Protein Transphosphorylation During the Mutual Interaction between Phytochrome A and a Nuclear Isoform of Nucleoside Diphosphate Kinase Is Regulated by Red Light
A. Hetmann, M. Wujak*, and S. Kowalczyk
Nicolaus Copernicus University, Faculty of Biology and Environment Protection, Department of Biochemistry, 1 Lwowska St, 87-100 Toruń, Poland; E-mail: mag_wuj@umk.pl* To whom correspondence should be addressed.
Received May 25, 2016; Revision received June 30, 2016
The nuclear isoform of nucleoside diphosphate kinase isoenzyme NDPK-In undergoes strong catalytic activation upon its interaction with the active form of phytochrome A (Pfr) in red light. The autophosphorylation or intermolecular transphosphorylation of NDPK-In leads to the formation of phosphoester bonds stable in acidic solution. The phosphate residue of the phosphamide bond in the active center of NDPK-In can also be transferred to serine and threonine residues localized in other proteins, including phytochrome A. Phytochrome A, similarly to NDPK-In, undergoes autophosphorylation on serine and threonine residues and can phosphorylate some potential substrate proteins. The physical interaction between phytochrome A in the Pfr form and NDPK-In results in a significant increase in the kinase activity of NDPK-In. The results presented in this work indicate that NDPK-In may function as a protein kinase regulated by light.
KEY WORDS: nucleoside diphosphate kinase, phytochrome, signal transduction, transphosphorylationDOI: 10.1134/S0006297916100126
Abbreviations: AK, adenylate kinase; CAT, catalase; ER, endoplasmic reticulum; MAP kinase, mitogen activated protein kinase; MBP, myelin basic protein; NDPK, NDP kinase (nucleoside diphosphate kinase); NDPK-In, nuclear isoform of NDPK; phy A, phytochrome A; PIF/PIL, phytochrome interacting factor/PIF like; PKS1, phytochrome kinase substrate 1.
Nucleoside diphosphate kinase (ATP:nucleoside diphosphate
phosphotransferase, NDPK, NDP kinase; EC 2.7.4.6) transfers an
γ-phosphate residue from nucleoside triphosphates, mainly ATP, to
nucleoside-5′-diphosphates, thereby playing a key role in the
maintenance of physiological concentrations of both ribo- and
deoxyribonucleoside triphosphates. The phosphotransfer reaction is
preceded by autophosphorylation of the enzyme, namely the formation of
a high-energy phosphamide linkage between the phosphate residue and the
nitrogen of the imidazole ring of histidine located in the catalytic
center of the enzyme:
N1TP + E ↔ E~P + N1DP,
E~P + N2DP → E + N2TP.
Initially, the role of NDP kinases was only bound to the metabolism of di- and trinucleotide phosphates. However, further studies, especially those demonstrating the evidence for transfer of phosphate residues from phosphohistidine to serine and threonine residues located within the enzyme itself or in other proteins, supported a more complex role of NDPK in plants [1]. Moreover, some NDP kinases exhibit other than transphosphorylase activity. The studies on eight human isoforms encoded by the NM23 genes [2, 3] as well as on NDP kinases from Drosophila melanogaster, Neurospora crassa, and Saccharomyces cerevisiae demonstrated that some of the isoforms act as 3′→5′ DNA exonucleases, whereas others function as transcriptional factors and DNA repair nucleases or bind and stabilize G-quadruplex DNA [4-6].
Primarily, investigations of plant NDP kinases initiated in the 1990s were directed to identification of individual NDPK isoforms localized in different cellular compartments, and subsequently to cloning the NDPK-encoding genes in spinach, pea, tomato, Arabidopsis, and rice [6-8]. On the basis of studies performed so far, a belief became prevalent that in plants four types of NDP kinases are present, namely NDPK1 localized in the cytoplasm [9], NDPK2 present in plastids and chloroplasts [10, 11], NDPK3 localized in mitochondria [8, 12-16], and a novel NDPK type present in the endoplasmic reticulum (ER) [17]. The roles of these NDP kinases are also diverse, since some of the isoforms are involved in stress-activated signaling pathways, mostly accompanied by the production of active forms of oxygen. Low and high temperature, changes in salinity, pathogen attacks, wounding, as well as hydrogen peroxide activate the expression of at least two NDPK genes (NDPK2 and NDPK1) in Arabidopsis and rice [18-23]. On the other hand, overexpression of NDPK confers increased stress tolerance in Arabidopsis [24, 25], potato [26], and alfalfa plants [27]. NDPK2 was found to interact with elements of signal transduction pathways activated by stress, such as MAP kinase (AtMPK3 and AtMPK6) [19, 22] and SOS2 protein kinase [21], whereas Arabidopsis NDPK1 binds to three isoforms of AtCAT catalase [18]. Interplay was also identified between mitochondrial NDPK3 [13, 14] and adenine nucleotide translocase [15], adenylate kinase (AK) [28], and an 86-kDa protein synthesized under thermal stress conditions [29]. The interaction between NDPK3 and AK affects the activity of both kinases, leading to the activation of NDPK3 and the inhibition of adenylate kinase [28]. Moreover, recombinant pea NDPK3 was shown to possess nuclease activity both towards plasmid DNA and tRNA, as well as 3′-UTR mRNA [16].
In the 1990s for the first time, the link between NDPK activity and responses of plants to red, blue, and UV light was observed [30-32]. Red light stimulates NDPK-catalyzed phosphorylation in etiolated pea and rice seedlings [33-36]. Several independent studies confirmed a physical interaction between NDPK2 and phytochromes A and B, followed by a marked increase of NDPK2 catalytic activity [37-40].
Our previous studies resulted in the purification of two well-separated NDPK isoforms from etiolated oat seedlings where only one of them (designated as NDPK-In) was strongly activated by red light in the presence of phytochrome A (phy A) isolated from oats [41]. This study investigated the phosphotransferase activities of both phytochrome A and NDPK-In in etiolated oat seedlings due to mutual interactions of the proteins.
MATERIALS AND METHODS
Plant material. The NDPK-In isoenzyme was purified from nuclei isolated from 5-day-old etiolated oat seedlings of the Chwat variety grown at 25°C according to the method described by Hetmann and Kowalczyk [41]. Phytochrome A was purified from etiolated oat seedlings according to the method described previously [42] with small modifications introduced by Hetmann and Kowalczyk [41].
