Plastid Genome of Seseli montanum: Complete Sequence and Comparison with Plastomes of Other Members of the Apiaceae Family
T. H. Samigullin1*, M. D. Logacheva1, E. I. Terenteva2, G. V. Degtjareva2, and C. M. Vallejo-Roman1
1Lomonosov Moscow State University, Belozersky Institute of Physico-Chemical Biology, 119991 Moscow, Russia; fax: +7 (495) 939-0338; E-mail: samigul@belozersky.msu.ru, maria.log@gmail.com, vallejo@genebee.msu.ru2Lomonosov Moscow State University, Biological Faculty, 119991 Moscow, Russia; E-mail: el.terenteva@mail.ru, degavi@mail.ru
* To whom correspondence should be addressed.
Received May 28, 2016; Revision received June 17, 2016
This work reports the complete plastid (pt) DNA sequence of Seseli montanum L. of the Apiaceae family, determined using next-generation sequencing technology. The complete genome sequence has been deposited in GenBank with accession No. KM035851. The S. montanum plastome is 147,823 bp in length. The plastid genome has a typical structure for angiosperms and contains a large single-copy region (LSC) of 92,620 bp and a small single-copy region (SSC) of 17,481 bp separated by a pair of 18,861 bp inverted repeats (IRa and IRb). The composition, gene order, and AT-content in the S. montanum plastome are similar to that of a typical flowering plant pt DNA. One hundred fourteen unique genes have been identified, including 30 tRNA genes, four rRNA genes, and 80 protein genes. Of 18 intron-containing genes found, 16 genes have one intron, and two genes (ycf3, clpP) have two introns. Comparative analysis of Apiaceae plastomes reveals in the S. montanum plastome a LSC/IRb junction shift, so that the part of the ycf2 (4980 bp) gene is located in the LSC, but the other part of ycf2 (1301 bp) is within the inverted repeat. Thus, structural rearrangements in the plastid genome of S. montanum result in an enlargement of the LSC region by means of capture of a large part of ycf2, in contrast to eight Apiaceae plastomes where the complete ycf2 gene sequence is located in the inverted repeat.
KEY WORDS: plastid genome, genome comparative analysis, Seseli montanum, ApiaceaeDOI: 10.1134/S0006297916090078
Abbreviations: CTAB, cetyltrimethylammonium bromide; IR, inverted repeat; JL, junction between LSC and IR; JS, junction between SSC and IR; LSC, large single-copy region; SSC, small single-copy region.
Chloroplasts (plastids of photosynthetic plants) are important
components of a plant cell. Their genome encodes genes required for
implementation of the main function – photosynthesis –
while their enzymatic systems participate in synthesis of fatty acids,
amino acids, and pigments. The structure of plastid chromosome is the
most conserved among the three genomes of a plant cell, it remains
preserved for many millions of years. In photosynthetic flowering
plants, in most cases the plastid genome is a circular double-stranded
DNA molecule of about 140-150 kb in length. Comparative study of
plastid genomes revealed that their organization is similar in the
majority of analyzed plants [1] and is
characterized by the presence of two extended inverted repeats (IRa and
IRb) that divide the plastome into two unequal parts – the
large (LSC) and small (SSC) single-copy regions [2]. Variability of gene composition and order is
obvious when comparing evolutionarily distant plant species [3], however, the plastome architecture is preserved in
the majority of angiosperms except for some heterotrophic (parasitic
and mycoheterotrophic) plants and several evolutionary lineages of
autotrophs. For instance, a number of taxa of the large complex
Fabaceae family lost one of the inverted repeats (IR-lacking clade) [4]. Inverted repeat boundaries are also variable,
structural rearrangements lead to changes in boundaries of the inverted
repeats so that they may differ even among closely related species of
angiosperms, though remaining within the same genome regions [5-9]. In dicotyledons (basal
angiosperms and eudicots), typically, the LSC/IRa (JLa) junction is
located near the trnH-GUG gene, the SSC/IRa (JSa) junction is within
the ycf1 gene, and the SSC/IRb (JSb) junction is situated upstream of
the ndhF gene, while the LSC/IRb (JLb) junction resides within or near
the rps19 gene of the S10 operon. Small (below 100 bp) shifts of
inverted repeat junctions are observed frequently, while large (over 1
kb) shifts of the junctions in angiosperms occur much more rarely [10] and in some cases may be considered as
synapomorphies.
