REVIEW: Features of Protein–Protein Interactions in the Cyanobacterial Photoprotection Mechanism
N. N. Sluchanko1,2*, Y. B. Slonimskiy1,3, and E. G. Maksimov2
1Bach Institute of Biochemistry, Federal Research Center “Fundamentals of Biotechnology”, Russian Academy of Sciences, 119071 Moscow, Russia; E-mail: nikolai.sluchanko@mail.ru2Lomonosov Moscow State University, Faculty of Biology, Biophysics Department, 119991 Moscow, Russia
3Lomonosov Moscow State University, Faculty of Biology, Biochemistry Department, 119991 Moscow, Russia
* To whom correspondence should be addressed.
Received August 21, 2017; Revision received September 11, 2017
Photoprotective mechanisms of cyanobacteria are characterized by several features associated with the structure of their water-soluble antenna complexes – the phycobilisomes (PBs). During energy transfer from PBs to chlorophyll of photosystem reaction centers, the “energy funnel” principle is realized, which regulates energy flux due to the specialized interaction of the PBs core with a quenching molecule capable of effectively dissipating electron excitation energy into heat. The role of the quencher is performed by ketocarotenoid within the photoactive orange carotenoid protein (OCP), which is also a sensor for light flux. At a high level of insolation, OCP is reversibly photoactivated, and this is accompanied by a significant change in its structure and spectral characteristics. Such conformational changes open the possibility for protein–protein interactions between OCP and the PBs core (i.e., activation of photoprotection mechanisms) or the fluorescence recovery protein. Even though OCP was discovered in 1981, little was known about the conformation of its active form until recently, as well as about the properties of homologs of its N and C domains. Studies carried out during recent years have made a breakthrough in understanding of the structural-functional organization of OCP and have enabled discovery of new aspects of the regulation of photoprotection processes in cyanobacteria. This review focuses on aspects of protein–protein interactions between the main participants of photoprotection reactions and on certain properties of representatives of newly discovered families of OCP homologs.
KEY WORDS: orange carotenoid protein, carotenoids, photoprotection, fluorescence, energy transfer, photosynthesis, phycobilisomesDOI: 10.1134/S000629791713003X
Abbreviations: APC, allophycocyanin; APC660, APC with the fluorescence maximum at 660 nm; ApcC (or Lc), PBs core linker polypeptide with molecular weight of 7.8 kDa; Apc D, E, F, allophycocyanin-like proteins with fluorescence maximum at about 680 nm; Apo-OCP, apo form of OCP; ETC, electron transport chain; FRP, fluorescence recovery protein; HCP, helical carotenoid protein homologous to the N-domain of OCP; LCM, PBs core–membrane linker polypeptide; NTE, N-terminal segment of OCP; NTF2, nuclear transport factor 2; OCP, orange carotenoid protein; OCPO, orange form of OCP; OCPR, red form of OCP; OCP-W288A, OCP with W288A amino acid substitution, an analog of the activated form of OCP; PBs, phycobilisome; PC, phycocyanin; PS I(II), photosystem I(II); RC, reaction center of a photosystem; RCP, red carotenoid protein (OCP N-terminal fragment); ROS, reactive oxygen species; TE, terminal emitter.
Photosynthesis plays a central role in the energetics of living systems
because it is the primary source of all energy used by living organisms
in their vital processes. Photosynthesis is a complex set of reactions
occurring over time periods from 10–15 s (light
absorption) up to 104 s (formation of photosynthesis
products). The concept of primary photosynthetic processes includes
photophysical and photochemical stages of photosynthesis –
light absorption, energy migration, charge separation, and the
formation of electrochemical potential in the reaction centers of
photosystems. Studies carried out in recent decades have shown very
high efficiency of primary photosynthetic processes due to specifics of
their structural organization [1-9].
Light-harvesting, or the so-called antenna, complexes contain pigments having strictly defined positions with orientations relative to each other. In addition, due to interactions of pigments with different amino acid residues of proteins, many different spectral forms are present in the antenna complexes, and therefore the pigments form a broad total absorption spectrum [10]. Due to strong overlapping of the absorption spectra of individual pigments after excitation, an equilibrium is quickly established within the antenna, both between the pigments in the antenna (within fractions of picoseconds) and between the antenna and the reaction center. With the participation of pigments having different spectral characteristics, energy migrates to the longest wavelength pigments of the reaction center, where excitation energy is captured and converted into the energy of separated charges [6].
Excited states of molecules are reactive, and many pigments can interact with molecular oxygen during the lifetime of its excited state, resulting in the formation of reactive oxygen species (ROS) [10]. Properties of ROS and their role in oxidative reactions are well known. The formation of ROS inevitably accompanies photosynthesis and respiration, but in addition to regulatory functions, ROS can pose a threat to a living cell. Several enzyme systems (superoxide dismutase, catalase, peroxidase, etc.) can effectively inactivate newly formed free radicals [11]. Photosynthetic antenna complexes are particularly at risk, since intense energy flux occurs through them to be stored in the form of energy of chemical bonds. Moreover, the oxidation rate of the pool of electron acceptors in the electron transport chain (ETC) is significantly lower than the rate of their reduction [12]. Therefore, at high intensity of light flux, photosynthetic cells are inevitably in a situation where light-harvesting antenna complexes are temporarily not needed. In this case, antenna complexes pose a threat to the photosynthetic apparatus and cell metabolism, since replacing damaged pigment–protein complexes requires a significant amount of energy and time. In addition to spatial removal of an antenna from a reaction center (so-called “state transitions”) [13-15], other fundamentally different photoprotection mechanisms can be realized. The latter allow a temporary reduction in the energy flux from antenna complexes to reaction centers of the photosystems.