Autophosphorylation of NDPK-In and phytochrome A. The reaction mixture in a final volume of 150 µl contained 60 mM Hepes-KOH, pH 7.5, 8 mM KCl, 2 mM MgCl2, and 0.08 mM ATP plus 0.8 µCi [γ32P]ATP or 0.16 mM ATP plus 0.8 µCi [γ32P]ATP. The reaction was initiated by adding 50 µl of NDPK-In purified from nuclei or 100 µl of purified phy A to the final concentration of 60.0 and 50.5 µg/ml, respectively. Phytochrome A was illuminated with red (660 nm) or far-red (730 nm) light prior to its addition to the reaction mixture. The autophosphorylation activity of phy A was performed under dim green light. The samples were mixed, and then 5 or 10 µl of a solution was spotted at fixed time intervals on Whatman 3MM discs (15 mm in diameter). The discs were immediately placed on the surface of 10% trichloroacetic acid (TCA) solution containing 1% tetrasodium diphosphate cooled to 4°C. The TCA solution was changed 5 times every 30 min to completely remove substances of a low molecular weight including [γ32P]ATP. After that, the discs with denatured proteins immobilized on filter paper were dried in 96% ethanol and subsequently placed in scintillation vials with 2 ml of scintillation fluid (EcoliteTM(+)). Radioactivity was measured with a WALLAC 1409 scintillation counter. The Pr form of phy A refers to the biologically inactive form of phy A, which upon irradiation converts to the biologically active Pfr form.
Phosphorylation of substrate proteins by NDPK-In, by phy A, and by interacting phy A and NDPK-In. The above reaction mixture contained additionally 10 µg of one of the protein substrates for protein kinases: dephosphorylated casein, histone III-SS (H III-SS), or myelin basic protein (MBP). The reaction was initiated by the addition of purified NDPK-In or phy A illuminated for 5 min with red or far-red light or both kinases (phy A and NDPK-In) depending on the type of analyzed interactions. The final concentrations of NDPK-In and phy A were 22.5 and 50.5 µg/ml, respectively. The samples were subsequently processed as described above.
SDS-PAGE, immunoblotting, and immunochemical identification of phosphorylated amino acids. The reaction mixture in a final volume of 80 µl contained: 50 mM Hepes-KOH, pH 7.5, 25 mM KCl, 6.25 mM MgCl2, 2 mM ATP, 1 µg of histone III-SS or 2 µg of myelin basic protein or 4 µg of dephosphorylated casein, and 4.5 µg of NDPK-In isolated from oat seedlings. Control samples (without ATP) were prepared by mixing all components and adding to each of them 40 µl of solution for denaturation (IVC) containing 2.5% SDS, 50% glycerol and 1.43 M β-mercaptoethanol in 75 mM Tris-HCl, pH 6.82. The analyzed samples were prepared by mixing all components except for ATP. The reaction was initiated by adding ATP. All samples were incubated for 15 min at room temperature. The phosphorylation reaction was terminated by adding 40 µl of IVC solution to the samples. The samples were denatured for 30 min at 37°C and subsequently separated by SDS-PAGE. SDS-PAGE was performed according to the method of Ogita and Markert [43] in a Mini Protean II electrophoresis cell (Bio-Rad, USA) using a 10 or 16% (w/v) resolving gel. The molecular mass standard was the 10 kDa Protein Ladder (Gibco, USA). The separated proteins were transferred electrophoretically to a nitrocellulose membrane in buffer containing 50 mM Tris, 380 mM glycine, 0.1% (w/w) SDS, and 20% (v/v) methanol. Positions of the protein markers were visualized by staining with Ponceau S. The blot was incubated with biotinylated anti-phosphoserine (Clone PSR-45; Sigma, USA), anti-phosphothreonine (Clone PTR-8; Sigma), or anti-phosphotyrosine (Clone PT-66; Sigma) monoclonal antibodies or with primary antibodies against phy A. The positions of phosphorylated amino acids were detected using goat anti-biotin-IgG antibodies (Sigma) conjugated to alkaline phosphatase or using anti-rabbit IgG conjugated to alkaline phosphatase [44].
Immunoprecipitation of phy A from oat seedling homogenate. Five grams of 5-day-old etiolated seedlings was homogenized and portions of the homogenate were exposed to red or far-red light for 15 min. A 50 µl aliquot of polyclonal antibodies raised in rabbits against phy A was added to 1 ml of homogenate to the final concentration of 2 µg/ml. After 30 min incubation on ice, protein A immobilized on 250 µm acrylic beads (Sigma) was used for immunoprecipitation. The pellet obtained after centrifugation was suspended in 100 mM glycine-HCl buffer, pH 2.5. Acrylic beads were centrifuged, and the resulting supernatant was analyzed by SDS-PAGE and immunoblotting as described above.
RESULTS
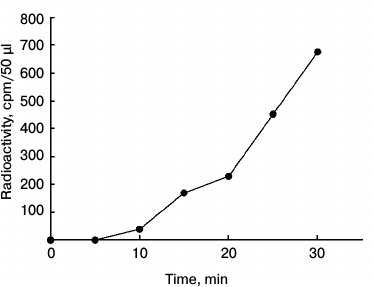
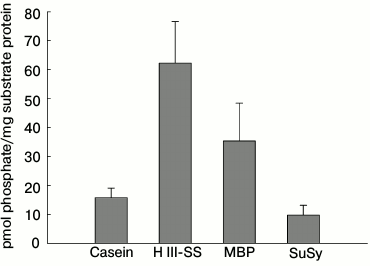
Autophosphorylation of NDPK-In and transfer of the phosphate residue to some substrate proteins. In the first stage of the investigations, the autophosphorylation and/or transphosphorylation reaction of the purified NDPK-In isoform was analyzed. We subsequently checked whether NDPK-In transfers a phosphate residue from ATP to some substrate proteins, including phy A. Finally, the interaction between phy A and NDPK-In was analyzed in reactions of phosphorylation of potential substrate proteins regulated by red and far-red light. The results presented in Fig. 1 indicate that the purified NDPK-In undergoes autophosphorylation. The increase in radioactivity during the 30 min incubation proves that a phosphate residue is transferred from the histidine imidazole ring to hydroxyl residues of amino acids, as only phosphoester bonds are stable in an acidic mixture (10% TCA solution). The possibility of transferring a phosphate residue from phosphohistidine to hydroxyl residues of serine, threonine, or tyrosine (autophosphorylation or transphosphorylation) also suggests the possible transfer of a phosphate residue from phosphohistidine to hydroxyl residues in other substrate proteins. To test this possibility, the reaction mixture used to follow NDPK-In autophosphorylation contained additionally one of four proteins as potential acceptors of phosphate residues: dephosphorylated casein, histone III-SS (H III-SS), myelin basic protein (MBP), sucrose synthase (SuSy). Changes in the level of γ32P in the analyzed proteins are shown in Fig. 2 (depicted as a difference between total radioactivity and radioactivity derived from NDPK-In autophosphorylation). Though all four proteins can be phosphorylated by NDPK-In, histone III-SS is definitely the best acceptor of phosphate residues.
