA protospacer, and replaced “nontarget” single DNA strand. The next stage requires the effector Cas3 protein [18, 19] that contains the core SF2 family helicase domain with RecA motifs and the N-terminal HD nuclease domain [20]. Cas3 introduces breaks into the replaced DNA strand of the R-loop, thereby initiating target degradation [21, 22]. The 3D structures of some Cas3 proteins and their complexes with short single-stranded DNA fragments, presumably DNA degradation products, have been solved [23-25].
In Type I systems, prevention of an autoimmune response, i.e. discrimination between the protospacer in the target DNA and the CRISPR array spacer, occurs by the following mechanism. Cse1, one of the Cascade complex components, recognizes a short protospacer-adjacent motif (PAM) that precedes the protospacer [26], such recognition being an obligatory condition for the interaction with the target DNA [18, 26, 27]. On one hand, this prevents autoimmune response against the CRISPR array spacer, because the repeat sequence differs from the PAM sequence. On the other hand, it narrows the possibility for target recognition. Thus, mutations in PAMs adjacent to protospacers completely matching crRNA spacers allow bacteriophages to avoid CRISPR interference [28-30].
Type III. Type III systems are characterized by the presence of the Cas10 protein. Cas10 contains a Palm domain that is similar to the RNA-recognizing domains of polymerases (table). There are two families of multi-subunit interference complexes – Cas10–Csm and Cas10–Cmr – that are typical for Subtype III-A and III-B systems, respectively [13]. Type III CRISPR-Cas systems are active against both DNA [8, 31-33] and RNA [33-38] protospacers: they first recognize and cleave the RNA transcript and then the corresponding template DNA [33]. Cas10 is responsible for the DNase activity [33], while Csm and Cmr display ribonuclease activity [33, 35, 36]. Active transcription of the target sequence is an obligatory condition for CRISPR interference mediated by Type III systems [32, 39]. It prevents CRISPR interference against prophages that can result in host cell death [32].
The crRNA maturation in Type III systems is not completely understood. It is known that the Cas6 ribonuclease, which is not a component of the interference complex, cleaves the CRISPR array primary transcript into intermediates composed of individual spacers flanked by partial repeats on both ends. The last eight nucleotides of the 5-flanking repeat [8] were named crRNA tag [40]. Partially matured crRNA is incorporated by an unknown mechanism into the Csm or Cmr complex, where its 3-terminus undergoes additional processing [41]. Unlike Type I systems, in which an autoimmune response depends on PAM recognition, autoimmune response in Type III systems is determined by complementary interactions between the crRNA tag and the target sequence. Full complementarity of the crRNA tag and a fragment of the CRISPR array repeat prevent CRISPR interference. In all other cases, CRISPR interference is possible [33, 37, 42].
Type IV. The existence of Type IV CRISPR-Cas systems was predicted through bioinformatic searches [13]. Their characteristic feature is the presence of the csf1 gene (table) usually associated with the cas5 and cas7 genes. The occurrence of these operons in the absence of association with CRISPR arrays suggests they have functions other than adaptive immunity. However, the mechanisms of action and the functions of Type IV CRISPR-Cas systems require further study and experimental verification.
Class 2. Type II. The interference complex of Type II CRISPR-Cas systems includes the Cas9 protein and two RNAs – crRNA and trans-encoded small RNA (tracrRNA), the latter containing a sequence complementary to the CRISPR repeat [43] (table). The tracrRNA pairs with the pre-crRNA and directs its cleavage by RNase III to produce mature crRNA [43]. Similarly to Type I systems, the interaction of crRNA with the target protospacers requires the presence of PAM [44-46]. Recognition of PAM and the protospacer, local unwinding of DNA target, R-loop formation, and introduction of breaks into target DNA are provided by the activity of Cas9 in complex with the two above-mentioned RNAs. The simplicity of the CRISPR interference mechanism in the Type II systems and the possibility of combining crRNA and tracrRNA into a single guiding RNA provided the basis for rapid development of Cas9-mediated techniques for eukaryotic genome editing [47]. Cas9 has two domains – HNH and RuvC [48]. Each of these domains cleaves one of the protospacer strands in the R-loop. The breaks in both chains are located across from each other, so that the cleavage produces a DNA molecule with blunt ends [10, 43, 49-52].
Type V. The typical feature of Type V CRISPR-Cas systems is the presence of the Cpf1 protein and its homologs (table). This relatively large protein resembles Cas9 of the Type II system, because it contains a RuvC-like nuclease domain, but it differs from Cas9 by the absence of the HNH nuclease domain [13, 53]. Only recently, the functional activity of some Type V CRISPR-Cas systems has been confirmed experimentally [54, 55]. It appeared that, similarly to Type II systems, Type V systems require PAM for target recognition. In some Type V systems, crRNA maturation does not require tracrRNA and RNase III and is presumably catalyzed by the Cpf1 protein itself. Cleavage of two target DNA strands occurs with a shift, which results in the formation of “sticky” single-stranded ends 4-5 nucleotides long [55]. The possibility of crRNA-guided genome editing has been demonstrated for at least two Cpf1 proteins – from Acidaminococcus sp. BV3L6 and Lachnospiraceae bacterium ND2006 [55].
Enormous interest in Class 2 systems due to their application in genome editing and the necessity for perfecting existing techniques has stimulated the search for other systems in which CRISPR interference could be performed by a single protein. In 2015, bioinformatic analysis predicted the existence of Type VI CRISPR-Cas systems (table) [54], although the mechanisms of action and the functionality of these systems still have to be investigated. However, such studies are extremely important, because they can identify new genome editing tools that (i) consist of smaller genes/proteins, which will make their packaging into viral particles for delivery into cells possible; (ii) have higher specificity. Increasing the number of PAMs recognized by different types of CRISPR systems will allow choosing an appropriate tool for editing specific genome sequences. It is also of importance that new systems avoid intellectual property problems that have not been resolved for Type II systems.