The general principle of the functioning of photoprotective mechanisms in higher plants and cyanobacteria is the introduction of a special molecule into the structure of the antenna complex that can intercept the flux of excitation energy due to so-called nonphotochemical quenching1. This process reduces the probability of ROS formation, and this is the fundamental difference between photoprotection mechanisms and other antioxidant systems aimed at counteracting already formed ROS.
1 This term is used to describe any energy dissipation process that is opposite to the photochemical quenching that results in storing excitation energy in the form of separated charges or reduced equivalents. Nevertheless, among plant physiologists, nonphotochemical quenching is often and undeservedly identified only as chlorophyll fluorescence quenching by carotenoids of the xanthophyll cycle by thermal dissipation of energy. Even though cyanobacteria are preferred, relatively simple objects for studies of photosynthetic processes, their photoprotection mechanism, as well as a whole set of the features of its functioning that is largely based on protein–protein interactions, was discovered less than 15 years ago and retains many mysteries. It should be noted, however, that this area of research is developing extremely rapidly, so it is reasonable to expect new discoveries and updates in the very near future.
In this review, we consider modern concepts of the structural and functional organization of pigment–protein and protein–protein complexes involved in regulation of nonphotochemical quenching of cyanobacterial antenna complexes – the phycobilisomes. In cyanobacteria, like in higher plants and green algae, the role of the functional molecule – of excitation energy quencher – is performed by carotenoids, but structural features of cyanobacterial water-soluble antenna complexes have contributed to the emergence of a water-soluble protein system that determines functioning of the photoprotective mechanism.
STRUCTURE OF LIGHT-HARVESTING ANTENNA COMPLEXES IN
CYANOBACTERIA
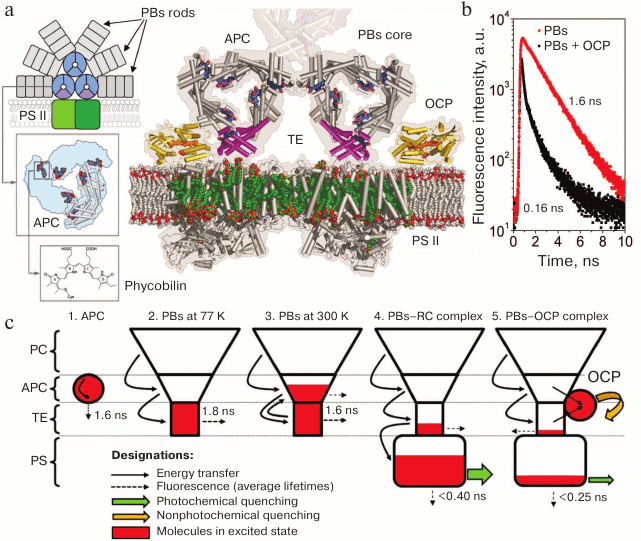
The photosynthetic apparatus of cyanobacteria, just as in many other photosynthetic organisms, is comprised of two interacting photosystems – PS I and PS II. Light is harvested by the core antennae of reaction centers (RCs), which contain chlorophyll [16]. In cyanobacteria, additional light harvesters are phycobilisomes (PBs) – multi-protein complexes located on the surface of the thylakoid membrane adjacent to PS I and PS II [17-19]. PBs consist of chromophore-containing phycobiliproteins and linker polypeptides devoid of chromophores [20-22]. Phycobiliprotein molecules usually contain two types of subunits – α and β – each being covalently bound to a phycobilin chromophore that is a linear tetrapyrrole [20]. Cyanobacterium Synechocystis sp., strain PCC6803 (hereinafter referred to as Synechocystis 6803), is characterized by PBs of a semi-disk shape, with the major phycobiliprotein components being C-phycocyanin and allophycocyanin [17-20, 23]. The PBs core, which is adjacent to the stromal surface of the thylakoid membrane, is in the center of a semi-disk and is made up of three cylinders, which consist of conjoined allophycocyanin trimers (Fig. 1). The coupling of PBs with the photosynthetic membrane is provided by the linker polypeptide LCM (CM meaning “core–membrane junction”) that consists of several domains and participates in the assembly of the PBs core [21, 24]. Six-sided cylinders diverge from the core in radial directions, and each consists of phycocyanin hexamer stacks (Fig. 1a). The light energy absorbed by phycobilins is transferred to the chlorophylls of PS I and PS II RCs, increasing photosynthesis efficiency by harvesting light in a spectral region where chlorophyll absorption is low [16, 18, 19, 25, 26].