Fig. 1. Autophosphorylation of the NDPK-In isoform. The reaction was initiated by adding 50 µl of NDPK-In purified from nuclei to 150 µl of the reaction mixture containing 60 mM Hepes-KOH, pH 7.5, 8 mM KCl, 2 mM MgCl2, 0.08 mM ATP, and 0.8 µCi [γ32P]ATP. The final concentration of NDPK-In was 60 µg/ml.
Fig. 2. Phosphorylation of selected substrate proteins by the NDPK-In isoform. The reaction mixture used in autophosphorylation experiments contained additionally 10 µg of one of the protein substrates for protein kinases: dephosphorylated casein, histone III-SS (H III-SS), myelin basic protein (MBP), or sucrose synthase (SuSy). The reaction was initiated by addition of the NDPK-In isoform to the final concentration of 22.5 µg/ml.
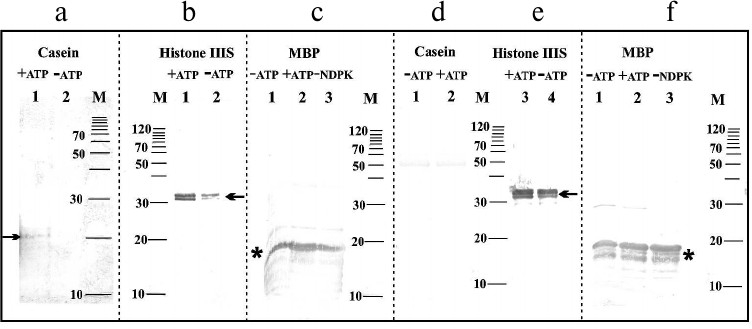
In the next stage of the experiments, an attempt was made to immunochemically identify amino acids in substrate proteins phosphorylated by NDPK-In. Figure 3a shows changes in the phosphoserine levels in dephosphorylated casein. The difference in the intensity of two bands with molecular weights of approximately 21 and 23 kDa (Fig. 3a) clearly indicates that the phosphate residue is transferred from NDPK-In to a serine residue in casein. Similar results were obtained for histone III-SS (data not shown). In histone III-SS, threonine residues are also phosphorylated (Fig. 3b) similarly as in dephosphorylated casein (data not shown). Tracking the transfer of phosphate residues to serine and threonine in MBP is practically impossible because of the high level of phosphorylation of the commercially available proteins (Fig. 3c). Similarly, a high level of phosphotyrosine in MBP and histone III-SS precludes following the possible transfer of phosphate to tyrosine in these proteins (Fig. 3, e and f). Obviously, tyrosine phosphorylation was not detected in dephosphorylated casein (Fig. 3d). It is worth noting that monoclonal antibodies do not identify NDPK-In on any of the blots (Fig. 3). The lack of bands at the 17-kDa level is due to the very small amount of NDPK-In in the analyzed samples and possibly the large excess of substrate proteins.
Fig. 3. Immunochemical identification of phosphoserine, phosphothreonine, and phosphotyrosine in substrate proteins phosphorylated by the NDPK-In isoform. a) Serine phosphorylation in dephosphorylated casein. Lanes: 1) medium with ATP; 2) medium without ATP. b) Threonine phosphorylation in histone III-SS. Lanes: 1) medium with ATP; 2) medium without ATP. c) Threonine phosphorylation in myelin basic protein. Lanes: 1) medium without ATP; 2) medium with ATP; 3) medium without NDPK-In. d) Tyrosine phosphorylation in dephosphorylated casein. Lanes: 1) medium without ATP; 2) medium with ATP. e) Tyrosine phosphorylation in histone III-SS. Lanes: 3) medium with ATP; 4) medium without ATP. f) Tyrosine phosphorylation in myelin basic protein. Lanes: 1) medium without ATP; 2) medium with ATP; 3) medium without NDPK-In; M) molecular weight standards. Histone III-SS and dephosphorylated casein are marked by one arrow, whereas myelin basic protein is marked by an asterisk.
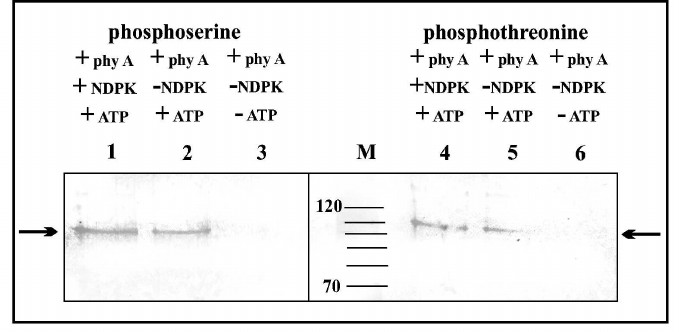
The results depicted in Fig. 4 indicate that the purified phy A preparation does not contain phosphoserine and phosphothreonine. Bands with low intensity are visible in samples containing only phy A and ATP (Fig. 4, lanes 2 and 5), constituting evidence for phy A autophosphorylation/transphosphorylation both on its serine and threonine residues. The most intense bands of phytochrome containing phosphoserine and phosphothreonine are visible in samples incubated with ATP + NDPK-In (Fig. 4, lanes 1 and 4).
Fig. 4. Immunochemical identification of phosphoserine and phosphothreonine in phy A. Lanes: 1-3) polypeptides containing phosphoserine; 4-6) polypeptides containing phosphothreonine; M) molecular weight standard. Arrows indicate phy A polypeptides identified by antibodies against phosphoserine or phosphothreonine.
The molecular weight of polypeptides separated using SDS-PAGE indicates that purified phy A (126 kDa in size) undergoes partial degradation giving two distinct phosphorylated bands with molecular weights of 118 and 100 kDa.
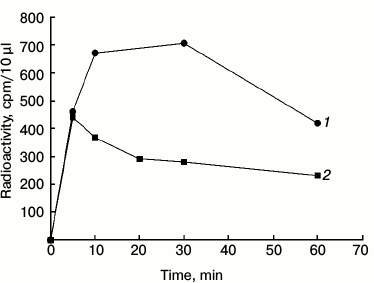
Autophosphorylation of phy A in the Pfr and Pr forms. The changes in the level of protein-bound 32P remaining on the disc are shown in Fig. 5. The radioactivity of the phytochrome increases within 5 min regardless of the length of the illumination. However, after a dozen minutes distinct differences in the level of labeling are visible. The radioactivity level of phy A in the Pfr form increases up to about 10 min and then remains at an almost unchanged level for 30 min. Phytochrome A in the Pr form attains maximal labeling within 5 min, after which the radioactivity level declines slowly. The disappearance of the signal of the labeled phytochrome may be due to its dephosphorylation or degradation followed by elution of products from the filter paper disc by the TCA solution.