CRISPR ADAPTATION
CRISPR adaptation is one of the few mechanisms of directed genome modification: a fragment of foreign DNA (protospacer) integrates into a CRISPR array to become a new spacer. Simultaneously, a CRISPR repeat is duplicated. The protein involved in CRISPR adaptation is Cas1. Cas1 is common for all CRISPR-Cas systems. It is also the most evolutionarily conserved component of these systems [56]. As a rule, cas1 and cas2 genes are located close to each other, and the encoded proteins form a stable complex [57-59]. Cas1 is an endonuclease [60, 61]; Cas2 displays nuclease activity toward both RNA and DNA in vitro [62-64]. However, CRISPR adaptation in vivo requires Cas1 nuclease activity only [57]. The Cas1–Cas2 complex is indispensable and sufficient for CRISPR adaptation in vivo, as demonstrated in E. coli [65]. The promoter region (leader) of the CRISPR array provides its transcription with the formation of pre-crRNA and plays an important role in CRISPR adaptation: novel spacers are integrated adjacent to the repeat that is proximal to the leader sequence [65-67]. Polar insertion of new spacers allows reconstruction of the chronological order of spacer incorporation.
In CRISPR-Cas systems that require the presence of PAMs for target DNA recognition, selection of protospacers for acquisition should involve a specific mechanism of PAM identification [68], because only such spacers will be functional in CRISPR interference. Type II systems require Cas9 for PAM recognition and CRISPR adaptation [69, 70]. In Type I systems, the adaptational Cas1–Cas2 complex was found to be capable of recognizing PAMs during protospacer selection [65].
The mechanism of preference for foreign DNA over the course of protospacer acquisition remains a major unresolved question. Recently, Levy et al. showed that intermediates of double-strand break reparation by RecBCD-mediated homologous recombination act as hotspots of protospacer selection [71]. They suggested that integration of a protospacer from the chromosome is prevented by stalling the RecBCD complex at Chi-sites that are frequent in the E. coli genome but quite rare in plasmids and bacteriophages. This work revealed an important relation between CRISPR adaptation and mechanisms of genome stability in cells.
Another mechanism for targeting adaptational complexes to foreign DNA is DNA recognition by the interference complex. Thus, it has been demonstrated that expression of the CRISPR interference complex increased the efficiency of CRISPR adaptation [72]: new spacers were selected from the target DNA strand that was recognized by crRNA in the content of the effector complex [72-76]. This process, in which existing crRNA determines specific selection of new spacers, was called primed CRISPR adaptation. The molecular mechanism of primed CRISPR adaptation remains unknown.
Recently, new details of spacer integration into CRISPR arrays have been revealed in experiments performed in vivo [59] and in vitro [57]. It was found that the Cas1–Cas2 complex introduces a single-strand break exactly at the leader–repeat junction and catalyzes nucleophilic attack of the 3-OH end of the incoming spacer onto the 5-end of the first repeat. Similarly, the other strand is nicked at the first repeat–spacer junction and the 5-end of the repeat strand is joined to the 3-end of the new spacer. As a result, the incorporated spacer is flanked by the single-stranded repeat sequences that get filled later.
The two-step mechanisms of spacer insertion into a CRISPR array resemble the process of transposon integration into a genome. Koonin and Krupovic proposed a hypothesis on modular composition of CRISPR-Cas systems, according to which the CRISPR adaptation module appears on the basis of mobile genomic elements and gains the ability for integration of sequences (spacers) into the genome. In the course of evolution, the adaptation module has been combined with other modules that provided CRISPR interference, i.e. target DNA degradation [77].
APPLICATIONS OF CRISPR-Cas SYSTEMS
Practical interest in CRISPR-Cas systems originated from their ability to recognize almost any unique DNA locus. This recognition can be directed toward specific DNA sequences via complementary interactions between crRNA spacers and protospacers of the target molecules. It underlies the development of Cas9-mediated techniques of genome editing that have revolutionized biotechnology in the past three years. However, the potential of CRISPR-Cas systems as sources for new biotechnological tools is not yet exhausted. Genome editing with Class 2 CRISPR-Cas systems has been described in several reviews [78, 79], and this is why in this article we focus on other approaches to the application of CRISPR-Cas systems.
Gene expression regulation. A CRISPR interference complex capable of recognizing a protospacer in the regulatory or encoding regions of a gene, but lacking nuclease activity can serve as an efficient tool for gene expression regulation, as demonstrated for Type II CRISPR-Cas systems. A Cas9 protein with impaired nuclease activity suppressed transcription of bacterial genes at the initiation or elongation stages when the protospacer was located in the promoter or coding regions, respectively [80, 81]. The observed suppression might result from screening the target locus within the promoter region or physically stopping the elongation complex by having Cas9 tightly bound to the transcribed DNA template. This type of repression is successfully used for expression control in eukaryotic systems. The nuclease Cas3-deficient multi-subunit Type I CASCADE complex from E. coli can also be used for transcription repression [82, 83]. The opposite effect, transcription activation, can be achieved using Cas9 with impaired nuclease activity in combination with some activator domains. Such a combination provides a reversible increase in the levels of gene expression in bacteria, yeast, mouse, and human cells [81, 84, 85].
