HuR Stabilizes lnc-Sox5 mRNA to Promote Tongue Carcinogenesis
Lifang Wang1,2#, Shucheng Ye2, Junye Wang2, Zhenfang Gu2, Yanhui Zhang2, Chunmei Zhang2, and Xuezhen Ma3#*
1Qingdao University, 266000 Qingdao, China2Department of Oncology, The Affiliated Hospital of Jining Medical University, Jining, Shandong, China
3Department of Oncology, The Second Affiliated Hospital of Medical College Qingdao University, Qingdao, Shandong, China; E-mail: maxuezhen1978@hotmail.com, npc_2016@163.com
# These authors contributed equally to this work.
* To whom correspondence should be addressed.
Received August 30, 2016; Revision received October 12, 2016
Long noncoding RNAs (lncRNAs) have been recently regarded as systemic regulators in multiple biological processes including tumorigenesis. In this study, we report an ultra-highly expressed lncRNA, lnc-Sox5, in tongue tumor tissues. The results imply that lnc-Sox5 may play vital role in tongue carcinoma progression. We observed that the growth of Tca8113 cells was suppressed by lnc-Sox5 downregulation. Additionally, lnc-Sox5 knockdown simultaneously increased Tca8113 cell apoptosis, but the cell cycle was arrested. RNA immunoprecipitation suggested that HuR directly bound to and stabilized lnc-Sox5 RNA. Consistently, HuR knockdown reduced the level of lnc-Sox5 in Tca8113 cells. However, overexpression of HuR induced more lnc-Sox5 in Tca8113 cells. Both lnc-Sox5 knockdown and HuR knockdown suppressed Tca8113 cell tumorigenesis in xenograft models. These results suggest that lnc-Sox5, which was stabilized by HuR, could regulate carcinogenesis of tongue cancer and may serve as a predicted target for tongue carcinoma therapies.
KEY WORDS: long non-coding RNA, lnc-Sox5, tongue carcinoma, cell cycle, apoptosis, HuR, RNA immunoprecipitationDOI: 10.1134/S0006297917040046
Head and neck squamous cell carcinoma (HNSCC) is the sixth most common cancer worldwide including tumors of oropharynx, oral cavity, hypopharynx, and larynx [1]. Tongue squamous cell carcinoma (TSCC) is the most common type of oral tumors and is characterized by high metastatic rate of lymph nodes [2]. Multiple factors including smoking, alcohol consumption [3, 4], and human papilloma virus (HPV) infection [5, 6] have been demonstrated to increase the risk of tongue squamous cell carcinoma. Despite great advances achieved in TSCC therapy, the mortality rate remains high. Therefore, it is urgent to clarify the mechanism and to identify new factors in tongue carcinogenesis.
Long noncoding RNAs (lncRNAs) are generally defined as un-translational transcripts composed of more than 200 nucleotides [7], and they are associated with many important cellular processes and pathogenesis. Numerous lncRNAs are involved in tongue carcinogenesis and metastasis, such as MALAT1 [8], HOTTIP [9], MEG3 [10], and UCA1 [11]. Despite many lncRNAs being expressed at higher or lower level in tongue tumor tissues than them in adjacent tissues, their role and mechanism in tumorigenesis are not clearly elucidated.
In this study, we identified an ultra-highly expressing lncRNA lnc-Sox5 in tongue squamous cell carcinoma. To investigate whether this abnormally expressing lncRNA regulates TSCC progression, we first specifically suppressed lnc-Sox5 in Tca8113 cells by siRNA transfection. Interestingly, the growth of Tca8113 cells was dramatically suppressed by lnc-Sox5 knockdown. Additionally, the apoptosis of Tca8113 cells was simultaneously enhanced by lnc-Sox5 knockdown. Furthermore, the cell cycle was also arrested at G1 to S phase by lnc-Sox5 suppression. To test whether lnc-Sox5 affected tongue cancer metastasis, transwell assays were performed for examining the invasion and migration of Tca8113 cells. These results suggested that both migration and invasion of Tca8113 cells were suppressed by lnc-Sox5 knockdown. A xenograft tumor model suggested that lnc-Sox5 promoted Tca8113 cell carcinogenesis. Moreover, with bioinformatics analysis we observed that there are several HuR binding sites in lnc-Sox5 mRNA. RNA immunoprecipitation (RIP) assay indicated that HuR directly bound to lnc-Sox5 mRNA. Taken together, we report here a new lncRNA, lnc-Sox5, to promote tumorigenesis of tongue squamous cell carcinoma.