Fig. 5. Autophosphorylation of phy A illuminated with (1) red and (2) far-red light. The reaction was initiated by adding 100 µl of purified phy A illuminated with red (660 nm) or far-red (730 nm) light to 150 µl of the reaction mixture containing 60 mM Hepes-KOH, pH 7.5, 8 mM KCl, 2 mM MgCl2, 0.16 mM ATP, and 0.8 µCi [γ32P]ATP. The final concentration of phy A was 50.5 µg/ml.
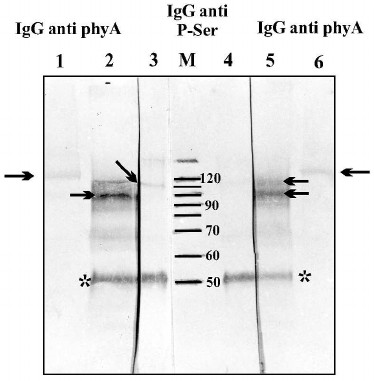
Autophosphorylation of phy A was also analyzed by immunochemical methods. One half of the homogenate from etiolated oat coleoptiles was left in the dark, and the second half was illuminated for 15 min with red light. After that, the phy A was immunoprecipitated by antibodies against phy A. Proteins of the immunological complex were separated in 10% SDS-polyacrylamide gel followed by transfer to a nitrocellulose membrane. Phosphorylated serine residues in the separated polypeptides were detected using monoclonal antibodies (Fig. 6, lanes 3 and 4). In parallel, proteins derived from the immunological complex (Fig. 6, lanes 2 and 5) and also purified phy A (not subjected to immunoprecipitation) (Fig. 6, lanes 1 and 6) were localized using antibodies against phy A. The intensity of the phytochrome band visualized by anti-phosphoserine antibodies is clearly higher in the sample obtained from the homogenate illuminated with red light (Fig. 6, lane 3) in comparison with the sample derived from the unilluminated homogenate (Fig. 6, lane 4). The presence of two polypeptides localized using antibodies against phy A shows that during the immunoprecipitation procedure, phy A is partially degraded (visible bands with molecular weights of 90 to 120 kDa) (Fig. 6, lanes 2 and 5). A single phy A band localized by antibodies is only visible in samples not subjected to immunoprecipitation (Fig. 6, lanes 1 and 6). The results of these experiments unequivocally indicate that red light stimulates autophosphorylation/transphosphorylation of phy A.
Fig. 6. Phytochrome A and proteins precipitated in an immunological complex localized on a nitrocellulose membrane by antibodies directed against phy A and against phosphoserine. Lanes: 1, 6) purified phy A localized by anti-phy A IgG; 2-5) polypeptides of an immunological complex precipitated from an illuminated (lanes 2 and 3) and unilluminated extract (lanes 4 and 5); 2, 5) polypeptides identified by IgG against phy A; 3, 4) polypeptides identified by antibodies against phosphoserine; M) molecular weight standard. The arrow indicates a phy A polypeptide and the asterisk marks the heavy IgG chain from rabbit.
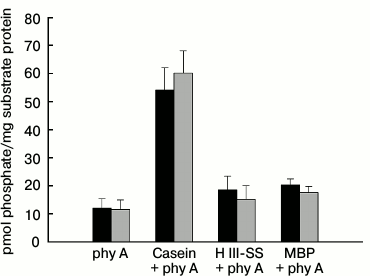
Interaction of phy A and NDPK-In during phosphorylation of selected substrate proteins. The phy A-catalyzed transfer of γ-phosphate of ATP was studied on three substrate proteins: dephosphorylated casein, histone III-SS, and MBP. The results presented in Fig. 7 reveal that the level of autophosphorylation/transphosphorylation of phytochrome in the Pr and Pfr forms differs slightly. However, almost five times more 32P remains on the disc in samples that besides phy A also contain dephosphorylated casein (Fig. 7), but only a slightly higher radioactivity was detected in samples containing histone III-SS and MBP. It is worth noting that no distinct differences were observed in the level of radioactivity between samples illuminated with red and far-red light.
Fig. 7. Phosphorylation of selected proteins by phy A purified from oat coleoptiles. The reaction was initiated by adding 100 µl of phy A to a mixture of 150 µl containing 10 µg of one of the substrate proteins, 0.08 mM ATP, and 0.8 µCi [γ32P]ATP. The final concentration of phy A was 50.5 µg/ml. The sample was illuminated with (■) red or (■) far-red light during the whole incubation period.
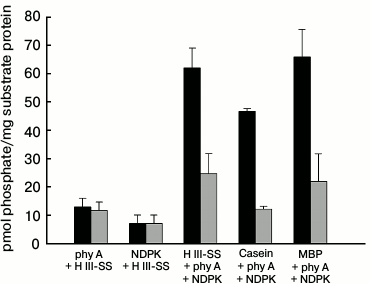
A similar procedure was used in experiments checking the possibility of the transfer of a phosphate residue to substrate proteins under conditions allowing the physical interaction of purified phy A with purified NDPK-In. Changes in the level of labeling of denatured proteins remaining on the discs are presented in Fig. 8. The radioactivity of samples containing histone III-SS (sample 3), dephosphorylated casein (sample 4), and MBP (sample 5) illuminated with red light was found to be distinctly higher in comparison with samples containing only phy A or NDPK-In (sample 1 and 2). The results are different for samples illuminated with far-red light, where the level of protein labeling in samples 3, 4, and 5 is close to the total radioactivity of samples 1 and 2. Thus, these results show that phy A in the Pfr form not only stimulates the catalytic activity of NDPK-In determined as the velocity of transfer of a phosphate residue from ATP to dCDP [41], but also increases the velocity of NDPK-In-catalyzed transfer of the phosphate residue from ATP to a substrate protein. Moreover, the results shown in Fig. 7 demonstrate that the phy A form (Pfr or Pr) does not exert a significant effect on the activity of phy A as a protein kinase, though it cannot be excluded that phy A phosphorylation by NDPK-In (Figs. 4 and 5) may affect the kinase activity of the photoreceptor. The answer to this question will be the aim of future studies.
Fig. 8. Effects of (■) red and (■) far-red light on the phosphorylation of selected substrate proteins by the phy A–NDPK-In system. Apart from 0.08 mM ATP and 0.8 µCi [γ32P]ATP, successive samples contained phy A and histone III-SS (sample 1), NDPK-In isoform and histone III-SS (sample 2), phy A–NDPK-In and histone III-SS (sample 3), phy A–NDPK-In and casein (sample 4), phy A–NDPK-In and myelin basic protein (sample 5). Samples were incubated for 15 min illuminating with red or far-red light.