Fig. 1. a) Schematic representation of a cyanobacterial photosynthetic membrane containing a dimer of PS II (shades of green) coupled with the phycobilisome (phycocyanin rods are shown in gray, and the components of the core cylinders are shown in blue and purple). Insets show the structure of one of the core cylinders and the associated chromophores. To the right of the scheme is a cross-section of the PBs on the level of terminal emitters (TE), PS II chlorophylls are shown in green, OCP is yellow and orange, and LCM is purple. The model is illustrative and is based on crystallographic structures of OCP, APC, and PS II (PDB 1M98, 1ALL, and 3KZI, respectively). b) The PBs fluorescence decay kinetics characteristic of in vivo (for mutants lacking PS I and II) and in vitro experiments, before and after activation of the OCP-dependent nonphotochemical quenching. Fluorescence lifetimes of the main components are given. c) Localization of excitation energy in the PBs–RC–OCP system according to the “energy funnel” model. 1) Isolated phycobiliproteins are characterized by mean lifetimes of the excited state of approximately 1.6 ns. 2-3) In a complex composed of several spectral forms of phycobiliproteins, transfer of excitation energy from short-wavelength to long-wavelength is observed. A small energy gap between the electron levels of APC and TE leads to the fact that at physiological temperatures reverse (uphill) energy transfer occurs. In PBs core, the number of TEs is an order of magnitude lower than that of APCs. At high temperatures, it is APC that makes the main contribution to PBs fluorescence, while at low temperatures the TEs fluoresce. 4) When a PBs is bound to a PS RC that performs charge separation (photochemical quenching), electron excitation energy of PBs is efficiently transferred to the chlorophylls of the photosystems. When the light flux increases, electron acceptors of the available RCs in photosystems are rapidly reduced and, accordingly, the efficiency of photochemical quenching decreases. At the same time, OCP activation and its binding to the core PBs site occur (5), resulting in the appearance of an additional channel for deactivation of the excitation.
As described above, phycobiliprotein molecules consist of α and β subunits in a 1 : 1 ratio, with molecular weights of approximately 16 and 18 kDa, respectively [27-29]. Phycobiliproteins are acidic proteins with isoelectric points in the range of 4.25-4.85 pH units. There are no disulfide bonds between the two subunits, and cysteine residues present in the α and β subunits participate in covalent binding of chromophore groups, phycobilins, via thioether bonds [30, 31]. Cysα-84 and Cysβ-84 residues are essential for chromophore attachment to the apoprotein [21]. Additional chromophore groups can be attached to an α or a β subunit around the 50th and the 150th amino acid residues. The number of chromophores in a (αβ)1 monomer determines the division of phycobiliproteins into classes, for example, an allophycocyanin (αβ)1 monomer contains two chromophores. Phycobilins are linear tetrapyrroles, whose chemical structure is based on four pyrrole rings connected by three single-carbon bridges (Fig. 1a).
Phycobiliproteins are characterized by a pronounced propensity for self-association [32]. Under nondenaturing conditions in solutions, it is impossible to obtain α and β phycobiliprotein subunits that are separated from each other since they form αβ heterodimers even at extremely low concentrations (~10–7 M), which are commonly (although less correctly) called (αβ)1 monomers. The surface of (αβ)1 monomers is hydrophobic, which ensures their stability only at low ionic strength [33]. Spontaneous oligomerization depends strongly on protein concentration, and (αβ)3 trimers and (αβ)6 hexamers of phycobiliproteins already predominate at 10–6 M concentrations in solutions.
According to X-ray diffraction data, the α and β polypeptide subunits contain two α-hairpins and eight α-helical stretches, six of which form the common globin structure of these proteins [17, 27, 34]. Chromophore groups are located in the pockets between the α helices. The remaining two α helices and two α-hairpins of each subunit form interacting domains through which the formation of (αβ)1 monomers occurs. In turn, monomers that have the shape of an arc form a hydrophobic contact between an α subunit of one monomer and a β polypeptide of the next one, closing into a ring-shaped trimer. All trimers, regardless of the phycobiliprotein type, have the shape of flat disks with a diameter of 11.5 nm and thickness of 3 nm with a triangular opening located in the center of the disk measuring 3 nm [31, 35] (see Fig. 1).
Allophycocyanins (APCs) take a special place among phycobiliproteins since their structure and photophysical properties provide effective capture of excitation energy from short-wavelength forms of phycobiliproteins – phycocyanin (PC) and phycoerythrin – and transfer excitation energy to the so-called terminal emitters (TEs) and chlorophylls of reaction centers (see Fig. 1). In several studies, the authors note the complex structure of the absorption spectra of APC trimers [28, 36], as well as the dependence of the shape of spectra on temperature [33]. The main maximum in the absorption spectrum (650 nm) is associated with the formation of trimers and the interaction of chromophores located in adjacent (αβ)1 monomers. When the temperature is increased up to 45°C, the interaction between chromophores is broken, while the contribution from a component with a maximum at 610 nm increases in the absorption spectra, corresponding to the absorption of monomers. APCs, like other phycobiliproteins, are water-soluble proteins, and the most important factor affecting the properties of APCs are complex interactions in the chromophore–protein–water system. Temperature changes is the most important factor affecting the energy of interaction in such systems [37-39].
Features of the structural organization of PBs and phycobiliproteins need to be considered when carrying out experiments. For example, at low temperatures the formation of ice crystals inside the trimer cavity may disrupt its structure causing effects comparable to monomerization, which is accompanied by a decrease in the quantum yield of fluorescence and distortions of the spectrum not related to changes in the efficiency of energy migration or to any other physiologically relevant processes [37-39]. Therefore, for proper interpretation and analysis of low-temperature PBs fluorescence spectra in vitro or in cyanobacterial cells, a proper experimental setup is required: optimized freezing rate, addition of a cryoprotectant, and utilization of time-resolved methods for estimating lifetimes and adequate normalization of fluorescence intensity. It should be noted that in vitro PBs complexes can dissociate into separate structural elements (rods and cores – see Fig. 1a), for example, in the presence of glycerol as a cryoprotectant [40]. Disruption of PBs integrity leads to an increase in PC quantum yield and, correspondingly, to a decrease in the contrast of changes in fluorescence intensity during the analysis of nonphotochemical quenching. Therefore, in some studies, researchers use PBs cores as a simplified and more stable model object.