MATERIALS AND METHODS
Cells and tissues. Human cell lines HEK-293, HeLa, and U266 were obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China) and Tb3.1, Tca8113, SCC-4, CTSC-3, and Cal27 cells were obtained from the Department of Oncology (Second Affiliated Hospital of Qingdao University). HEK-293 and HeLa cells were cultured in Dulbecco’s modified Eagle medium (DMEM) and U266, Tb3.1, Tca8113, SCC-4, CTSC-3, and Cal27 cells were cultured in RPMI-1640 medium supplemented with 10% (v/v) fetal bovine serum (FBS; Invitrogen, USA) at 37°C in a humidified atmosphere containing 5% CO2. All pairs of TSCC tissues and adjacent tongue tissues were collected from patients who underwent tumor resection at the Second Affiliated Hospital of Qingdao University. All samples were pathologically confirmed, and these patients did not receive any anticancer treatment before surgery. All tissues were acquired with approval of patients, and the protocol was reviewed and approved by the Ethics Committee of the Second Affiliated Hospital of Qingdao University. Fresh tissue samples were snap-frozen in liquid nitrogen and stored at –80°C until RNA or protein extraction.
Plasmids and retrovirus packaging. DNA fragments containing HuR complement sequence were inserted into pMSCV-PIG plasmid. The lnc-Sox5 and HuR siRNA were inserted into pMDH-PGK-EGFP2.0 (pMDH) vector. The sequence of siRNA to lnc-Sox5 and HuR are listed in the table. Retroviral supernatant was generated using standard procedures after calcium phosphate cotransfection of pMDH-siRNA and pAmpho viral packaging plasmid into HEK293T cells. pMDH-siRNA table expression cell lines were generated by virus infection and selected using flow cytometry (FACSAria III; BD Biosciences, USA).
Sequences of primers and siRNAs used in this study
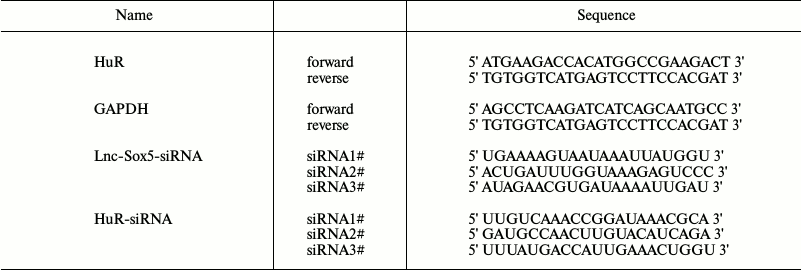
qRT-PCR. Total RNA was purified from TSCC tissues or Tca8113 cells following the manufacturer’s instructions. To detect the expression of lnc-Sox5, cDNA was synthesized using a Reverse Transcription Kit (Takara, China). qRT-PCR was performed using SYBR Green Master Mix (Roche, Switzerland). All primers are listed in the table. The mRNA levels were determined by the comparative Ct method after normalization to β-actin.
Western blot analysis. Tca8113 cell lysates were prepared with RIPA lysis buffer (Beyotime, China) containing protease inhibitor cocktail. Protein samples were loaded for SDS-PAGE and transferred to a nitrocellulose membrane. After blocking with 5% fat-free milk, the membrane was probed with primary anti-HuR (dilution 1 : 1000; Santa Cruz Biotechnology, USA) and anti-GAPDH (dilution 1 : 2000; Santa Cruz Biotechnology) antibody. After washing, the membrane was incubated with horseradish peroxidase-conjugated (HRP) secondary antibody for 1 h. The signal was visualized using the ECL detection system (Thermo Fisher, USA) and quantified by densitometry using Quantity One software (Bio-Rad, USA).