DISCUSSION
Ion-exchange chromatography of the protein extract from the nuclear fraction indicated the presence of two well-separated NDPK isoenzymes of which only one (NDPK-In) is activated by purified phy A [41]. The properties of purified NDPK-In suggest that it is a homolog of the Arabidopsis NDPK2 isoform strongly activated by phy A and B in the Pfr form [37-39]. Previous studies have demonstrated the participation of AtNDPK2 in the phytochrome-signaling pathway and suggested the localization of this NDPK isoform in the plasma membrane, the cytoplasm, or the nucleus [45-47]. In the context of these findings, it is worth mentioning that the results of our earlier experiments suggest that NDPK-In also occurs in the cytoplasm and is probably transported to the nucleus together with phytochromes [41]. The cytoplasmic localization of AtNDPK2 is also supported by the results of studies linking the role of this isoform with plant responses to various stress conditions, particularly the participation of NDPK2 in the functioning of the MAP kinase cascade, the SOS2 kinase, or the interaction of NDPK2 with small G proteins of the ROP family [19, 21, 22, 48]. Hydrogen peroxide was found to strongly activate the expression of the AtNDPK2 gene and to induce the phosphorylation of two kinases of the MAP kinase family, namely AtMPK3 and AtMPK6, which finally resulted in the phosphorylation of myelin basic protein [19]. In tomato, NDPK2/TAB2 interacts with LeMKK2/tMEK2, a kinase phosphorylating LeMPK3 [22]. Plants with AtNDPK2 overexpression demonstrate clearly higher tolerance for stress conditions such as cold, elevated salinity, or increased reduction of reactive oxygen species [19]. In this context, the results of experiments demonstrating the interaction of NDPK2 with protein kinase SOS2, a known member of signal pathways activated by various stress factors [21], are noteworthy, thus SOS2 kinase binds also catalases CAT2 and CAT3. Interestingly, the direct interaction between NDPK and CAT isoforms from pea and Arabidopsis has recently been reported [18, 23].
The studies mentioned above prompted us to test the hypothesis concerning the function of oat NDPK-In as a protein kinase regulated by phytochromes. The results confirmed that NDPK-In undergoes autophosphorylation or intermolecular transphosphorylation, leading to the formation of phosphoester bonds, which are stable in an acid environment. Our results are in agreement with observations made many years ago on pea in which NDPK was shown to be an 18-kDa protein phosphorylated in the presence of red light [31, 34-36]. These results are of a great importance as several years ago it was reported that AtNDPK2 undergoes autophosphorylation only on a histidine residue and the autophosphorylation/transphosphorylation found previously on a serine residue is an artifact linked to protein denaturation [49]. At this point, it is worth considering that other NDPK isoforms also undergo autophosphorylation on serine residues. Heat shock was found to cause a strong labeling of the NDPK1 isoform with 32P in sugar beet cell cultures [50, 51]. Cytosolic NDPK1 of Solanum chacoense undergoes autophosphorylation on the serine-117 residue [9], and mitochondrial NDPK3 autophosphorylation leads to the formation of a phosphoester bond on at least the serine-119 residue [52].
Interesting experiments performed on various Salmonella typhimurium NDPK mutants indicated that serine phosphorylation is not due to autophosphorylation but rather to intermolecular phosphorylation [53]. In the light of these results, it was justified to check whether the phosphate residue can be transferred from histidine in NDPK to nitrogen in the histidine imidazole ring or to a serine, threonine, or tyrosine residue of another protein. The results of our experiments have confirmed that all four analyzed proteins can be phosphorylated by NDPK-In, though histone III-SS turned out to be the best acceptor of phosphate residues. Phosphate is transferred to serine and threonine residues, while tyrosine residues probably are not acceptors of phosphate residues from NDPK-In. It is worth emphasizing that NDPK-In also phosphorylates purified phy A. The intensity of the bands of intact phy A and also polypeptides formed due to its partial degradation is very clear in the case of phosphoserine identification and slightly weaker in the case of phosphothreonine. Moreover, our results clearly indicate that phy A also undergoes autophosphorylation/transphosphorylation. In experiments performed by other authors, at least three serine residues were found to be phosphorylated in phy A. Serine residues at positions 7 and 598 are phosphorylated in vivo, whereas serine residues at positions 17 and 598 may be phosphorylated in vitro. The serine residue at position 7 is phosphorylated in both forms of phy A, while the phosphorylation of the Ser598 residue occurs only in the Pfr form [54].
It should be pointed out that NDPK activity analyzed as the activity of a protein kinase had been observed earlier in respect to such substrate proteins as histone or MBP [36, 50]. The presumed natural plant substrate protein phosphorylated by NDPK3 in Brassica campestris is protein kinase SRK participating in self-incompatibility [55]. In investigations performed on animal material, it was demonstrated that a phosphate residue from phosphohistidine in NDPK can be transferred to a histidine residue in citrate lyase [56] and to nitrogen of the imidazole ring in protein beta of heterotrimeric G proteins, which is particularly interesting [57]. However, most reports concern transfer of a phosphate residue to a serine residue [1, 2].
The possibility of transferring phosphate residues from NDPK-In to phy A demonstrated in the above experiments turned our attention to the possible interaction of the two proteins during phosphorylation of specific substrate proteins. It should be emphasized here that already in the 1990s some animal protein kinases activated by cAMP and a protein kinase activated by Ca2+ and phospholipids [58-61] were found to be able to phosphorylate phy A [54], and in more recent investigations phy A itself was shown to be a serine/threonine protein kinase [62, 63]. Single reports have also appeared suggesting that the natural substrate proteins for phytochromes are: cryptochrome – a blue light receptor [64], cytoplasmic protein PKS1 [65], and AUX/IAA – a repressor protein of the auxin pathway [66]. Nonetheless, the kinase function of phytochromes was only confirmed in the studies on the interactions of phytochromes with transcription factors PIF/PIL [47]. The results of these investigations prove that at least some PIF/PIL proteins are phosphorylated by phytochromes in the cell nucleus, leading to their ubiquitination and proteolytic degradation in proteasomes [67-69]. Moreover, it was shown that the kinase activity of phytochromes is probably linked with its N-terminal domain since this part of the photoreceptor can act as a native phytochrome upon dimerization followed by transport to the cell nucleus [70]. Thus, phytochromes would form a hitherto unknown family of protein kinases.