DISCOVERY OF THE MECHANISM OF PHYCOBILISOME FLUORESCENCE
QUENCHING
The fully sequenced genome and the capacity for photoheterotrophic growth have made the unicellular cyanobacterium Synechocystis sp. PCC6803 a convenient object for studying protein–protein interactions and regulation of light harvesting and heat dissipation of energy in pigment–protein PBs complexes [32]. Since in vitro experiments with PBs are associated with several methodological limitations (see below), preparation of Synechocystis 6803 mutants lacking one or several components of the photosynthetic apparatus has enabled investigation of many functions and features of PBs in vivo. For example, interesting data were obtained using the CK and PAL mutants [25]. In PBs of the CK mutant, deletion of four genes, those of both the α and β phycocyanin subunits as well as two linker polypeptides, cause these proteins to be absent. In the PAL mutant, as a result of additional deletions in the genes encoding APC subunits, the phycobiliprotein antenna is missing completely, which is confirmed by data from immunological analysis and fluorescence spectroscopy [41]. Comparison of the rate of reduction of the primary quinone electron acceptor in PS II showed that the PBs core increases the effective absorption cross-section of PS II by 2.5-fold, and the presence of additional cylinders from PC (so-called rods) in PBs increases light-harvesting efficiency of PS II by an order of magnitude [16, 19, 25, 34].
The idea that cyanobacteria can thermally dissipate absorbed light energy, just as chloroplasts of higher plants do, has existed for a long time. In particular, when studying cyanobacterial adaptation to yellow light, in 1986 it was shown that under such conditions, the dissipation of about 35% of the energy absorbed by PBs was observed [42]. However, the exact mechanism and the spectrum of activation of this process were revealed only 15 years later.
When studying changes in the state of ETC (state transitions) and the effect of membrane fluidity on it, the process of nonradiative (thermal) energy dissipation was rediscovered. Under exposure to intense blue light, researchers observed a reversible thermal energy dissipation mechanism that was independent of temperature and protein synthesis or ubiquinone reduction inhibitors [43]. Therefore, this process was not due to disruption of the PS II components (the process was reversible even in the presence of translation inhibitors) or to changes in the ETC state (oxidation of ubiquinol pool did not affect the process). However, the nature and mechanism of this process remained completely unclear.
It should be noted that strong overlapping of fluorescence spectra of the chlorophyll of the photosystems and PBs (at room temperature), as well as the presence of intermediate processes associated with the change in quantum yield of PS II chlorophyll fluorescence, complicate estimations of the effects of various experimental factors on PBs fluorescence [25]. Therefore, the creation of mutants lacking photosystems was an important step in studying the regulation of thermal dissipation of energy in PBs. The use of a Synechocystis mutant lacking PS II allowed Rakhimberdieva et al. in the laboratory of N. V. Karapetyan at the Bach Institute of Biochemistry to carry out a detailed analysis of the action (activation) spectrum of thermal dissipation of energy [44, 45]. It was shown for the first time that intense blue-green light caused reversible quenching of 40% of the fluorescence of PBs, and the shape of the action spectrum corresponded to characteristic S0–S2 transitions in the absorption spectrum of carotenoids. These data indicated the existence of some unknown form of a carotenoid that interacted (in a controlled manner) with water-soluble PBs and, accordingly, this form should have also been water-soluble, but that did not correspond to the physicochemical properties of carotenoids.
Even though a water-soluble protein containing a carotenoid with unknown physiological activity had been discovered and characterized long before the experiments of Rakhimberdieva, it took several more years to determine the function of the orange carotenoid protein (OCP). This step was taken in a study by Diana Kirilovsky’s group [46], which showed that inactivation of gene slr1963, encoding the OCP protein in Synechocystis sp. PCC6803, results in a complete disappearance of PBs fluorescence quenching in cyanobacterial cells under the intense blue-green light. The results of these experiments were undoubtedly a breakthrough in the understanding of the regulation of photoprotective mechanisms in cyanobacterial cells.
In subsequent studies by Kirilovsky’s group, the presence or absence of PBs fluorescence quenching in vivo became the key criterion for evaluating the efficiency of protein–protein interactions between OCP and a region of PBs [47-49]. However, the complexity of the mechanism of OCP functioning ultimately resulted in the fact that the results of these experiments required additional in vitro studies (see the following sections of this review).
Even in early studies, it was noted that adaptation of cyanobacterial cells to high-intensity blue-green light causes not only a decrease in PBs fluorescence intensity but also a change in the shape of the fluorescence spectrum. After activation of the energy dissipation mechanism, fluorescence intensity in the 660-680 nm range becomes significantly lower, which was interpreted as the interaction of the quencher with a certain site located exactly in the PBs core [50]. This idea (more specifically, the impossibility and ineffectiveness of OCP interaction with phycocyanin rods) was supported by all research groups, which narrowed the search for the protein–protein interaction site of OCP and PBs to the PBs core consisting of APC and terminal emitters.