Transwell assays. Boyden chambers were used for cell migration assays. Tca8113 cells (2·105 cells/well) were placed in the upper transwell chambers (8 µm pore size) 24 h after siRNA transfection. The lower chambers were filled with 0.5 ml RPMI-1640 containing 5% FBS. Cells were incubated for 24 h at 37°C in 5% CO2. Nonmigratory cells were carefully scraped with a cotton swab. Cells remaining on the bottom surface were counted after staining with crystal violet. The number of cells in five random fields was counted for cell migration analysis. The same Boyden chambers were used for cell invasion assay. The upper surface of the filter was coated with 20 µl Matrigel (0.3 mg/ml; BD Biosciences). Cells were incubated for 24 h at 37°C in 5% CO2. Cells were counted as described above.
Cell proliferation assay. Tca8113 cells were seeded in 96-well plates (4·103 cells per well) and transfected with lnc-Sox5 siRNAs or mimics control and cultured at 37°C in an incubator containing 5% CO2. After culturing for 24 h, cells were tested for viability using the 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) assay. Briefly, cells were incubated with MTT at final concentration 0.5 mg/ml (Sigma, USA) for 4 h. The supernatant was discarded, and the precipitated formazan was dissolved in dimethyl sulfoxide. Absorbance was measured at 570 nm with a microplate reader (Molecular Devices, USA).
Flow cytometric analysis. To determine the effects of lnc-Sox5 on apoptosis and the cell cycle, Tca8113 cells were transfected with lnc-Sox5 siRNA and then subjected to flow cytometric analysis using an Annexin V Apoptosis Detection Kit (Becton Dickinson, USA) or staining with Annexin V-fluorescein isothiocyanate (FITC), propidium iodide (PI) for 25 min, and then analyzed by flow cytometry (Thermo Fisher, USA). FACS data were analyzed using FlowJo software (Tree Star, Inc.).
Bioinformatics analysis. About 118,777 transcripts were downloaded from LNCipedia (www.lncipedia.org). All these transcripts containing AT-rich elements were selected as the candidates for HuR target.
RNA immunoprecipitation (RIP). Tca8113 cells were collected and washed with PBS, and the pellets were disintegrated in RIP buffer on ice for 30 min and centrifuged at 10,000g for 5 min. Anti-HuR antibodies (5 µg) were added to the supernatant and incubated for 4 h at 4°C. Protein A beads (40 µl) were added and incubated for another 2 h at 4°C. The beads were pelleted at 1600g for 30 s, the supernatant was removed, and the beads were resuspended in 500 ml of RIP buffer. The RIP wash was repeated thrice, followed by one wash in PBS. Coprecipitated RNAs were isolated by resuspending the beads in TRIzol RNA extraction reagent (500 µl) according to the manufacturer’s instructions. Purified RNA was subject to quantitative PCR for lnc-Sox5 determination as described above.
Animal tumor model. Six-to-eight-week-old BALB/c (nu/nu) mice were purchased from Shanghai SLAC Laboratory Animal Co (China). The mice were maintained in a barrier facility at the Animal Center of Chongqing Medical University. Stably expressing lnc-Sox5 siRNA or scramble control cells were implanted subcutaneously (s.c.) into the right flank of the mice. Tumor volume was measured three times per week. All groups of mice were sacrificed, and tumors were weighted at the endpoint of these experiments.
Statistical analysis. Comparison between groups was performed using Student’s t-test and one-way ANOVA. SPSS 16.0 (SPSS, USA) was used for all statistical analyses. Experiments were performed at least three times. All results are described as means ± standard error of mean (S.E.M); p values less than 0.05 were considered as significant differences. All tests were done using Prism 6 software (Graphpad Prism 6).