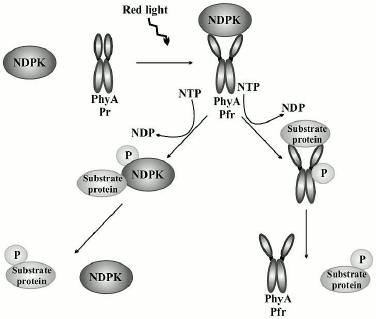
In our studies, phy A was shown to phosphorylate all analyzed proteins with a clear preference for dephosphorylated casein. Similarly to the results obtained by other authors, no clear correlations were found between kinase activity and phy A [62-65]. The results of our studies concerning the effect of red light and far-red light on phosphorylation of substrate proteins by the system of phy A/NDPK-In provide evidence that red light activates the phosphorylation of all three analyzed proteins. Thus, these results indicate that phy A in the Pfr form not only stimulates the catalytic activity of NDPK-In defined as the velocity of the transfer of a phosphate residue from ATP to dCDP [41], but also activates NDPK-In function determined as the velocity of the transfer of a phosphate residue from ATP to a substrate protein. Figure 9 demonstrates a possible mechanism of protein transphosphorylation mediated by the interplay between phy A and NDPK-In. This interesting discovery encourages further investigations encompassing, for instance, attempts to identify other natural substrate proteins phosphorylated by the NDPK-In upon its mutual interaction with phytochromes.
Fig. 9. A proposed mechanism of protein transphosphorylation upon mutual interaction between NDPK-In and phy A stimulated by red light. Red light promotes the interaction between phy A and NDPK-In resulting in their auto- or transphosphorylation and the subsequent stimulation of their protein kinase activities.
REFERENCES
1.Mehta, A., and Orchard, S. (2009) Nucleoside
diphosphate kinase (NDPK, NM23, AWD): recent regulatory advances in
endocytosis, metastasis, psoriasis, insulin regulatory, fetal erythroid
lineage and heart failure; translational medicine exemplified, Mol.
Cell. Biochem., 329, 3-15.
2.Kimura, N., Shimada, N., Ishijima, Y., Fukuda, M.,
Takagi, Y., and Ishikawa, N. (2003) Nucleoside diphosphate kinases in
mammalian signal transduction systems: recent development and
perspective, J. Bioenerg. Biomembr., 35, 41-47.
3.Postel, E. H. (2003) Multiple biochemical
activities of NM23/NDP kinase in gene regulation, J. Bioenerg.
Biomembr., 35, 31-40.
4.Timmons, L., and Shearn, A. (2000) Role of
AWD/nucleoside diphosphate kinase in Drosophila development,
J. Bioenerg. Biomembr., 32, 293-300.
5.Kopylov, M., Bass, H. W., and Stroupe, M. E. (2015)
The maize (Zea mays L.) Nucleoside Diphosphate Kinase 1
(ZmNDPK1) gene encodes a human NM23-H2 homologue that binds and
stabilizes G-quadruplex DNA, Biochemistry, 54,
1743-1757.
6.Hasunuma, K., Yabe, N., Yoshida, Y., Ogura, Y., and
Hamada, T. (2003) Putative functions of nucleoside diphosphate kinase
in plants and fungi, J. Bioenerg. Biomembr., 35,
57-65.
7.Hammargren, J., Sundstrom, J., Johansson, M.,
Bergman, P., and Knorpp, C. (2007) On the phylogeny, expression and
targeting of plant nucleoside diphosphate kinases, Physiol.
Plant., 129, 79-89.
8.Kihara, A., Saburi, W., Wakuta, S., Kim, M. H.,
Hamada, S., Ito, R., Imai, R., and Matsui, H. (2011) Physiological and
biochemical characterization of three nucleoside diphosphate kinase
isozymes from rice (Oryza sativa L.), Biosci. Biotechnol.
Biochem., 75, 1740-1745.
9.Dorion, S., Matton, D. P., and Rivoal, J. (2006)
Characterization of a cytosolic nucleoside diphosphate kinase
associated with cell division and growth in potato, Planta,
224, 108-124.
10.Lubeck, J., and Soll, J. (1995) Nucleoside
diphosphate kinase from pea chloroplasts: purification, cDNA cloning
and import into chloroplasts, Planta, 196, 668-673.
11.Bolter, B., Sharma, R., and Soll, J. (2007)
Localisation of Arabidopsis NDPK2 – revisited,
Planta, 226, 1059-1065.
12.Escobar Galvis, M. L., Hakansson, G., Alexciev,
K., and Knorpp, C. (1999) Cloning and characterization of a pea
mitochondrial NDPK, Biochimie, 81, 1089-1096.
13.Struglics, A., and Hakansson, G. (1999)
Purification of a serine and histidine phosphorylated mitochondrial
nucleoside diphosphate kinase from Pisum sativum, Eur. J.
Biochem., 262, 765-773.
14.Sweetlowe, L. J., Mowday, B., Hebestreit, H. F.,
Leaver, C. J., and Millar, A. H. (2001) Nucleoside diphosphate kinase
III is localised to the inter-membrane space in plant mitochondria,
FEBS Lett., 508, 272-276.
15.Knorpp, C., Johansson, M., and Baird, A. M.
(2003) Plant mitochondrial nucleoside diphosphate kinase is attached to
the membrane through interaction with the adenine nucleotide
translocator, FEBS Lett., 555, 363-366.
16.Hammargren, J., Salinas, T., Marechal-Drouard,
L., and Knorpp, C. (2007) The pea mitochondrial nucleoside diphosphate
kinase cleaves DNA and RNA, FEBS Lett., 58,
3507-3511.
17.Dorion, S., and Rivoal, J. (2015) Clues to the
functions of plant NDPK isoforms, Naunyn Schmiedeberg’s Arch.
Pharmacol., 388, 119-132.
18.Fukamatsu, Y., Yabe, N., and Hasunuma, K. (2003)
Arabidopsis NDK1 is a component of ROS signaling by interacting
with three catalases, Plant Cell Physiol., 44,
982-989.
19.Moon, H., Lee, B., Choi, G., Shin, D., Prasad, D.
T., Lee, O., Kwak, S. S., Kim, D. H., Nam, J., Bahk, J., Hong, J. C.,
Lee, S. Y., Cho, M. J., Lim, C. O., and Yun, D. J. (2003) NDP kinase 2
interacts with two oxidative stress-activated MAPKs to regulate
cellular redox state and enhances multiple stress tolerance in
transgenic plants, Proc. Natl. Acad. Sci. USA, 100,
358-363.
20.Cho, S. M., Shin, S. H., Kim, K. S., Kim, Y. C.,
Eun, M. Y., and Cho, B. H. (2004) Enhanced expression of a gene
encoding a nucleoside diphosphate kinase 1 (OsNDPK1) in rice plants
upon infection with bacterial pathogens, Mol. Cell, 18,
390-395.
21.Verslues, P. E., Batelli, G., Grillo, S., Agius,
F., Kim, Y. S., Zhu, J., Agarwal, M., Katiyar-Agarwal, S., and Zhu, J.
K. (2007) Interaction of SOS2 with nucleoside diphosphate kinase 2 and
catalases reveals a point of connection between salt stress and
H2O2 signaling in Arabidopsis thaliana,
Mol. Cell. Biol., 27, 7771-7780.