Apiaceae is one of families where large shifts of the inverted repeat junctions were observed. It was shown earlier that in representatives of a large Apioid superclade the inverted repeats may be both extended (~1 kb) or shortened (to 16 kb), while outside the superclade such large-scale variations were not observed [11, 12]. This superclade comprises over 10 tribes and several large clades of unclear relationship [13, 14], data on complete plastid genomes may help to resolve them. As an increased variability of junctions of the inverted repeats is often accompanied with structural rearrangements in other plastome regions [15], comprehensive study of gene composition, their order, and mechanisms of plastid genome evolution in Apiaceae is of special interest. At present, the complete plastid genome sequences of 17 Apiaceae are known. Twelve of these belong to the superclade that does not depict the structural diversity of plastomes with variable inverted repeat junctions, as the superclade includes about 450 species.
This work aimed to determine and annotate the complete sequence of the plastid genome of Seseli montanum L. (Selineae tribe, Apiaceae family) using a high-throughput sequencing method, followed by its structural analysis and comparison with the longest and shortest plastomes of other Apiaceae.
MATERIALS AND METHODS
Total DNA of S. montanum was isolated from a herbarium specimen (MW, 2013 collection, Russia) using the CTAB-method [16] with modifications. One microgram (1 µg) of total DNA was used for preparation of a DNA library. Ultrasonic disintegration of DNA was carried out with a Covaris S220 apparatus (Covaris, USA). Terminus repair, adenylation, and adapter ligation with subsequent PCR were performed using an Illumina TruSeq DNA Sample Prep Kit (Illumina, USA). After PCR, concentration of DNA fragments was determined using a Qubit fluorometer (Invitrogen, USA) and a real-time PCR thermal cycler (Agilent, USA). Libraries were sequenced with HiSeq 2000 (Illumina) with read length of 101 bp from the terminus of each fragment.
A plastome sequence was assembled de novo with the Genomics Workbench v. 5.5 software. Contigs were joined by PCR with primers for contig ends (Table S1 in Supplement to this paper available on the site of the journal http://protein.bio.msu.ru/biokhimiya and Springer site Link.springer.com) and subsequent sequencing with the Sanger technology. CPGAVAS software was used for automatic annotation [17] with further manual correction including alignment of certain sequenced regions with sequences available in GenBank using BLAST, in silico translation of regions where presence of protein-encoding genes was proposed, and search for tRNA genes using the tRNAscan-SE [18] and ARAGORN [19] programs. The plastid genome map was visualized using the OGDraw software [20]. The search for dispersed repeated sequenced of >20 bp in length was carried out with the REPuter program [21].
RESULTS AND DISCUSSION
As a result of de novo assembly of the S. montanum plastid genome, four contigs with 732× coverage were obtained that were used to compose a complete genome sequence. The sequence has been annotated and deposited in GenBank (accession number KM035851).
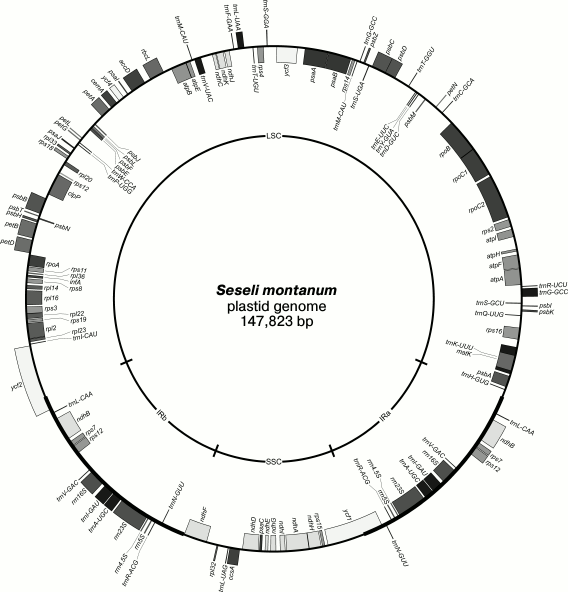
The S. montanum plastid genome is presented in Fig. 1 as a circular molecule 147,823 bp in length. The plastome has a structure that is typical for land plants: a large single-copy (LSC) region 92,620 bp in length and a small single-copy (SSC) region 17,481 bp in length. These regions are separated by two 18,861-bp inverted repeats (IRa and IRb). After annotation, 114 unique genes were identified in the plastome including 30 tRNA genes, 4 rRNA genes, and 80 protein genes. The composition and order of the genes in the S. montanum plastome are also typical for dicotyledons. Eighteen genes have introns: 16 have one intron, and two genes (ycf3, clpP) contain two introns. Similarly to other plastid genomes, the S. montanum plastome is AT-rich (62.43%) (Table S2 in Supplement). The search for dispersed repeated sequences revealed 12 direct repeats 29, 26, 25, and 24 bp in length, while in carrot (Daucus) the largest repeat is 60 bp in length.