RESULTS
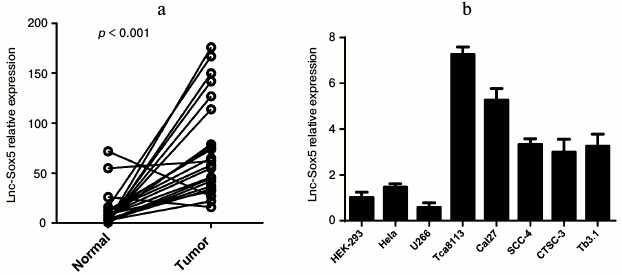
Aberrant lnc-Sox5 expression in tongue squamous cell carcinoma and tongue cancer cell lines. To explore abnormally expressing lncRNAs in tongue squamous cell carcinoma, we examined the level of several lncRNAs in TSCC tissues (data not shown). We observed that the expression of lnc-Sox5 was significantly increased in TSCC. Therefore, 22 pairs of clinic TSCC tissues and matched adjacent normal tissues were subjected to quantitative PCR (q-PCR) for lnc-Sox5 expression. Interestingly, almost all matched samples showed significantly increased level of lnc-Sox5 in tumor tissues compared with their normal control, whereas only three samples showed downregulated or equivalent lnc-Sox5 level in TSCC tissues (Fig. 1a). To investigate whether lnc-Sox5 played a functional role in TSCC progression, we subsequently detected the expression of lnc-Sox5 by q-PCR in several TSCC cell lines, including Tca8113, Tb3.1, SCC-4, CTSC-3, Cal27 and other cancer cell lines, such as 293T, HeLa, and U266 cells (Fig. 1b). Consistently, lnc-Sox5 was found to be expressed at high level in tongue cancer cell lines. These results suggested that the aberrant lnc-Sox5 might act in a potential oncogenic role in TSCC progression.
Fig. 1. Lnc-Sox5 expression in tumor tissue samples and tongue cancer cell lines. a) Expression of lnc-Sox5 in tongue tumor samples and adjacent normal samples detected by qRT-PCR. b) Expression of lnc-Sox5 in tongue cancer cell lines and other control cell lines detected by qRT-PCR. qRT-PCR results are normalized to β-actin. Data are shown as mean ± S.E.M.
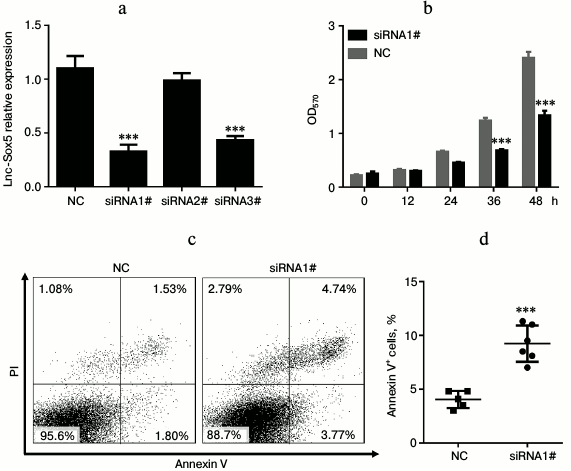
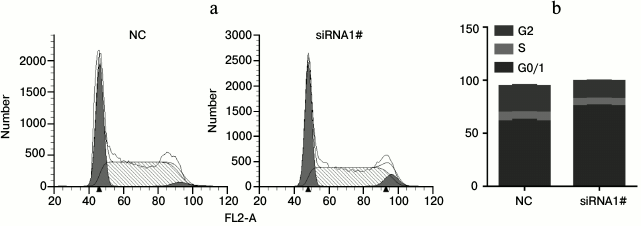
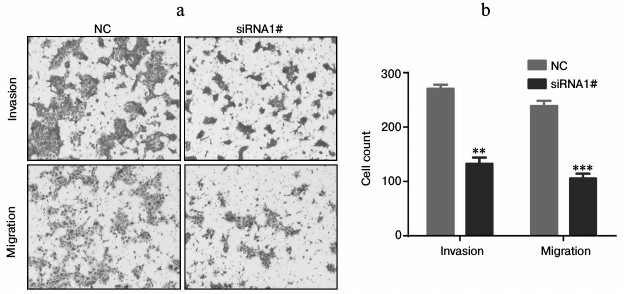
Lnc-Sox5 impaired Tca8113 cell apoptosis and cell cycle in vitro. To extensively investigate the oncogenic role of lnc-Sox5 in TSCC, we selected Tca8113 cells for further experimental analysis because lnc-Sox5 was found to be expressed at the highest level in this cell line (Fig. 1b). First, we specifically suppressed the expression of lnc-Sox5 by siRNA transfection. As the result described, the first siRNA (siRNA1#) had the best inhibitory efficacy in Tca8113 cells (Fig. 2a). MTT result showed that the cell growth was significantly suppressed by lnc-Sox5 knockdown (Fig. 2b). Additionally, the apoptotic rate of Tca8113 cells was increased by lnc-Sox5 knockdown (Fig. 2, c and d). Flow cytometry showed that the cell cycle was also retarded by lnc-Sox5 suppression (Fig. 3, a and b). These results suggested that lnc-Sox5 promoted cell growth in Tca8113 cells. To clearly understand the oncogenic role of lnc-Sox5 in tongue cancer, the migration and invasion of Tca8113 cells in vitro were also investigated. The invasion and migration of Tca8113 cells were analyzed by transwell assays. Interestingly, both invasion and migration of Tca8113 cells were decreased after lnc-Sox5 inhibition (Fig. 4, a and b). Taken together, these data suggested that lnc-Sox5 acted as an oncogene in tongue cancer progression.