The decrease in PC fluorescence intensity is related to the highly efficient transfer of excitation energy from short-wavelength (PCs) to long-wavelength protein forms (TEs). To analyze these processes, studies were carried out using high-resolution time-resolved spectrofluorimetry and theoretical calculations [51-56]. It was found that activation of OCP-dependent quenching in the absence of photosystems in vivo, as well as in in vitro experiments, results in a synchronous and comparable decrease in PBs fluorescence intensity and mean lifetimes by ~90%. This indicates that OCP-dependent fluorescence quenching is due to capture of PBs excitation energy by a quencher molecule (a carotenoid). The lifetime of the carotenoid excited state (S2) in organic media or in OCP does not exceed 10 ps [52, 57-59], which is two orders of magnitude less than the characteristic lifetime of the excited state of phycobiliproteins (~1.6 ns). Thus, the transfer of excitation energy from PBs chromophores to OCP results in the disappearance of the excited state due to thermal dissipation, and the energy transfer rate determines fluorescence quenching efficiency. It is important to note that, according to published data [57, 59], both the initial and the photoactivated forms of OCP have practically the same excited state lifetimes; therefore, even the physiologically inactive OCPO form is an excellent potential excitation quencher. This emphasizes the role of protein–protein interactions between OCP and PBs as a prerequisite for the activation of photoprotection mechanisms, and the trigger of these interactions is, in turn, a change in protein–chromophore interactions in the OCP molecule (see also below).
In one study [54], it was shown that the transition between states (quenched and non-quenched PBs) is characterized by a smooth change in lifetimes and contributions of components of PBs fluorescence decay kinetics. This indicates a gradual change in the distance between the donor and acceptor of excitation energy, and not a change in the fraction of quenched PBs due to instantaneous formation/dissociation of the OCP–PBs complex. This fact was interpreted as manifestation of the intermolecular interaction between OCP and its binding site on the phycobilisome, which stimulated further studies of conformational changes in OCP during its photoactivation in vitro.
As noted above, the energy interaction between OCP and PBs may be considered within the framework of Förster theory. However, calculation of the distance between the energy donor and acceptor based on changes in the quantum yield of the donor is a non-trivial task, since the S0–S1 transition is forbidden by symmetry rules; therefore, it is impossible to identify the position of S1 in the OCP absorption spectra. Studies of OCP by femtosecond absorption spectroscopy revealed an S1 level in the 714-nm range (1.74 eV) [60]. At the same time, the S1 level of terminal emitters corresponds to the energy of ~1.81 eV (685 nm), which is much lower than the S2 level of a carotenoid in the OCPR form (510 nm, 2.43 eV). Thus, the fluorescence spectrum of PBs overlaps with S0–S1 absorption much better than with S0–S2 of OCP. Based on experimental data on the efficiency of PBs fluorescence quenching during their interaction with OCP [54, 56] and of the available models of OCP–PBs complexes [61-64], the inverse problem can be solved and the value of the overlap integral of the OCP absorption spectrum and PBs core fluorescence can be estimated. The calculations demonstrate that the efficiencies of energy migration observed in experiments cannot be achieved if the transfer is carried out only to the S2 level of a carotenoid. This is due to the small value of the overlap integral of the PBs core fluorescence spectrum and the absorption spectrum of the carotenoid S0–S2. A consequence of this is a low rate of energy migration, which would not allow OCP to compete with RCs of the photosystems for the energy flux. Since the observed rates of energy migration from PBs to OCP are much higher, it can be assumed that other electron levels of OCP are also involved in the energy interaction. Quantum mechanical calculations can be used to estimate theoretically the contribution of the energy interaction between the excited state of PBs and the electron levels of OCP, and to calculate the corresponding rate constants of energy migration [55]. However, when constructing the model, it is necessary to rely on experimental data.
When interpreting kinetic data, strong overlapping of the absorption and fluorescence spectra of APC and terminal emitters should be taken into account, which at room temperature leads to the uphill transfer of excitation energy from terminal emitters to APC [53, 54]. It should be noted that evaluation of the effect of OCP-dependent quenching of PBs fluorescence on energy transfer processes within PBs using spectrofluorimetry with picosecond time resolution and subsequent global data analysis yields little information, since a decrease in the lifetime of the fluorescence of terminal emitters, which are considered to be acceptors of excitation energy in the model, affects the shape of the front of fluorescence increase and, accordingly, the apparent energy transfer rate constants [56], which makes the comparison inaccurate. Formation of a complex between OCP and PBs might not affect the rate of energy migration between different spectral forms of PC, at the intensities of excitation light fluxes ruling out simultaneous excitation of several pigments. The effects observed under normal conditions (reduction in fluorescence lifetimes for all forms) indicate effective discharge of the excited state of chromophores in the PBs core located closest to the site of contact between OCP and PBs and the carotenoid; thus, the reduction in fluorescence lifetimes of chromophores that are far from the core is due to a decrease in the probability of reverse energy transfer and to the contribution of fluorescence of long-wavelength forms.