There are numerous prerequisites for creating CRISPR-based tools for gene expression regulation at the post-transcriptional level. In 2013, Sampson et al. identified a new activity of the Type II CRISPR-Cas system in Francisella novicida that was not defense against foreign DNA [86]. They showed that Cas9, tracrRNA, and an additional RNA named scaRNA interacted with a transcript of the lipoprotein gene and caused its degradation. As a result, the cells became highly virulent. The mechanisms of pre-crRNA processing present considerable interest for post-transcriptional regulation, because the corresponding components of the CRISPR-Cas systems can recognize repeat fragments in a content of RNA molecule and introduce specific breaks in them. The possibility of RNA stability regulation was demonstrated for the I-F Subtype system protein, Cas6f, also known as Csy4 [87, 88]. It is possible that molecular tools for post-transcriptional regulation of gene expression will develop further in the near future, since such regulation is especially important for a prokaryotic cell, in which efficient RNA interference has not yet been achieved.
Cell selection. Activation of CRISPR interference could be used for directed manipulation of the content of bacterial communities. Bikard et al. showed that delivery of crRNA bearing a spacer matching a protospacer of the virulence-providing genes into Staphylococcus aureus cells possessing the Type II CRISPR-Cas system selectively inhibited the growth of virulent cells [89]. In the same work, the delivery of crRNA complementary to the protospacer of an antibiotic resistance-encoding plasmid resulted in the loss of cell resistance to the antibiotic. Similarly, induction of expression of components of the Subtype I-E CRISPR interference system in E. coli caused selective death of cells containing a protospacer complementary to the crRNA spacer in their genomes [90, 91].
Strain subtyping. Profiling of CRISPR array spacers by PCR or restriction analysis for identifying microorganism strains has been used for years and started a long time before elucidation of the functions and mechanisms of CRISPR-Cas systems [92, 93]. The main condition for profiling is a relatively stable composition of CRISPR arrays due to low levels of CRISRP adaptation. Since no expression of cas genes has been observed in E. coli and closely related genera Yersinia and Salmonella, at least in laboratory conditions [66, 94], profiling their spacers still remains quite useful, especially for subtyping of pathogenic strains [95, 96]. At present, profiling is mostly performed by high-throughput sequencing [97].
In strains with high levels of CRISPR adaptation, profiling of spacers allows prediction of strain resistance to certain bacteriophages based on the already existent spacers. This might be important for selecting bacteriophages for phage therapy and for following interactions of bacteria with bacteriophages [28, 98, 99], in particular, in a course of therapy.
Design of strains with a required set of spacers. Creation of industrial microorganisms with resistance to bacteriophage infection determined by their CRISPR arrays is highly important in biotechnology. It is also often required to express crRNA with particular properties in bacterial cells, for example, for studying CRISPR-Cas system interactions with various bacteriophages. A new simple approach for rapid creation of such strains has been suggested based on the phenomenon of primed adaptation [100]. Escherichia coli cells bearing inducible cas genes were transformed with a plasmid that contained a fragment of bacteriophage genome, against which new spacers should be obtained, and a protospacer partially complementary to an existing spacer of the bacterial CRISPR array. After induction of cas genes, interactions between the spacer in the content of crRNA and the priming protospacer provided preferential selection of new spacers from the same plasmid. Over 50% of the cells acquired the “phage” spacer.
Despite enormous progress in understanding CRISPR-Cas system-mediated adaptive immunity in prokaryotes during the last decade, many questions still remain unanswered, such as the mechanisms of selection and cleavage of protospacers during CRISPR adaptation, evolutionary origin of CRISPR-Cas systems, their role in population structure and in cell interactions with plasmids and bacteriophages, prediction of novel CRISPR-Cas systems in bioinformatic searches and mechanisms of their action, and approaches to the regulation of CRISPR-Cas system activity. The elucidation of these problems will broaden our understanding of fundamental principles of biology and help in the development of biotechnological tools for applications other than genome editing.
This work was supported by the Russian Foundation for Basic Research (project No. 16-04-00767).
REFERENCES
1.Ishino, Y., Shinagawa, H., Makino, K., Amemura, M.,
and Nakatura, A. (1987) Nucleotide sequence of the iap gene,
responsible for alkaline phosphatase isoenzyme conversion in
Escherichia coli, and identification of the gene product, J.
Bacteriol., 169, 5429-5433.
2.Mojica, F. J. M., Diez-Villasenor, C., Soria, E.,
and Juez, G. (2000) Biological significance of a family of regularly
spaced repeats in the genomes of Archaea, Bacteria and mitochondria,
Mol. Microbiol., 36, 244-246.
3.Mojica, F. J. M., Diez-Villasenor, C.,
Garcia-Martinez, J., and Soria, E. (2005) Intervening sequences of
regularly spaced prokaryotic repeats derive from foreign genetic
elements, J. Mol. Evol., 60, 174-182.
4.Bolotin, A., Quinquis, B., Sorokin, A., and Dusko
Ehrlich, S. (2005) Clustered regularly interspaced short palindrome
repeats (CRISPRs) have spacers of extrachromosomal origin,
Microbiology, 151, 2551-2561.
5.Pourcel, C., Salvignol, G., and Vergnaud, G. (2005)
CRISPR elements in Yersinia pestis acquire new repeats by
preferential uptake of bacteriophage DNA, and provide additional tools
for evolutionary studies, Microbiology, 151, 653-663.
6.Tang, T.-H., Bachellerie, J.-P., Rozhdestvensky,
T., Bortolin, M.-L., Huber, H., Drungowski, M., Elge, T., Brosius, J.,
and Huttenhofer, A. (2002) Identification of 86 candidates for small
non-messenger RNAs from the archaeon Archaeoglobus fulgidus,
Proc. Natl. Acad. Sci. USA, 99, 7536-7541.
7.Jansen, R., Van Embden, J. D. A., Gaastra, W., and
Schouls, L. M. (2002) Identification of genes that are associated with
DNA repeats in prokaryotes, Mol. Microbiol., 43,
1565-1575.