Fig. 2. Lnc-Sox5 inhibition suppressed cell growth and induced cell apoptosis in Tca8113 cells. a) Tca8113 cells were transfected with lnc-Sox5 siRNAs and the inhibitory efficacy was determined by qRT-PCR (n = 3). b) Growth of Tca8113 cells was detected by MTT assay every 12 h after siRNA transfection (n = 4). c, d) Flow cytometric analysis for the apoptotic rate of Tca8113 cells at 24 h after siRNA transfection (n = 6). NC represents the random RNA sequence and acts as the negative control (herein and in Figs. 3-5). Data are shown as mean ± S.E.M; *** p < 0.001.
Fig. 3. Loss of lnc-Sox5 retarded Tca8113 cell cycle at G0/1 to S phase. a, b) Tca8113 cells were stained with PI as described above, and the cell cycle was determined by flow cytometry after lnc-Sox5 siRNA1#.
Fig. 4. Invasion and migration of Tca8113 cells were suppressed by lnc-Sox5 dysregulation. a, b) Invasion and migration of Tca8113 cells were detected by transwell assay after siRNA transfection (n = 5). Data are shown as mean ± S.E.M.; ** p < 0.01, *** p < 0.001.
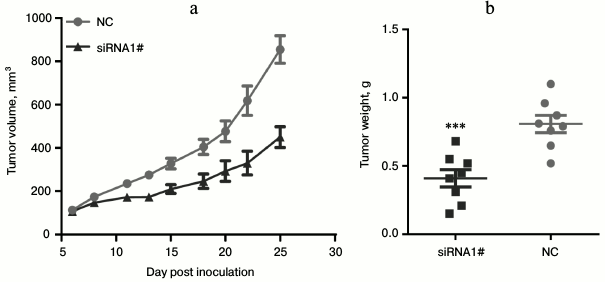
Lnc-Sox5 knockdown suppressed tongue tumor growth. To extensively explore the oncogenic role of lnc-Sox5 in tongue cancer progression, we subsequently generated a stably lnc-Sox5 knockdown Tca8113 cell line by infecting cells with retrovirus expressing siRNA1. Furthermore, we used this cell line to construct a xenograft mouse model, and the tumor volume was detected every two days when the tumor volume reached 100 mm3. As expected, lnc-Sox5 knockdown significantly suppressed the tumorigenesis of Tca8113 cells (Fig. 5a). Consistent to the tumor volume, tumor weight was also significant reduced by lnc-Sox5 knockdown (Fig. 5b). According to these results, we conclude that lnc-Sox5 plays an oncogene in tongue cancer progression.
Fig. 5. a, b) Lnc-Sox5 knockdown suppressed carcinogenesis of Tca8113 cells in a xenograft model. Mice were injected with lnc-Sox5 siRNA stably expressing Tca8113 cells (n = 8) or vector Tca8113 cells (n = 8) in the right side of the axillae. Data are shown as mean ± S.E.M.; *** p < 0.001.