22.Xing, T., Rampitsch, C., Sun, S., Romanowski, A.,
Conroy, C., Stebbing, J. A., and Wang, X. (2008) TAB2 a nucleoside
diphosphate kinase is a component of the tMEK2 disease resistance
pathway in tomato, Physiol. Mol. Plant Pathol., 73,
33-39.
23.Haque, M. E., Yoshida, Y., and Hasunuma, K.
(2010) ROS resistance in Pisum sativum cv. Alaska: the
involvement of nucleoside diphosphate kinase in oxidative stress
responses via the regulation of antioxidants, Planta,
232, 367-382.
24.Kim, Y. H., Kim, M. D., Choi, Y. I., Park, S. C.,
Yun, D. J., Noh, E. W., Lee, H. S., and Kwak, S. S. (2011) Transgenic
poplar expressing Arabidopsis NDPK2 enhances growth as well as
oxidative stress tolerance, Plant Biotech. J., 9,
334-347.
25.Liu, H., Weisman, D., Tang, L., Tan, L., Zhang,
W. K., Wang, Z. H., Huang, Y. H., Lin, W. X., Liu, X. M., and
Colon-Carmona, A. (2015) Stress signaling in response to polycyclic
aromatic hydrocarbon exposure in Arabidopsis thaliana involves a
nucleoside diphosphate kinase, NDPK, Planta, 241,
95-107.
26.Kim, M. D., Kim, Y. H., Kwon, S. Y., Yun, D. J.,
Kwak, S. S., and Lee, H. S. (2010) Enhanced tolerance to methyl
viologen-induced oxidative stress and high temperature in transgenic
potato plants overexpressing the CuZnSOD, APX and
NDPK2 genes, Physiol. Plant., 140, 153-162.
27.Wang, Z., Li, H., Ke, Q., Jeong, J. C., Lee, H.
S., Xu, B., Deng, X. P., Lim, Y. P., and Kwak, S. S. (2014) Transgenic
alfalfa plants expressing AtNDPK2 exhibit increased growth and
tolerance to abiotic stress, Plant. Physiol. Biochem.,
84, 67-77.
28.Johansson, M., Hammargren, J., Uppsall, E.,
MacKenzie, A., and Knorpp, C. (2008) The activities of nucleoside
diphosphate kinase and adenylate kinase are influenced by their
interaction, Plant Sci., 174, 192-199.
29.Escobar Galvis, M. L., Marttila, S., Hakansson,
G., Forsberg, J., and Knorpp, C. (2001) Heat stress response in pea
involves interaction of mitochondrial nucleoside diphosphate kinase
with a novel 86-kilodalton protein, Plant Physiol., 126,
69-77.
30.Ito, K., Hamada, T., and Hasunuma, K. (1995) Blue
light signal transmission to 15 kDa proteins in the crude membrane
fraction from the stem section of etiolated pea seedlings, J.
Photochem. Photobiol., 28, 223-227.
31.Hamada, T., Hasunuma, K., and Komatsu, S. (1999)
Phosphorylation of proteins in the stem section of etiolated rice
seedling irradiated with red light, Biol. Pharm. Bull.,
22, 122-126.
32.Zimmermann, S., Baumann, A., Jaekel, K., Marbach,
I., Engelberg, D., and Frohnmeyer, H. (1999) UV-responsive genes of
Arabidopsis revealed by similarity to the Gcn4-mediated UV
response in yeast, J. Biol. Chem., 274, 17017-17024.
33.Hamada, T., and Hasunuma, K. (1994)
Phytochrome-mediated light signal transmission to the phosphorylation
of proteins in the plasma membrane and the soluble fraction of
etiolated pea stem sections, J. Photochem. Photobiol. B,
24, 163-167.
34.Hamada, T., Tanaka, N., Noguchi, T., Kimura, N.,
and Hasunuma, K. (1996) Phytochrome regulates phosphorylation of a
protein with characteristics of a nucleoside diphosphate kinase
activity in the crude membrane fraction from stem section of etiolated
pea seedlings, J. Photochem. Photobiol. B, 33,
143-151.
35.Tanaka, N., Ogura, T., Noguchi, T., Hirano, H.,
Yabe, N., and Hasunuma, K. (1998) Phytochrome-mediated light signal are
transduced to nucleoside diphosphate kinase in Pisum sativum L.
cv. Alaska, J. Photochem. Photobiol. B, 45, 113-121.
36.Ogura, T., Tanaka, N., Yabe, N., Komatsu, S., and
Hasunuma, K. (1999) Characterization of protein complexes containing
nucleoside diphosphate kinase with characteristics of light signal
transduction through phytochrome in etiolated pea seedlings,
Photochem. Photobiol., 69, 397-403.
37.Choi, G., Yi, H., Lee, J., Kwon, Y. K., Soh, M.
S., Shin, B., Luka, Z., Hahn, T. R., and Song, P. S. (1999) Phytochrome
signaling is mediated through nucleoside diphosphate kinase 2,
Nature, 401, 610-613.
38.Im, Y. J., Kim, J. I., Shen, Y., Na, Y., Han, Y.
J., Kim, S. H., Song, P. S., and Eom, S. H. (2004) Structural analysis
of Arabidopsis thaliana nucleoside diphosphate kinase-2 for
phytochrome-mediated light signaling, J. Mol. Biol., 343,
659-670.
39.Shen, Y., Kim, Y. I., and Song, P. S. (2005)
NDPK2 as a signal transducer in the phytochrome-mediated light
signaling, J. Biol. Chem., 280, 5740-5749.
40.Ryu, J. S., Kim, J. I., Kunkel, T., Kim, B. C.,
Cho, D. S., Hong, S. H., Kim, S. H., Fernandez, A. P., Kim, Y., Alonso,
J. M., Ecker, J. R., Nagy, F., Lim, P. O., Song, P. S., Schafer, E.,
and Nam, H. G. (2005) Phytochrome-specific type 5 phosphatase controls
light signal flux by enhancing phytochrome stability and affinity for a
signal transducer, Cell, 120, 395-406.
41.Hetmann, A., and Kowalczyk, S. (2009) Nucleoside
diphosphate kinase isoforms regulated by phytochrome A isolated from
oat coleoptiles, Acta Biochim. Pol., 56, 143-153.
42.Vierstra, R. D., and Quail, P. H. (1983)
Purification and initial characterization of 124-kilodalton
phytochrome, Biochemistry, 22, 2498-2505.
43.Ogita, Z. I., and Markert, C. L. (1979) A
miniaturized system for electrophoresis on polyacrylamide gels,
Anal. Biochem., 99, 233-241.
44.Harlow, E., and Lane, D. (1988) in Antibodies.
A Laboratory Manual, Cold Spring Harbor Laboratory Press, New
York.