8.Marraffini, L. A., and Sontheimer, E. J. (2008)
CRISPR interference limits horizontal gene transfer in staphylococci by
targeting DNA, Science, 322, 1843-1845.
9.Brouns, S. J. J., Jore, M. M., Lundgren, M.,
Westra, E. R., Slijkhuis, R. J. H., Snijders, A. P. L., Dickman, M. J.,
Makarova, K. S., Koonin, E. V., and Van der Oost, J. (2008) Small
CRISPR RNAs guide antiviral defense in prokaryotes, Science,
321, 960-964.
10.Barrangou, R., Fremaux, C., Deveau, H., Richards,
M., Moineau, S., Romero, D. A., and Horvath, P. (2007) CRISPR provides
acquired resistance against viruses in prokaryotes, Science,
315, 1709-1712.
11.Sorek, R., Kunin, V., and Hugenholtz, P. (2008)
CRISPR-a widespread system that provides acquired resistance against
phages in bacteria and archaea, Nat. Rev. Microbiol., 6,
181-186.
12.Makarova, K. S., Haft, D. H., Barrangou, R.,
Brouns, S. J. J., Charpentier, E., Horvath, P., Moineau, S., Mojica, F.
J. M., Wolf, Y. I., Yakunin, A. F., Van der Oost, J., and Koonin, E. V.
(2011) Evolution and classification of the CRISPR-Cas systems, Nat.
Rev. Microbiol., 9, 467-477.
13.Makarova, K. S., Wolf, Y. I., Alkhnbashi, O. S.,
Costa, F., Shah, S. A., Saunders, S. J., Barrangou, R., Brouns, S. J.
J., Charpentier, E., Haft, D. H., Horvath, P., Moineau, S., Mojica, F.
J., Terns, R. M., Terns, M. P., White, M. F., Yakunin, A. F., Garrett,
R. A., Van der Oost, J., Backofen, R., and Koonin, E. V. (2015) An
updated evolutionary classification of CRISPR-Cas systems, Nat. Rev.
Microbiol., 13, 722-736.
14.Jore, M. M., Lundgren, M., Van Duijn, E.,
Bultema, J. B., Westra, E. R., Waghmare, S. P., Wiedenheft, B., Pul,
U., Wurm, R., Wagner, R., Beijer, M. R., Barendregt, A., Zhou, K.,
Snijders, A. P., Dickman, M. J., Doudna, J. A., Boekema, E. J., Heck,
A. J., Van der Oost, J., and Brouns, S. J. (2011) Structural basis for
CRISPR RNA-guided DNA recognition by Cascade, Nat. Struct. Mol.
Biol., 18, 529-536.
15.Semenova, E., Kuznedelov, K., Datsenko, K. A.,
Boudry, P. M., Savitskaya, E. E., Medvedeva, S., Beloglazova, N.,
Logacheva, M., Yakunin, A. F., and Severinov, K. (2015) The Cas6e
ribonuclease is not required for interference and adaptation by the
E. coli type I-E CRISPR-Cas system, Nucleic Acids Res.,
43, 6049-6061.
16.Lintner, N. G., Kerou, M., Brumfield, S. K.,
Graham, S., Liu, H., Naismith, J. H., Sdano, M., Peng, N., She, Q.,
Copie, V., Young, M. J., White, M. F., and Lawrence, C. M. (2011)
Structural and functional characterization of an archaeal clustered
regularly interspaced short palindromic repeat (CRISPR)-associated
complex for antiviral defense (CASCADE), J. Biol. Chem.,
286, 21643-21656.
17.Plagens, A., Tjaden, B., Hagemann, A., Randau,
L., and Hensel, R. (2012) Characterization of the CRISPR/Cas subtype
I-A system of the hyperthermophilic crenarchaeon Thermoproteus
tenax, J. Bacteriol., 194, 2491-2500.
18.Westra, E. R., Van Erp, P. B. G., Kunne, T.,
Wong, S. P., Staals, R. H. J., Seegers, C. L. C., Bollen, S., Jore, M.
M., Semenova, E., Severinov, K., De Vos, W. M., Dame, R. T., De Vries,
R., Brouns, S. J., and Van der Oost, J. (2012) CRISPR immunity relies
on the consecutive binding and degradation of negatively supercoiled
invader DNA by Cascade and Cas3, Mol. Cell, 46,
595-605.
19.Hochstrasser, M. L., Taylor, D. W., Bhat, P.,
Guegler, C. K., Sternberg, S. H., Nogales, E., and Doudna, J. A. (2014)
CasA mediates Cas3-catalyzed target degradation during CRISPR
RNA-guided interference, Proc. Natl. Acad. Sci. USA, 111,
6618-6623.
20.Jackson, R. N., Lavin, M., Carter, J., and
Wiedenheft, B. (2014) Fitting CRISPR-associated Cas3 into the helicase
family tree, Curr. Opin. Struct. Biol., 24, 106-114.
21.Mulepati, S., and Bailey, S. (2013) In
vitro reconstitution of an Escherichia coli RNA-guided
immune system reveals unidirectional, ATP-dependent degradation of DNA
target, J. Biol. Chem., 288, 22184-22192.
22.Sinkunas, T., Gasiunas, G., Fremaux, C.,
Barrangou, R., Horvath, P., and Siksnys, V. (2011) Cas3 is a
single-stranded DNA nuclease and ATP-dependent helicase in the
CRISPR/Cas immune system, EMBO J., 30, 1335-1342.
23.Huo, Y., Nam, K. H., Ding, F., Lee, H., Wu, L.,
Xiao, Y., Farchione, M. D., Zhou, S., Rajashankar, K., Kurinov, I.,
Zhang, R., and Ke, A. (2014) Structures of CRISPR Cas3 offer
mechanistic insights into Cascade-activated DNA unwinding and
degradation, Nat. Struct. Mol. Biol., 21, 771-777.