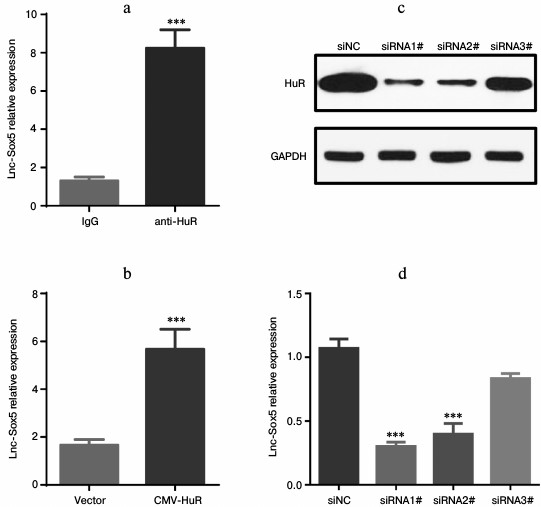
Lnc-Sox5 expression is associated with and stabilized by HuR. Lnc-Sox5 contributed to tongue cancer carcinoma, prompting us to investigate the regulatory mechanism of controlling lnc-Sox5 expression in tongue cancer cells. HuR is a RNA-binding protein that directly binds to messenger RNA frequently containing AU- or U-rich sequence elements (ARE). Thus, we first explored the sequence of lnc-Sox5 mRNA. Surprisingly, there were many AU-rich sequences distributed in the full length of lnc-Sox5 (http://www.lncipedia.org/db/transcript/lnc-SOX5). Therefore, we subsequently detected the interaction between HuR and lnc-Sox5 in Tca8113 cells by RNA-immunoprecipitation (RIP). Lysate from Tca8113 cells were immunoprecipitated by anti-HuR antibody or IgG antibody, and the lnc-Sox5 mRNA was validated by qRT-PCR. According to the RT-qPCR results, lnc-Sox5 level in anti-HuR immunoprecipitated lysate was significantly higher than in the IgG control group (Fig. 6a). To extensively confirm whether HuR is bound to lnc-Sox5 in Tca8113 cells, we next overexpressed (Fig. 6b) or specifically inhibited HuR expression in Tca8113 cells (Fig. 6, c and d). RT-qPCR data indicated that overexpressing HuR could increase the level of lnc-Sox5, while suppressed HuR expression could reduce the expression of lnc-Sox5 in Tca8113 cells. Therefore, these results suggest that lnc-Sox5 is physically associated with and stabilized by HuR.
Fig. 6. HuR stabilized lnc-Sox5 mRNA in Tca8113 cells. a) The HuR–mRNA complex was pulled down by anti-HuR antibody, and the level of lnc-Sox5 was detected by qRT-PCR, IgG being the control. b) lnc-Sox5 mRNA level was detected in Tca8113 cells when HuR was upregulated by transfecting with HuR-expressing plasmid (CMV-HuR). c) HuR was inhibited by transfecting with siRNA, and the inhibitory efficacy was determined by Western blot assay. d) Expression of lnc-Sox5 was detected by qRT-PCR in Tca8113 cells with HuR siRNAs transfection. siNC represents the random RNA sequence and acts as the negative control. Data are shown as mean ± S.E.M.; *** p < 0.001.
DISCUSSION
LncRNAs have been shown to play a critical role in multiple biological progressions including tumorigenesis [12, 13]. To date, several lncRNAs have been reported to act as oncogene or suppressor of tongue cancer. LncRNA HOTTIP is located at the 5′ tip of the HOXA locus and coordinates the activation of multiple 5′ HOXA genes, which plays an important role in multiple cancers [14]. Zhang reported that lncRNA HOTTIP is correlated with progression and prognosis in tongue carcinoma [9]. Additionally, it has been reported that lncRNA UCA1 is ultra-highly expressed in tongue squamous cell carcinoma and is correlated with tongue cancer metastasis [11]. Jia showed that miR-26a and lncRNA MEG3 expression were both strongly reduced in TSCC, and combined low expression levels of both miR-26a and MEG3 emerged as an independent prognostic factor for poor clinical outcome in TSCC patients [10]. In this study, we first investigated the role of lnc-Sox5 in human tongue cancer. We observed that lnc-Sox5 expression was upregulated in tumor tissues from 22 tongue cancer patients. To explore whether it acted as an oncogene or a suppressor in human tongue cancer, the lnc-Sox5 level was specifically suppressed by siRNA transfection in tongue cancer cells. We observed lnc-Sox5 knockdown significantly suppressed the growth of Tca8113 cells. Moreover, both cell apoptosis and cell cycle were impaired by lnc-Sox5 knockdown. Consistently, we also observed the invasion and migration of Tca8113 cells were suppressed by lnc-Sox5 knockdown. These results suggest that lnc-Sox5 is an oncogene in human tongue cancer. We are still studying the targets of lnc-Sox5 in tongue cancer cells.