Unfortunately, the complexity of measuring and interpreting data on fluorescence lifetimes (as in the case of many other objects) led to the fact that data on the presence or absence of PBs quenching in vivo for several interesting OCP mutants were obtained using steady-state spectroscopic methods [47, 49, 65]. Therefore, the question of the ability of various spectral forms of OCP mutants to quench fluorescence still remains open and should be carefully dissected in vitro. However, it appears that the site of the OCP interaction with PBs is located in the N domain of the OCP, since in the absence of the C domain, the N-terminal OCP fragment, called RCP (Red Carotenoid Protein) [66], is capable of effective PBs fluorescence quenching [67]. Therefore, it is believed that after its activation, OCP interacts with the PBs core [68] through the N-terminal domain [67, 69] (see below). This is also in line with the data indicating that the amino acid residue R155 of the N-terminal domain, which forms a salt bridge with E244 of the C-terminal domain, is required for the interaction with PBs. The R155L or R155E substitutions prevented PBs fluorescence quenching during OCP photoactivation, while the E244L substitution did not [69]. Intriguingly, a similar replacement, R155K, did not prevent PBs fluorescence quenching but significantly destabilized the interaction of OCPR with PBs, leading to the rapid recovery of PBs fluorescence after termination of photoactivation [69]. However, it should be noted that the R155 residue is not fully conserved in OCP and may be replaced, for example, by a glutamine residue, which may explain the less stable connection between the domains in such OCP variants and the higher rate of their photoactivation [70].
Despite the importance of the described discoveries, many aspects of the mechanism of PBs fluorescence quenching under the influence of OCP are still not understood and require further study both at the level of the entire system and at the level of its individual components.
PARTICIPANTS OF NONPHOTOCHEMICAL QUENCHING IN
CYANOBACTERIA
OCP – a sensor and effector of nonphotochemical quenching of phycobilisome fluorescence. The very fact of OCP participation in PBs photoprotection has been clearly shown and there are no doubts regarding it. However, the specific mechanism of OCP photoactivation and its protein–protein interaction with the PBs is still a subject of scientific debate. Surprisingly, the fact that OCP is a photoactive protein and can undergo a transition from the orange (OCPO) to the red (OCPR) form under the action of light was found only after the establishment of its structure, function, and many physicochemical properties [47, 71], i.e. 27 years after its discovery and description in 1981 [66]. Although the existence of various spectral forms of OCP became known during its first isolation from cells, the effect of light on the spectral characteristics of OCP was not examined until 2008, for unknown reasons.
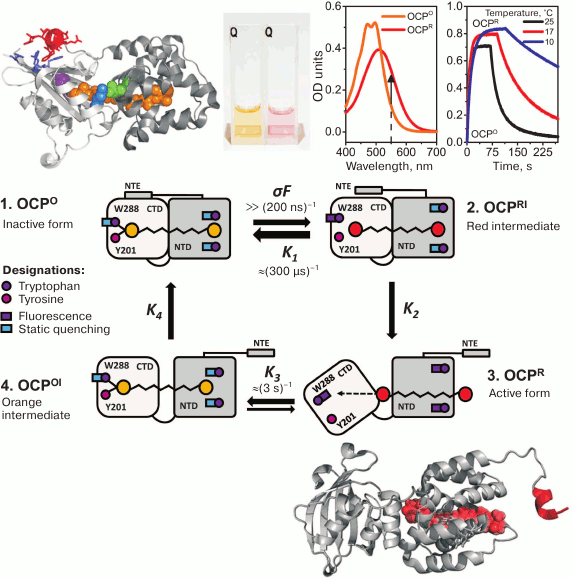
Undoubtedly, determination of the atomic structure of OCP by X-ray diffraction [72, 73] was one of the most important events in the history of studies of photoprotective mechanisms. It turned out that OCP is a water-soluble two-domain protein (35 kDa), in the inner cavity of which a molecule of an asymmetric keto-carotenoid (3-hydroxyechinenone) is located. The structure of the N domain of this protein is unique for OCP, whereas the C domain belongs to the superfamily of the so-called NTF2-like proteins (nuclear transport factor 2). The two domains are connected by an extended linker, have an extensive interdomain interface, and are additionally stabilized by the interaction between a short N-terminal α-helix (NTE) with the C domain (Fig. 2).
Fig. 2. Photocyclic transformations of OCP. On the top, from left to right – the crystal structure of the orange form of OCP (PDB 3MG1): the carotenoid is shown in orange, NTE is red, the W288 residue is purple, residues 155 and 244 forming the interdomain salt bridge are green and blue, respectively; a photo of cuvettes with OCP solutions before and after photoactivation; OCP absorption spectra before and after photoactivation, the arrow shows the direction of changes in optical density at 550 nm; the kinetics of OCP absorption changes during adaptation to high-intensity blue light and after switching off the light at different temperatures. In the center: a schematic representation of the four main stages of the OCP photocycle (see explanation in the text). On the bottom is a model of the active red form of OCP obtained based on RCP and OCP structures (PDB 4XB4 and 3MG1, respectively) and on a low-resolution OCPR structure obtained based on small-angle X-ray scattering data [77].
Despite the abundance of experimental data obtained so far, the complete sequence of conformational changes during OCP photoactivation is unknown. Usually, when discussing the physiological role of OCP, two stages of the photocycle are distinguished: (i) a stable orange form, OCPO, which does not quench PBs fluorescence, and (ii) a red activated (or signaling) form, OCPR, the interaction with which results in quenching of PBs fluorescence [47, 74, 75]. Since these two forms differ in the absorption spectra and visually by the colors of their samples (Fig. 2), in the literature the red color of OCP samples started to be associated with the active state, and vice versa. However, this assumption is not always correct, since the color (absorption) of an OCP sample depends on the conjugation length of the π-system in the polyene chain of the carotenoid. The spatial arrangement of conjugated double bonds is determined by protein–chromophore interactions, whereas the possibility of interaction with PBs and fluorescence quenching is dictated by protein–protein interactions between the PBs core and OCP. In other words, both red forms of OCP that do not quench PBs fluorescence (possibly due to an impaired interaction with PBs) and orange forms capable of quenching can exist [49, 65, 69]. As noted above, spectral characteristics of a carotenoid do not have much effect on its lifetime in an excited state and, accordingly, its capability for non-radiative dissipation of excitation energy. To induce PBs fluorescence quenching, proximity of OCP and its binding site in the PBs core is required, i.e. formation of a complex made possible due to specific protein–protein interactions. All existing models of the OCP–PBs complex show that the minimum distance between chromophores exceeds 15Å, which excludes the possibility of energy transfer by the Dexter mechanism and makes migration of excitation energy via dipole–dipole interactions ineffective [76].