24.Zhao, H., Sheng, G., Wang, J., Wang, M.,
Bunkoczi, G., Gong, W., Wei, Z., and Wang, Y. (2014) Crystal structure
of the RNA-guided immune surveillance Cascade complex in Escherichia
coli, Nature, 515, 147-150.
25.Gong, B., Shin, M., Sun, J., Jung, C.-H., Bolt,
E. L., Van der Oost, J., and Kim, J.-S. (2014) Molecular insights into
DNA interference by CRISPR-associated nuclease-helicase Cas3, Proc.
Natl. Acad. Sci. USA, 111, 16359-16364.
26.Sashital, D. G., Wiedenheft, B., and Doudna, J.
A. (2012) Mechanism of foreign DNA selection in a bacterial adaptive
immune system, Mol. Cell, 46, 606-615.
27.Sinkunas, T., Gasiunas, G., Waghmare, S. P.,
Dickman, M. J., Barrangou, R., Horvath, P., and Siksnys, V. (2013)
In vitro reconstitution of Cascade-mediated CRISPR immunity in
Streptococcus thermophilus, EMBO J., 32,
385-394.
28.Andersson, A. F., and Banfield, J. F. (2008)
Virus population dynamics and acquired virus resistance in natural
microbial communities, Science, 320, 1047-1050.
29.Deveau, H., Barrangou, R., Garneau, J. E.,
Labonte, J., Fremaux, C., Boyaval, P., Romero, D. A., Horvath, P., and
Moineau, S. (2008) Phage response to CRISPR-encoded resistance in
Streptococcus thermophilus, J. Bacteriol., 190,
1390-1400.
30.Paez-Espino, D., Sharon, I., Morovic, W., Stahl,
B., Thomas, B. C., Barrangou, R., and Banfield, F. (2015) CRISPR
immunity drives rapid phage genome evolution in Streptococcus
thermophilus, MBio, 6, 1-9.
31.Garrett, R., Shah, S., Erdmann, S., Liu, G.,
Mousaei, M., Leon-Sobrino, C., Peng, W., Gudbergsdottir, S., Deng, L.,
Vestergaard, G., Peng, X., and She, Q. (2015) CRISPR-Cas adaptive
immune systems of the Sulfolobales: unravelling their complexity and
diversity, Life, 5, 783-817.
32.Goldberg, G. W., Jiang, W., Bikard, D., and
Marraffini, L. A. (2014) Conditional tolerance of temperate phages via
transcription-dependent CRISPR-Cas targeting, Nature,
514, 633-637.
33.Samai, P., Pyenson, N., Jiang, W., Goldberg, G.
W., Hatoum-Aslan, A., and Marraffini, L. A. (2015) Co-transcriptional
DNA and RNA cleavage during type III CRISPR-Cas immunity, Cell,
161, 1164-1174.
34.Hale, C. R., Zhao, P., Olson, S., Duff, M. O.,
Graveley, B. R., Wells, L., Terns, R. M., and Terns, M. P. (2009)
RNA-guided RNA cleavage by a CRISPR RNA–Cas protein complex,
Cell, 139, 945-956.
35.Staals, R. H. J., Zhu, Y., Taylor, D. W.,
Kornfeld, J. E., Sharma, K., Barendregt, A., Koehorst, J. J., Vlot, M.,
Neupane, N., Varossieau, K., Sakamoto, K., Suzuki, T., Dohmae, N.,
Yokoyama, S., Schaap, P. J., Urlaub, H., Heck, A. J., Nogales, E.,
Doudna, J. A., Shinkai, A., and Van der Oost, J. (2014) RNA targeting
by the type III-A CRISPR-Cas Csm complex of Thermus
thermophilus, Mol. Cell, 56, 518-530.
36.Tamulaitis, G., Kazlauskiene, M., Manakova, E.,
Venclovas, C., Nwokeoji, A. O., Dickman, M. J., Horvath, P., and
Siksnys, V. (2014) Programmable RNA shredding by the type III-A
CRISPR-Cas system of Streptococcus thermophilus, Mol.
Cell, 56, 506-517.
37.Zebec, Z., Manica, A., Zhang, J., White, M. F.,
and Schleper, C. (2014) CRISPR-mediated targeted mRNA degradation in
the archaeon Sulfolobus solfataricus, Nucleic Acids Res.,
42, 5280-5288.
38.Zhang, J., Rouillon, C., Kerou, M., Reeks, J.,
Brugger, K., Graham, S., Reimann, J., Cannone, G., Liu, H., Albers,
S.-V., Naismith, J. H., Spagnolo, L., and White, M. F. (2012) Structure
and mechanism of the CMR complex for CRISPR-mediated antiviral
immunity, Mol. Cell, 45, 303-313.
39.Deng, L., Garrett, R. A., Shah, S. A., Peng, X.,
and She, Q. (2013) A novel interference mechanism by a type IIIB
CRISPR-Cmr module in Sulfolobus, Mol. Microbiol.,
87, 1088-1099.
40.Carte, J., Wang, R., Li, H., Terns, R. M., and
Terns, M. P. (2008) Cas6 is an endoribonuclease that generates guide
RNAs for invader defense in prokaryotes, Genes Dev., 22,
3489-3496.
41.Hatoum-Aslan, A., Samai, P., Maniv, I., Jiang,
W., and Marraffini, L. A. (2013) A ruler protein in a complex for
antiviral defense determines the length of small interfering CRISPR
RNAs, J. Biol. Chem., 288, 27888-27897.
42.Marraffini, L. A., and Sontheimer, E. J. (2010)
Self versus non-self discrimination during CRISPR RNA-directed
immunity, Nature, 463, 568-571.