HuR is a member of the ELAV (embryonic lethal abnormal vision) family of RNA-binding proteins (RBPs), and it binds or stabilizes AU-rich element (ARE)-containing mRNAs [15]. An ARE is commonly present in the untranslated region of many protooncogenes, growth factors, and cytokine mRNAs [16, 17]. Multiple copies of the sequence AU often exist in the ARE and they target ARE-mRNAs for rapid degradation [16]. However, numerous proteins, including HuR, are known to interact with AREs and modulate either the stabilization or destabilization of ARE-mRNAs [18]. To date, it has been reported that HuR also could bind to and stabilize or destabilize lncRNA mRNAs that contained AU-rich elements [19-21]. In this study, we observed there were many AU-rich fragments in lnc-Sox5 mRNA. RNA immunoprecipitation suggested that HuR directly interacted with lnc-Sox5 mRNA.
In this study, we identified very highly expressing lncRNA, lnc-Sox5, in tumor tissues from 22 tongue cancer patients. To investigate whether this abnormally expressing lncRNA regulated tongue cancer progression, the expression of lnc-Sox5 was first suppressed by siRNA. MTT assays showed that the growth of Tca8113 was dramatically suppressed by lnc-Sox5 knockdown. Additionally, lnc-Sox5 knockdown also induced more apoptotic Tca8113 cells. Furthermore, the cell cycle was also arrested at G1 to S phase in Tca8113 cells. To test whether lnc-Sox5 affects tongue cancer metastasis, we also determined the invasion and migration in lnc-Sox5 suppressed Tca8113 cells. Transwell assays indicated that both migration and invasion of Tca8113 cells were suppressed by lnc-Sox5 knockdown. A xenograft tumor model suggested lnc-Sox5 promoted tongue carcinogenesis. We also observed that there were several HuR binding sites in lnc-Sox5 mRNA. RNA immunoprecipitation showed that HuR directly bound to and therefore stabilized lnc-Sox5 mRNA. The direct targets of lnc-Sox5 in Tca8113 cells are still under study. These results suggest that lnc-Sox5, which was stabilized by HuR, may serve as a prediction target for tongue carcinoma therapies.
Acknowledgements
This work was supported in part by grants from the Medicine and Healthcare Development Plan in Science and Technology of Shandong Province (Project No. 2015WSB34005 to X. Z. Ma).
REFERENCES
1.Siegel, R., Naishadham, D., and Jemal, A. (2012)
Cancer statistics, CA Cancer J. Clin., 62, 10-29.
2.Jemal, A., Siegel, R., Ward, E., Murray, T., Xu,
J., and Thun, M. J. (2007) Cancer statistics, CA Cancer J.
Clin., 57, 43-66.
3.Nisar, Y. B. (2006) Association of smoking with
lymph nodes metastasis in early stages of squamous cell carcinoma of
tongue, J. Coll. Physicians Surg. Pak., 16, 312; author
reply 312.
4.Yamazaki, H., Inoue, T., Yoshida, K., Kotsuma, T.,
Yoshioka, Y., Koizumi, M., Furukawa, S., Kakimoto, N., Shimizutani, K.,
and Nishimura, T. (2009) Assessment of influence of smoking, drinking,
leukoplakia and dental irritation on local control of early oral tongue
carcinoma treated with brachytherapy: age and dental factors are
potential prognostic factors, Tumori, 95, 461-466.
5.Sgaramella, N., Coates, P. J., Strindlund, K.,
Loljung, L., Colella, G., Laurell, G., Rossiello, R., Muzio, L. L.,
Loizou, C., Tartaro, G., Olofsson, K., Danielsson, K., Fahraeus, R.,
and Nylander, K. (2015) Expression of p16 in squamous cell carcinoma of
the mobile tongue is independent of HPV infection despite presence of
the HPV-receptor syndecan-1, British J. Cancer,
113, 321-326.
6.Zhang, H., Zhang, Y., Zhao, H., Niyaz, H., Liu, P.,
Zhang, L., Zhang, S., Reheman, Y., Bao, Y., and Chen, X. (2016) HPV
infection and prognostic factors of tongue squamous cell carcinoma in
different ethnic groups from geographically closed cohort in Xinjiang,
China, Biochem. Res. Int., 7498706.