In the literature, OCP mutants are described that lack the ability to photoconvert, but nevertheless several such mutants (Y44S, E34A, P126V/Y129F) exhibit the ability to quench PBs fluorescence [49, 65, 69]. On the other hand, absorption spectroscopy with nanosecond time resolution revealed that the red form appears upon excitation of OCP by a short (7 ns) laser pulse in the S0–S2 region during less than 100 ns [78], which is several orders of magnitude faster than any possible conformational changes in the protein structure [79-82]. Therefore, this intermediate of the photocycle spectrally corresponds to the red (and active) form of OCP, but is analogous to the orange (inactive) form in terms of its protein matrix structure. The relaxation kinetics of the red form (in the time range from 100 µs to 10 s) is approximated by a sum of two components with characteristic times of ~200 µs and ~3 s, with comparable activation barriers, which indicates the presence of two spectrally identical red forms of OCP, the transition of which into the orange form requires similar changes in protein–chromophore interactions, but the number of attempts required for the transition differs by ~10,000-fold [78]. This hierarchy of times is determined by two requirements for a photoactive construction: (i) the need to maintain the stability of the inactive form of OCP sufficient to ensure that the protein does not undergo transition into the active form at normal (for photosynthesis) insolation levels, and (ii) the lifetime of the active form should be sufficient to find and connect to the site within the PBs core before OCP spontaneously transforms into an inactive form. According to certain data, the quantum yield of the formation of the active form does not exceed ~0.2% [51, 71, 78]. Apparently, the stability of the orange form and the lifetime of the active red form are important for maintaining the correct balance between the photosynthetic and photoprotective processes and, therefore, for the energetic state of a cell.
Several methods have shown convincingly that OCP photoactivation is accompanied by decompaction of the protein molecule [77, 83, 84] and displacement of the carotenoid molecule into the N domain [85, 86] (Fig. 2). Such structural changes might be necessary to stabilize the active signal state of OCP, in which it is able to bind PBs and cause fluorescence quenching. According to available information from different laboratories, the sequence of events resulting in the formation of the active form of OCP can be described as follows.
When a quantum of light is absorbed by a carotenoid molecule, it transits to an excited state. The energy of absorbed quantum is sufficient for photoisomerization (rotation of the β-ionone ring in the C domain of OCP) and straightening of the polyene chain [71, 87, 88]. Both these events, in particular, the rotation of the ring group from the cis to trans position, result in an increase in the conjugation length and a change in the absorption spectrum of the carotenoid, i.e. in the formation of a red spectral form. Upon rotation of the β-ionone ring in the C domain, hydrogen bonds between the keto group in the ring and tyrosine-201 and tryptophan-288 residues are disrupted [71, 88]. These residues are highly conserved, and their replacement by alanines results in loss of OCP photoactivity, and therefore the presence of these hydrogen bonds is required for stabilization of the orange form of OCP [83, 84]. Disruption of hydrogen bonds in the C domain leads to a change in the hydrophobicity of the local environment of W288, which is located in the inner cavity of the C domain and is somehow conjugated to the hydrophobic residues that form the interface of the interaction of the C domain with the α-helical NTE, which stabilizes the interaction of the N and C domains. Therefore, changes in the environment of W288 appear to transmit to the outside of the C domain where NTE is bound [89, 90] (see Fig. 2 and the following sections). This leads to a disruption of the contact of NTE with the C domain and, probably, even to partial unfolding of NTE [91, 92]. Comparison of the OCP and RCP structures shows that the major differences in these proteins, corresponding to the stable orange form and to the N domain of the red active form of OCP, respectively, shows that the largest structural changes are in the region between NTE and nearby α-helices [77, 85, 88, 93]. Thus, disruption of the structure and mobility of the NTE segment, which is not attached to the C domain, might transfer a signal to the N domain of OCP. A change in the distance between α-helices in the N domain might result in weakening of stacking interactions between the conserved residues of tryptophan-101 and -41 and the ring of the carotenoid molecule. If in the orange form of OCP W41 blocks movements of the carotenoid, then due to the change in the conformation of the N domain and the rotation of the α-helix containing W41, a certain kind of a tunnel is formed, along which the carotenoid moves by more than 10 Å into the N domain [85]. These changes may destabilize the structure of the protein, which cannot remain compact due to the lack of several hydrogen bonds in the interface between its domains, and the disruption of protein–chromophore interactions results in an increase in the mutual mobility of the N and C domains and their separation. It can be assumed that this OCPR form is physiologically active, since in this state the surface of the N domain and the embedded carotenoid are the most accessible for interaction with PBs, and the C domain is available for interaction with the fluorescence recovery protein, or FRP (see below).