43.Deltcheva, E., Chylinski, K., Sharma, C. M.,
Gonzales, K., Chao, Y., Pirzada, Z. A., Eckert, M. R., Vogel, J., and
Charpentier, E. (2011) CRISPR RNA maturation by trans-encoded
small RNA and host factor RNase III, Nature, 471,
602-607.
44.Anders, C., Niewoehner, O., Duerst, A., and
Jinek, M. (2014) Structural basis of PAM-dependent target DNA
recognition by the Cas9 endonuclease, Nature, 513,
569-573.
45.Szczelkun, M. D., Tikhomirova, M. S., Sinkunas,
T., Gasiunas, G., Karvelis, T., Pschera, P., Siksnys, V., and Seidel,
R. (2014) Direct observation of R-loop formation by single RNA-guided
Cas9 and Cascade effector complexes, Proc. Natl. Acad. Sci. USA,
111, 9798-9803.
46.Sternberg, S. H., Redding, S., Jinek, M., Greene,
E. C., and Doudna, J. A. (2014) DNA interrogation by the CRISPR
RNA-guided endonuclease Cas9, Nature, 507, 62-67.
47.Jiang, W., and Marraffini, L. A. (2015)
CRISPR-Cas: new tools for genetic manipulations from bacterial immunity
systems, Annu. Rev. Microbiol., 69, 209-228.
48.Makarova, K. S., Grishin, N. V., Shabalina, S.
A., Wolf, Y. I., and Koonin, E. V. (2006) A putative
RNA-interference-based immune system in prokaryotes: computational
analysis of the predicted enzymatic machinery, functional analogies
with eukaryotic RNAi, and hypothetical mechanisms of action, Biol.
Direct, 1, 7.
49.Garneau, J. E., Dupuis, M.-E., Villion, M.,
Romero, D. A., Barrangou, R., Boyaval, P., Fremaux, C., Horvath, P.,
Magadan, A. H., and Moineau, S. (2010) The CRISPR/Cas bacterial immune
system cleaves bacteriophage and plasmid DNA, Nature,
468, 67-71.
50.Sapranauskas, R., Gasiunas, G., Fremaux, C.,
Barrangou, R., Horvath, P., and Siksnys, V. (2011) The Streptococcus
thermophilus CRISPR/Cas system provides immunity in Escherichia
coli, Nucleic Acids Res., 39, 9275-9282.
51.Gasiunas, G., Barrangou, R., Horvath, P., and
Siksnys, V. (2012) Cas9-crRNA ribonucleoprotein complex mediates
specific DNA cleavage for adaptive immunity in bacteria, Proc. Natl.
Acad. Sci. USA, 109, 2579-2586.
52.Jinek, M., Chylinski, K., Fonfara, I., Hauer, M.,
Doudna, J. A., and Charpentier, E. (2012) A programmable
dual-RNA-guided, Science, 337, 816-821.
53.Schunder, E., Rydzewski, K., Grunow, R., and
Heuner, K. (2013) First indication for a functional CRISPR/Cas system
in Francisella tularensis, Int. J. Med. Microbiol.,
303, 51-60.
54.Shmakov, S., Abudayyeh, O. O., Makarova, K. S.,
Wolf, Y. I., and Jonathan, S. (2015) Discovery of novel, diverse class
2 CRISPR-Cas systems independently evolved from different mobile
elements, Mol. Cell, 60, 385-397.
55.Zetsche, B., Gootenberg, J. S., Abudayyeh, O. O.,
Slaymaker, I. M., Makarova, K. S., Essletzbichler, P., Volz, S. E.,
Joung, J., Van der Oost, J., Regev, A., Koonin, E. V., and Zhang, F.
(2015) Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas
system, Cell, 163, 759-771.
56.Koonin, E. V., and Makarova, K. S. (2013)
CRISPR-Cas: evolution of an RNA-based adaptive immunity system in
prokaryotes, RNA Biol., 10, 679-686.
57.Nunez, J. K., Kranzusch, P. J., Noeske, J.,
Wright, A. V., Davies, C. W., and Doudna, J. A. (2014) Cas1–Cas2
complex formation mediates spacer acquisition during CRISPR-Cas
adaptive immunity, Nat. Struct. Mol. Biol., 21,
528-534.
58.Nunez, J. K., Harrington, L. B., Kranzusch, P.
J., Engelman, A. N., and Doudna, J. A. (2015) Foreign DNA capture
during CRISPR-Cas adaptive immunity, Nature, 527,
535-538.
59.Arslan, Z., Hermanns, V., Wurm, R., Wagner, R.,
and Pul, U. (2014) Detection and characterization of spacer integration
intermediates in type I-E CRISPR-Cas system, Nucleic Acids Res.,
42, 7884-7893.
60.Wiedenheft, B., Zhou, K., Jinek, M., Coyle, S.
M., Ma, W., and Doudna, J. A. (2009) Structural basis for DNase
activity of a conserved protein implicated in CRISPR-mediated genome
defense, Structure, 17, 904-912.
61.Babu, M., Beloglazova, N., Flick, R., Graham, C.,
Skarina, T., Nocek, B., Gagarinova, A., Pogoutse, O., Brown, G.,
Binkowski, A., Phanse, S., Joachimiak, A., Koonin, E. V., Savchenko,
A., Emili, A., Greenblatt, J., Edwards, A. M., and Yakunin, A. F.
(2011) A dual function of the CRISPR-Cas system in bacterial antivirus
immunity and DNA repair, Mol. Microbiol., 79,
484-502.