Fig. 1. Map of the Seseli montanum plastid genome. LSC, large single-copy region; SSC, small single-copy region; IRa, inverted repeat “a”; IRb, inverted repeat “b”. Boxes outside the circle correspond to the genes expressed counter clockwise, genes shown inside the circle are expressed clockwise.
Comparison of genome lengths (table) shows that smaller plastomes are typical for representatives of the Selineae tribe (this tribe includes Seseli, Ostericum, and Angelica), and the S. montanum plastome corresponds well to this trait, it being 147,823 bp in length. Over 11-kb difference in plastome lengths in analyzed Apiaceae is largely related to variable length of the inverted repeat IR. It is believed that a positive role of inverted repeats is in an increase of dosage of genes encoding ribosomal components and possibility of correction of IR sequence using a second IR as a reference, which reduces nucleotide substitution rate and negative effect of deleterious mutations in the IR. As the plastid genome is not a single-copy molecule, any neighboring copy could be used for a sequence correction. However, this way of gene conversion is not utilized as it follows from an acceleration in the substitution rate of the formerly IR-genes in IR-lacking genomes [22]. Regardless of the role and reasons for emergence of an IR, the viability of plants whose plastomes lack inverted repeats (for instance, Fabaceae) suggests that the presence of the IR is not vital. Nevertheless, inverted repeats are preserved in the vast majority of photosynthetic plants. Furthermore, while in Charophyceae and nonvascular land plants the composition of inverted repeats is usually restricted to rRNA and tRNA genes [10] (though, some representatives possess extended repeats), inverted repeats in angiosperms include several additional genes. Thus, one may suppose a tendency for extension of inverted repeats along with plastome evolution from ancestral Charophyceae to modern angiosperms. Considering this tendency, examples of repeat junction shifts and especially decrease in their lengths deserve special attention.
Comparison of structural features of 11 Apiaceae plastomes
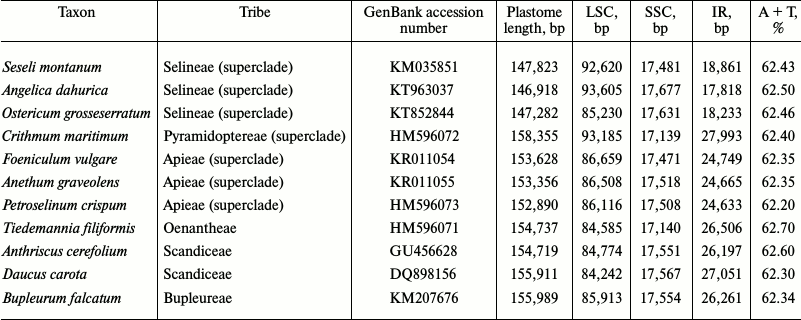
Angelica has the shortest inverted repeat and the smallest plastome among the analyzed Apiaceae. At the same time, it contains the largest LSC. Plastome lengths in Apiaceae vary from 146,918 bp (Angelica) to 158,355 bp (Crithmum), and these variations are largely caused by different locations of one of the junctions of inverted repeat IRb. The length of the small single-copy region of genomes in Apiaceae varies from 17,139 (Crithmum) to 17,677 bp (Angelica). This range is relatively small compared to the difference in lengths of inverted repeats and LSC: the length of inverted repeats varies from 17,818 (Angelica) to 27,993 bp (Crithmum), while the LSC length is between 84,242 (Daucus) and 93,605 bp (Angelica).