7.Diederichs, S. (2014) The four dimensions of
noncoding RNA conservation, Trends Genet., 30,
121-123.
8.Liang, J., Liang, L., Ouyang, K., Li, Z., and Yi,
X. (2016) MALAT1 induces tongue cancer cells’ EMT and inhibits
apoptosis through Wnt/beta-catenin signaling pathway, J. Oral
Pathol. Med., doi: 10.1111/jop.12466.
9.Zhang, H., Zhao, L., Wang, Y. X., Xi, M., Liu, S.
L., and Luo, L. L. (2015) Long non-coding RNA HOTTIP is correlated with
progression and prognosis in tongue squamous cell carcinoma, Tumor
Biol. J. Int. Soc. Oncodev. Biol. Med., 36, 8805-8809.
10.Jia, L. F., Wei, S. B., Gan, Y. H., Guo, Y.,
Gong, K., Mitchelson, K., Cheng, J., and Yu, G. Y. (2014) Expression,
regulation and roles of miR-26a and MEG3 in tongue squamous cell
carcinoma, Int. J. Cancer, 135, 2282-2293.
11.Fang, Z., Wu, L., Wang, L., Yang, Y., Meng, Y.,
and Yang, H. (2014) Increased expression of the long non-coding RNA
UCA1 in tongue squamous cell carcinomas: a possible correlation with
cancer metastasis, Oral Surg. Oral Med. Oral Pathol. Oral
Radiol., 117, 89-95.
12.Flippot, R., Malouf, G. G., Su, X., Mouawad, R.,
Spano, J. P., and Khayat, D. (2016) Cancer subtypes classification
using long non-coding RNA, Oncotarget, doi:
10.18632/oncotarget.10213.
13.Xu, Q., Deng, F., Qin, Y., Zhao, Z., Wu, Z.,
Xing, Z., Ji, A., and Wang, Q. J. (2016) Long non-coding RNA regulation
of epithelial-mesenchymal transition in cancer metastasis, Cell
Death Dis., 7, e2254.
14.Li, Z., Zhao, L., and Wang, Q. (2016)
Overexpression of long non-coding RNA HOTTIP increases chemoresistance
of osteosarcoma cell by activating the Wnt/beta-catenin pathway, Am.
J. Transl. Res., 8, 2385-2393.
15.Ma, W. J., Cheng, S., Campbell, C., Wright, A.,
and Furneaux, H. (1996) Cloning and characterization of HuR, a
ubiquitously expressed Elav-like protein, J. Biol. Chem.,
271, 8144-8151.
16.Chen, C. Y., and Shyu, A. B. (1995) AU-rich
elements: characterization and importance in mRNA degradation,
Trends Biochem. Sci., 20, 465-470.
17.Jacobson, A., and Peltz, S. W. (1996)
Interrelationships of the pathways of mRNA decay and translation in
eukaryotic cells, Annu. Rev. Biochem., 65,
693-739.
18.Grammatikakis, I., Abdelmohsen, K., and Gorospe,
M. (2016) Posttranslational control of HuR function, Wiley
Interdiscip. Rev. RNA, doi: 10.1002/wrna.1372.
19.Chai, Y., Liu, J., Zhang, Z., and Liu, L. (2016)
HuR-regulated lncRNA NEAT1 stability in tumorigenesis and progression
of ovarian cancer, Cancer Med., 5, 1588-1598.
20.Kim, J., Abdelmohsen, K., Yang, X., De, S.,
Grammatikakis, I., Noh, J. H., and Gorospe, M. (2016) LncRNA
OIP5-AS1/cyrano sponges RNA-binding protein HuR, Nucleic
Acids Res., 44, 2378-2392.
21.Noh, J. H., Kim, K. M., Abdelmohsen, K., Yoon, J.
H., Panda, A. C., Munk, R., Kim, J., Curtis, J., Moad, C. A., Wohler,
C. M., Indig, F. E., de Paula, W., Dudekula, D. B., De, S., Piao, Y.,
Yang, X., Martindale, J. L., De Cabo, R., and Gorospe, M. (2016) HuR
and GRSF1 modulate the nuclear export and mitochondrial localization of
the lncRNA RMRP, Genes Dev., 30, 1224-1239.