62.Nam, K. H., Ding, F., Haitjema, C., Huang, Q.,
DeLisa, M. P., and Ke, A. (2012) Double-stranded endonuclease activity
in Bacillus halodurans clustered regularly interspaced short
palindromic repeats (CRISPR)-associated Cas2 protein, J. Biol.
Chem., 287, 35943-35952.
63.Samai, P., Smith, P., and Shuman, S. (2010)
Structure of a CRISPR-associated protein Cas2 from Desulfovibrio
vulgaris, Acta Crystallogr. Sect. F Struct. Biol. Cryst.
Commun., 66, 1552-1556.
64.Beloglazova, N., Brown, G., Zimmerman, M. D.,
Proudfoot, M., Makarova, K. S., Kudritska, M., Kochinyan, S., Wang, S.,
Chruszcz, M., Minor, W., Koonin, E. V., Edwards, A. M., Savchenko, A.,
and Yakunin, A. F. (2008) A novel family of sequence-specific
endoribonucleases associated with the clustered regularly interspaced
short palindromic repeats, J. Biol. Chem., 283,
20361-20371.
65.Yosef, I., Goren, M. G., and Qimron, U. (2012)
Proteins and DNA elements essential for the CRISPR adaptation process
in Escherichia coli, Nucleic Acids Res., 40,
5569-5576.
66.Pougach, K., Semenova, E., Bogdanova, E.,
Datsenko, K. A., Djordjevic, M., Wanner, B. L., and Severinov, K.
(2010) Transcription, processing and function of CRISPR cassettes in
Escherichia coli, Mol. Microbiol., 77,
1367-1379.
67.Wei, Y., Chesne, M. T., Terns, R. M., and Terns,
M. P. (2015) Sequences spanning the leader-repeat junction mediate
CRISPR adaptation to phage in Streptococcus thermophilus,
Nucleic Acids Res., 43, 1749-1758.
68.Mojica, F. J. M., Diez-Villasenor, C.,
Garcia-Martinez, J., and Almendros, C. (2009) Short motif sequences
determine the targets of the prokaryotic CRISPR defense system,
Microbiology, 155, 733-740.
69.Wei, Y., Terns, R. M., and Terns, M. P. (2015)
Cas9 function and host genome sampling in type II-A CRISPR-Cas
adaptation, Genes Dev., 29, 356-361.
70.Heler, R., Samai, P., Modell, J. W., Weiner, C.,
Goldberg, G. W., Bikard, D., and Marraffini, L. A. (2015) Cas9
specifies functional viral targets during CRISPR-Cas adaptation,
Nature, 519, 199-202.
71.Levy, A., Goren, M. G., Yosef, I., Auster, O.,
Manor, M., Amitai, G., Edgar, R., Qimron, U., and Sorek, R. (2015)
CRISPR adaptation biases explain preference for acquisition of foreign
DNA, Nature, 520, 505-510.
72.Datsenko, K. A., Pougach, K., Tikhonov, A.,
Wanner, B. L., Severinov, K., and Semenova, E. (2012) Molecular memory
of prior infections activates the CRISPR/Cas adaptive bacterial
immunity system, Nat. Commun., 3, 945.
73.Fineran, P. C., Gerritzen, M. J. H., Suarez-Diez,
M., Kunne, T., Boekhorst, J., Van Hijum, S. A. F. T., Staals, R. H. J.,
and Brouns, S. J. J. (2014) Degenerate target sites mediate rapid
primed CRISPR adaptation, Proc. Natl. Acad. Sci. USA,
111, 1629-1638.
74.Li, M., Wang, R., and Xiang, H. (2014)
Haloarcula hispanica CRISPR authenticates PAM of a target
sequence to prime discriminative adaptation, Nucleic Acids Res.,
42, 7226-7235.
75.Swarts, D. C., Mosterd, C., Van Passel, M. W. J.,
and Brouns, S. J. J. (2012) CRISPR interference directs strand specific
spacer acquisition, PLoS One, 7, e35888.
76.Richter, C., Dy, R. L., McKenzie, R. E., Watson,
B. N. J., Taylor, C., Chang, J. T., McNeil, M. B., Staals, R. H. J.,
and Fineran, P. C. (2014) Priming in the type I-F CRISPR-Cas system
triggers strand-independent spacer acquisition, bi-directionally from
the primed protospacer, Nucleic Acids Res., 42,
8516-8526.
77.Koonin, E. V., and Krupovic, M. (2014) Evolution
of adaptive immunity from transposable elements combined with innate
immune systems, Nat. Rev. Genet., 16, 184-192.
78.Sternberg, S. H., and Doudna, J. A. (2015)
Expanding the biologist’s toolkit with CRISPR-Cas9, Mol.
Cell, 58, 568-574.
79.Doudna, J. A., and Charpentier, E. (2014) The new
frontier of genome engineering with CRISPR-Cas9, Science,
346, 1258096.
80.Qi, L. S., Larson, M. H., Gilbert, L. A., Doudna,
J. A., Weissman, J. S., Arkin, A. P., and Lim, W. A. (2013) Repurposing
CRISPR as an RNA-guided platform for sequence-specific control of gene
expression, Cell, 152, 1173-1183.
81.Bikard, D., and Marraffini, L. A. (2013) Control
of gene expression by CRISPR-Cas systems, F1000Prime Rep,
5, 47.
82.Luo, M. L., Mullis, A. S., Leenay, R. T., and
Beisel, C. L. (2015) Repurposing endogenous type I CRISPR-Cas systems
for programmable gene repression, Nucleic Acids Res., 43,
674-681.
83.Rath, D., Amlinger, L., Hoekzema, M.,
Devulapally, P. R., and Lundgren, M. (2014) Efficient programmable gene
silencing by Cascade, Nucleic Acids Res., 43,
237-246.