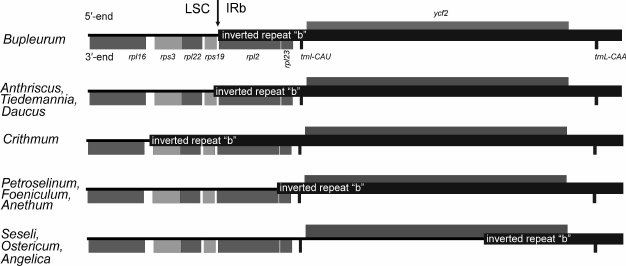
Comparison of Apiaceae plastomes revealed that the JSa junction is located within the ycf1 gene, the JLa junction is situated next to the trnH-GUG gene, while the JSb junction lies near the ndhF gene terminus, and only in Crithmum it resides within the 3′-end of the ndhF gene. At the same time, in plastomes of Anthriscus, Tiedemannia, and Daucus the JLb junction lies within the rps19 gene, while in Bupleurum the rps19 gene is not included already into the inverted repeat, and the LSC/IRb junction resides within the rpl2 gene in Petroselinum, Anethum, and Foeniculum (Fig. 2). In these eight plastomes, the entire ycf2 gene is included into the inverted repeat, in contrast to plastomes of Seseli, Angelica, and Ostericum, where the junction between the LSC and IRb divides the ycf2 gene into two parts. A major portion of the ycf2 gene (4980, 5772, and 5109 bp in Seseli, Angelica, and Ostericum, respectively) is located in the LSC, while part of the gene (1301, 560, and 1235 bp in length, respectively) remains within the inverted repeat IRb due to the JLb junction shift. Shifts of JLb junctions led to an increase in length of the large single-copy region and shortening the inverted repeat and the genome size in general. Reduction of IR length in Petroselinum, Anethum, and Foeniculum almost by 1.5 kb is due to localization of the rps19 gene and a major part of the rpl2 gene in the LSC. In contrast to plastomes of other representatives of Apioideae, the inverted repeat in Crithmum is 1.5 kb larger because of the rps19, rpl22, and rps3 genes and their intergenic spacers, which are now duplicated within the inverted repeats.
Fig. 2. Shift of junction between the large single-copy region (LSC) and the inverted repeat (IRb) in complete plastid genomes of Seseli, Angelica, Ostericum, Crithmum, Foeniculum, Anethum, Petroselinum, Tiedemannia, Anthriscus, Daucus, and Bupleurum.
It is still unknown how exactly the described repeat length changes occurred in the Apioideae. This process could proceed either gradually by small steps according to a proposed gene conversion mechanism [23] (in this case, “intermediate” variants of repeat junction location must exist), or independently in different lineages by a different mechanism that allows acquisition or loss of inverted repeat fragments of several kilobase pairs in length at once [23]. Further accumulation of data on plastome sequences of other Apioideae will provide new insights into the rules of genome organization, rate, manner, and mechanisms of structural rearrangements in plastid genomes.
Acknowledgements
This work was supported by the Russian Science Foundation (project No. 14-50-00029) and by the Russian Foundation for Basic Research (project No. 14-04-01486).
REFERENCES
1.Palmer, J. D. (1991) Plastid chromosome: structure
and evolution, in The Molecular Biology of Plastids (Bogorad,
L., and Vasil, I., eds.) Vol. 7A, pp. 5-53.
2.Kolodner, R., and Tewari, K. K. (1979) Inverted
repeats in chloroplast DNA from higher plants, Proc. Natl. Acad.
Sci. USA, 76, 41-45.
3.Jansen, R. K., and Ruhlman, T. A. (2012) Genomics
of chloroplasts and mitochondria, in The Advances in Photosynthesis
and Respiration (Bock, R., and Knoop, V., eds.) Vol. 35, pp.
103-126.
4.Lavin, M., Doyle, J. J., and Palmer, J. D. (1990)
Evolutionary significance of the loss of the chloroplast-DNA inverted
repeat in the Leguminosae subfamily Papilionoideae, Evolution,
44, 390-402.
5.Logacheva, M. D., Penin, A. A., Vallejo-Roman, C.
M., and Antonov, A. S. (2009) ITS phylogeny of West Asian Heracleum
species and related taxa of Umbelliferae-Tordylieae W. D. J. Koch, with
notes on evolution of their psbA–trnH sequences, Mol.
Biol., 43, 757-765.
6.Shi, C., Liu, Y., Huang, H., Xia, E.-H., Zhang,
H.-B., and Gao, L.-Z. (2013) Contradiction between plastid gene
transcription and function due to complex posttranscriptional splicing:
an exemplary study of ycf15 function and evolution in angiosperms,
PLoS One, 8, e59620.