45.Choi, G., Kim, J. I., Hong, S. W., Shin, B.,
Choi, G., Blakeslee, J. J., Murphy, A. S., Seo, Y. W., Kim, K., Koh, E.
J., Song, P. S., and Lee, H. (2005) A possible role for NDPK2 in the
regulation of auxin-mediated responses for plant growth and
development, Plant Cell Physiol., 46, 1246-1254.
46.Kevei, E., Schafer, E., and Nagy, F. (2007)
Light-regulated nucleo-cytoplasmic partitioning of phytochromes, J.
Exp. Bot., 58, 3113-3124.
47.Leivar, P., and Quail, P. H. (2011) PIFs: pivotal
components in a cellular signaling hub, Cell, 16,
19-28.
48.Shen, Y., Han, Y. J., Kim, J. I., and Song, P. S.
(2008) Arabidopsis nucleoside diphosphate kinase-2 as a plant
GTPase activating protein, BMB Rep., 41, 645-650.
49.Shen, Y., Kim, J. I., and Song, P. S. (2006)
Autophosphorylation of Arabidopsis nucleoside diphosphate kinase
2 occurs only on its active histidine residue, Biochemistry,
45, 1946-1949.
50.Moisyadi, S., Dharmasiri, S., Harrington, H. M.,
and Lukas, T. J. (1994) Characterization of a low molecular mass
authophosphorylating protein in cultured sugarcane cells and its
identification as a nucleoside diphosphate kinase, Plant
Physiol., 104, 1401-1409.
51.Dharmasiri, S., Harrington, H. M., and
Dharmasiri, N. (2010) Heat shock modulates phosphorylation status and
activity of nucleoside diphosphate kinase in cultured sugarcane cells,
Plant Cell Rep., 29, 1305-1314.
52.Johansson, M., MacKenzie-Hose, A., Andersson, I.,
and Knorpp, C. (2004) Structure and mutational analysis of a plant
mitochondrial nucleoside diphosphate kinase. Identification of residues
involved in serine phosphorylation and oligomerization, Plant
Physiol., 136, 3034-3042.
53.Dar, H. H., and Chakraborti, P. K. (2010)
Intermolecular phosphotransfer is crucial for efficient catalytic
activity of nucleoside diphosphate kinase, Biochem. J.,
430, 539-549.
54.Kim, J. I., Park, J. E., Zarate, X., and Song, P.
S. (2005) Phytochrome phosphorylation in plant light signaling,
Photochem. Photobiol. Sci., 4, 681-687.
55.Matsushita, Y., Suzuki, T., Kubota, R., Mori, M.,
Shimosato, H., Watanabe, M., Kayano, T., Nishio, T., and Nyunoya, H.
(2002) Isolation of a cDNA for a nucleoside diphosphate kinase capable
of phosphorylating the kinase domain of the self-incompatibility factor
SRK of Brassica campestris, J. Exp. Bot., 53,
765-767.
56.Wagner, P. D., and Vu, N. D. (1995)
Phosphorylation of ATP-citrate lyase by nucleoside diphosphate kinase,
J. Biol. Chem., 270, 21758-21764.
57.Wieland, T. (2007) Interaction of nucleoside
diphosphate kinase B with heterotrimeric G protein βγ
dimers: consequences on G protein activation and stability, Naunyn
Schmiedeberg’s Arch. Pharmacol., 374, 373-383.
58.Wong, Y. S., Cheng, H. C., Walsh, D. A., and
Lagarias, J. C. (1986) Phosphorylation of Avena phytochrome
in vitro as a probe of light-induced conformational changes,
J. Biol. Chem., 261, 12089-12097.
59.McMichael, R. W., and Lagarias, J. C. (1990)
Phosphopeptide mapping of Avena phytochrome phosphorylated by
protein kinase in vitro, Biochemistry, 29,
3872-3878.
60.Lapko, V. N., Jiang, X. Y., Smith, D. L., and
Song, P. S. (1997) Posttranslational modification of oat phytochrome A:
phosphorylation of a specific serine in a multiple serine cluster,
Biochemistry, 36, 10595-10599.
61.Lapko, V. N., Wells, T. A., and Song, P. S.
(1996) Protein kinase A-catalyzed phosphorylation and its effect on
conformation in phytochrome A, Biochemistry, 35,
6585-6594.
62.Yeh, K. C., and Lagarias, J. C. (1998) Eukaryotic
phytochromes: light-regulated serine/threonine protein kinases with
histidine kinase ancestry, Proc. Natl. Acad. Sci. USA,
95, 13976-13981.
63.Han, Y. J., Kim, H. S., Kim, Y. M., Shin, A. Y.,
Lee, S. S., Bhoo, S. H., Song, P. S., and Kim, J. I. (2010) Functional
characterization of phytochrome autophosphorylation in plant light
signaling, Plant Cell Physiol., 51, 596-609.
64.Ahmad, M., Jarillo, J. A., Smirnowa, O., and
Caschmore, A. R. (1998) The CRY1 blue light photoreceptor of
Arabidopsis interacts with phytochrome A in vitro,
Mol. Cell, 1, 939-948.
65.Fankhauser, C., Yeh, K. C., Lagarias, J. C.,
Zhang, H., Elich, T. D., and Chory, J. (1999) PKS1 a substrate
phosphorylated by phytochrome that modulates light signaling in
Arabidopsis, Science, 284, 1539-1541.
66.Colon-Carmona, A., Chen, D. L., Yeh, K. C., and
Abel, S. (2000) Aux/IAA proteins are phosphorylated by phytochrome
in vitro, Plant Physiol., 124, 1728-1738.
67.Al-Sady, B., Ni, W., Kircher, S., Schafer, E.,
and Quail, P. H. (2006) Photoactivated phytochrome induces rapid PIF3
phosphorylation prior to proteasome-mediated degradation, Mol.
Cell, 23, 439-446.
68.Shen, H. S., Zhu, L., Castillon, A., Majee, M.,
Downie, B., and Huq, E. (2008) Light-induced phosphorylation and
degradation of the negative regulator PHYTOCHROME-INTERACTING FACTOR1
from Arabidopsis depend upon its direct physical interactions
with photoactivated phytochromes, Plant Cell, 20,
1586-1602.
69.Shen, Y., Zhou, Z., Feng, S., Li, J., Tan-Wilson,
A., Qu, L. J., Wang, H., and Deng, X. W. (2009) Phytochrome A mediates
rapid red light-induced phosphorylation of Arabidopsis FAR-RED
ELONGATED HYPOCOTYL1 in a low fluence response, Plant Cell,
21, 494-506.
70.Bae, G., and Choi, G. (2008) Decoding of light
signals by plant phytochromes and their interacting proteins, Annu.
Rev. Plant Biol., 59, 281-311.