84.Cheng, A. W., Wang, H., Yang, H., Shi, L., Katz,
Y., Theunissen, T. W., Rangarajan, S., Shivalila, C. S., Dadon, D. B.,
and Jaenisch, R. (2013) Multiplexed activation of endogenous genes by
CRISPR-on, an RNA-guided transcriptional activator system, Cell
Res., 23, 1163-1171.
85.Gilbert, L. A., Larson, M. H., Morsut, L., Liu,
Z., Brar, G. A., Torres, S. E., Stern-Ginossar, N., Brandman, O.,
Whitehead, E. H., Doudna, J. A., Lim, W. A., Weissman, J. S., and Qi,
L. S. (2013) CRISPR-mediated modular RNA-guided regulation of
transcription in eukaryotes, Cell, 154, 442-451.
86.Sampson, T. R., Saroj, S. D., Llewellyn, A. C.,
Tzeng, Y. L., and Weiss, D. S. (2013) A CRISPR/Cas system mediates
bacterial innate immune evasion and virulence, Nature,
497, 254-257.
87.Lee, H. Y., Haurwitz, R. E., Apffel, A., Zhou,
K., Smart, B., Wenger, C. D., Laderman, S., Bruhn, L., and Doudna, J.
A. (2013) RNA–protein analysis using a conditional CRISPR
nuclease, Proc. Natl. Acad. Sci. USA, 110, 5416-5421.
88.Borchardt, E. K., Vandoros, L. A., Huang, M.,
Lackey, P. E., Marzluff, W. F., and Asokan, A. (2015) Controlling mRNA
stability and translation with the CRISPR endoribonuclease Csy4,
RNA, 21, 1921-1230.
89.Bikard, D., Euler, C. W., Jiang, W., Nussenzweig,
P. M., Goldberg, G. W., Duportet, X., Fischetti, V. A., and Marraffini,
L. A. (2014) Exploiting CRISPR-Cas nucleases to produce
sequence-specific antimicrobials, Nat. Biotechnol., 32,
1146-1150.
90.Gomaa, A. A., Klumpe, H. E., Luo, M. L., Selle,
K., Barrangou, R., and Beisel, C. L. (2014) Programmable removal of
bacterial strains by use of genome-targeting CRISPR-Cas systems,
MBio, 5, e00928-13.
91.Caliando, B. J., and Voigt, C. A. (2015) Targeted
DNA degradation using a CRISPR device stably carried in the host
genome, Nat. Commun., 6, 6989.
92.Kamerbeek, J., Schouls, L., Kolk, A., Van
Agterveld, M., Van Soolingen, D., Kuijper, S., Bunschoten, A.,
Molhuizen, H., Shaw, R., Goyal, M., and Van Embden, J. (1997)
Simultaneous detection and strain differentiation of Mycobacterium
tuberculosis for diagnosis and epidemiology, J. Clin.
Microbiol., 35, 907-914.
93.Van Soolingen, D., De Haas, P. E. W., Hermans, P.
W. M., Groenen, P. M. A., and Van Embden, J. D. A. (1993) Comparison of
various repetitive DNA elements as genetic markers for strain
differentiation and epidemiology of Mycobacterium tuberculosis,
J. Clin. Microbiol., 31, 1987-1995.
94.Westra, E. R., Pul, U., Heidrich, N., Jore, M.
M., Lundgren, M., Stratmann, T., Wurm, R., Raine, A., Mescher, M., Van
Heereveld, L., Mastop, M., Wagner, E. G., Schnetz, K., Van Der Oost,
J., Wagner, R., and Brouns, S. J. (2010) H-NS-mediated repression of
CRISPR-based immunity in Escherichia coli K12 can be relieved by
the transcription activator LeuO, Mol. Microbiol., 77,
1380-1393.
95.Delannoy, S., Beutin, L., and Fach, P. (2012) Use
of clustered regularly interspaced short palindromic repeat sequence
polymorphisms for specific detection of enterohemorrhagic
Escherichia coli strains of serotypes O26:H11, O45:H2, O103:H2,
O111:H8, O121:H19, O145:H28, and O157:H7 by real-time PCR, J. Clin.
Microbiol., 50, 4035-4040.
96.Shariat, N., Kirchner, M. K., Sandt, C. H.,
Trees, E., Barrangou, R., and Dudley, E. G. (2013) Subtyping of
Salmonella enterica serovar newport outbreak isolates by
CRISPR-MVLST and determination of the relationship between CRISPR-MVLST
and PFGE results, J. Clin. Microbiol., 51, 2328-2336.
97.Fabre, L., Zhang, J., Guigon, G., Le Hello, S.,
Guibert, V., Accou-Demartin, M., De Romans, S., Lim, C., Roux, C.,
Passet, V., Diancourt, L., Guibourdenche, M., Issenhuth-Jeanjean, S.,
Achtman, M., Brisse, S., Sola, C., and Weill, F. X. (2012) CRISPR
typing and subtyping for improved laboratory surveillance of Salmonella
infections, PLoS One, 7, e36995.
98.Sun, C. L., Thomas, B. C., Barrangou, R., and
Banfield, J. F. (2015) Metagenomic reconstructions of bacterial CRISPR
loci constrain population histories, ISME J., 10,
858-870.
99.Paez-Espino, D., Morovic, W., Sun, C. L., Thomas,
B. C., Ueda, K., Stahl, B., Barrangou, R., and Banfield, J. F. (2013)
Strong bias in the bacterial CRISPR elements that confer immunity to
phage, Nat. Commun., 4, 1430.
100.Strotksaya, A., Semenova, E., Savitskaya, E.,
and Severinov, K. (2015) Rapid multiplex creation of Escherichia
coli strains capable of interfering with phage infection through
CRISPR, Methods Mol. Biol., 1311, 147-159.