7.Bayly, M. J., Rigault, P., Spokevicius, A.,
Ladiges, P. Y., Ades, P. K., Anderson, C., Bossinger, G., Merchant, A.,
Udovicic, F., Woodrow, I. E., and Tibbits, J. (2013) Chloroplast genome
analysis of Australian eucalypts – Eucalyptus,
Corymbia, Angophora, Allosyncarpia and
Stockwellia (Myrtaceae), Mol. Phylogenet. Evol.,
69, 704-716.
8.Dong, W., Liu, H., Xu, C., Zuo, Y., Chen, Z., and
Zhou, S. (2014) A chloroplast genomic strategy for designing taxon
specific DNA mini-barcodes: a case study on ginsengs, BMC
Genet., 15, 138.
9.Cai, J., Ma, P.-F., Li, H.-T., and Li, D.-Z. (2015)
Complete plastid genome sequencing of four Tilia species
(Malvaceae): a comparative analysis and phylogenetic implications,
PLoS One, 10, e0142705.
10.Zhu, A., Guo, W., Gupta, S., Fan, W., and Mower,
J. P. (2016) Evolutionary dynamics of the plastid inverted repeat: the
effects of expansion, contraction, and loss on substitution rates,
New Phytol., 209, 1747-1756.
11.Plunkett, G. M., and Downie, S. R. (2000)
Expansion and contraction of the chloroplast inverted repeat in
Apiaceae subfamily Apioideae, Syst. Bot., 25,
648-667.
12.Downie, S. R., and Jansen, R. K. (2015)
Comparative analysis of whole plastid genomes from the Apiales:
expansion and contraction of the inverted repeat, mitochondrial to
plastid transfer of DNA, and identification of highly divergent
noncoding regions, Syst. Bot., 40, 336-351.
13.Valiejo-Roman, C. M., Terentieva, E. I.,
Samigullin, T. H., Pimenov M. G., Ghahremani-Nejad, F., and
Mozaffarian, V. (2006) Molecular data (nrITS-sequencing) reveal
relationships among Iranian endemic taxa of the Umbelliferae, Feddes
Repert., 117, 5-6.
14.Downie, S. R., Spalik, K., Katz-Downie, D. S.,
and Reduron, J.-P. (2010) Major clades within Apiaceae subfamily
Apioideae as inferred by phylogenetic analysis of nrDNA ITS sequences,
Plant Div. Evol., 128, 111-136.
15.Wicke, S., Schneeweiss, G. M., De Pamphilis, C.
W., Muller, K. F., and Quandt, D. (2011) The evolution of the plastid
chromosome in land plants: gene content, gene order, gene function,
Plant Mol. Biol., 76, 273-297.
16.Doyle, J. J., and Doyle, J. L. (1987) A rapid DNA
isolation procedure for small quantities of fresh leaf tissue,
Phytochem. Bull., 19, 11-15.
17.Liu, C., Shi, L, Zhu, Y., Chen, H., Zhang, J.,
Lin, X., and Guan, X. (2012) CpGAVAS, an integrated web server for the
annotation, visualization, analysis, and GenBank submission of
completely sequenced chloroplast genome sequences, BMC Genom.,
13, 715.
18.Lowe, T. M., and Eddy, S. R. (1997) tRNAscan-SE:
a program for improved detection of transfer RNA genes in genomic
sequence, Nucleic Acids Res., 25, 955-964.
19.Laslett, D., and Canback, B. (2004) ARAGORN, a
program to detect tRNA genes and tmRNA genes in nucleotide sequences,
Nucleic Acids Res., 32, 11-16.
20.Lohse, M., Drechsel, O., and Bock, R. (2007)
OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of
high-quality custom graphical maps of plastid and mitochondrial
genomes, Curr. Genet., 52, 267-274.
21.Kurtz, K., and Schleiermacher, Ch. (1999)
REPuter: fast computation of maximal repeats in complete genomes,
Bioinformatics, 15, 426-427.
22.Perry, A. S., and Wolfe, K. Y. (2002) Nucleotide
substitution rates in legume chloroplast DNA depend on the presence of
the inverted repeat, J. Mol. Evol., 55, 501-508.
23.Goulding, S. E., Olmstead, R. G., Morden, C. W.,
and Wolfe, K. H. (1996) Ebb and flow of the chloroplast inverted
repeat, Mol. Gen. Genet., 252, 195-206.
Supplementary Tables S1 and S2 (PDF)