The reverse transition from the active form of OCP to the inactive orange form requires restoration of protein–chromophore interactions. Simultaneous observations of the changes in carotenoid absorption and of fluorescence decay kinetics of the tryptophan residues in OCP show that the transition to the inactive form is also characterized by asynchronous changes in the states of the carotenoid and protein matrix [78]. The first event on the way to the dark inactive form might be the restoration of stacking interactions between the β-ring of the carotenoid molecule in the N domain and the tryptophan-41 and -101 residues. This event is accompanied by the restoration of static fluorescence quenching of these tryptophan residues that is observed experimentally [78]. Then a change in the absorption of the carotenoid occurs, which requires the formation of hydrogen bonds in the C domain between the carotenoid and the residues of tryptophan-288 and tyrosine-201. However, the process of reassociation of the N and C domains does not end here. The final stages of the transition to the inactive form are probably related to protein compaction and are accompanied by a change in the efficiency of excitation energy transfer between W101 and the carotenoid [78]. The large number of steps necessary to restore the inactive form of OCP, as well as the spatial distance between the N and C domains, result in the fact that this process is extremely slow, and its rate strongly depends on temperature. It is the factor of mutual domain arrangement that determines the probability of transition from the red form to the orange form. For example, the transition rate can be increased nonspecifically by orders of magnitude during stabilization of the OCP structure – due to a high concentration of kosmotropic ions, and specifically – under the action of FRP [78, 84, 90, 94-96].
Nevertheless, the OCPR–PBs complex demonstrated significant stability in an in vitro system [68], and spontaneous PBs fluorescence recovery under these conditions is extremely slow (~104 s). It should be noted that non-radiative energy transfer to the OCP’s carotenoid is a source of its excitation, which stimulates transition from the orange to the active red form [86]. Accordingly, the transfer of excitation energy from PBs contributes to the maintenance of a stable complex, since during the energy transfer the red form is replenished. This might adversely affect the efficiency of photosynthetic processes; therefore, to accelerate the recovery of the initial PBs parameters and the efficiency of light harvesting, another regulatory component is required that would ensure destabilization of the OCPR–PBs complex.
It should be clarified that special variants of OCP have been found very recently that are characterized by faster photoactivation and spontaneous relaxation, as well as by a lack of regulation by FRP (the so-called OCP2 variants, whose representatives may be present in various cyanobacteria either independently or together with the better-studied OCP1 variants) [70], but in this review, we only discuss the Synechocystis variant (OCP1) in more detail.
FRP – phycobilisome fluorescence recovery protein. In 2010, a new factor was found that determines PBs fluorescence recovery when insolation decreases [94]. During the study of Synechocystis 6803, the gene slr1964 was found that is in the close vicinity of the OCP gene (slr1963) and is responsible for this function – upon inactivation of slr1964, PBs fluorescence recovery was extremely long. The product of the slr1964 gene was termed fluorescence recovery protein (FRP). It turned out that FRP also directly affects OCP and accelerates its transition from the photoactivated red to the inactive orange form [94], which will be discussed in further detail in the following sections. It is noteworthy that some time ago there was a question about the length of the open reading frame for this gene. In FRP genes from Synechocystis and Microcystis species, an additional N-terminal sequence was found by bioinformatic methods, which is absent in FRP from other cyanobacterial species [94]. This elongated gene was expressed, and its product was characterized, leading to a few artifactual conclusions. In particular, it was postulated that FRP is strongly associated with membranes. Later, it was experimentally demonstrated that FRP from Synechocystis starts with the Met26 residue (according to the numbering of the extended reading frame product), i.e., the short form of the protein is expressed and functions. That particular form is present in other cyanobacteria, and it is a water-soluble protein [95]. Overexpression of this FRP form in Synechocystis (109 amino acid residues) resulted in cells not being able to induce the photoprotection mechanism [95], indicating the deactivating effect of FRP in relation to OCP and the need for a careful control of the expression level of both proteins for maintaining the photoprotection mechanism. Indeed, studies of these reactions in vitro and determination of corresponding rate constants were used to develop a model for nonphotochemical quenching of PBs fluorescence, which shows that FRP concentration in the cell is lower than that of OCP [97].
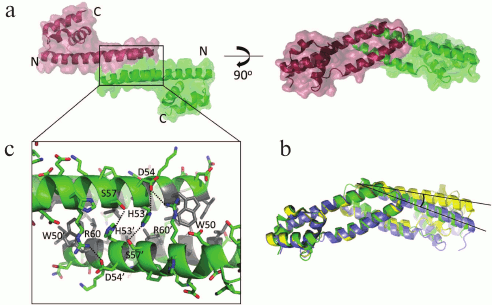
The 2.5-Å resolution structure of FRP from Synechocystis was determined in 2013 [96]. FRP was found to be an exclusively α-helical protein of an elongated shape (Fig. 3a), but in a crystal, two significantly differing conformations were present – dimeric and tetrameric, with subunits within a tetramer being arranged dissimilar to those in the dimeric conformation. It was suggested that the active form of FRP is a dimer, and tetramers are an inactive, storage form. Detection of FRP dimers and tetramers contradicted previously published data by the same authors that FRP is a trimer [94]. More recent biochemical studies have convincingly demonstrated that, at least in vitro, FRP is present as stable dimers [84, 98] with little propensity for spontaneous monomerization upon dilution, with tetrameric particles being detected only at extremely high protein concentrations [84, 98]. It can be speculated that the presence of the N-terminal His-tag, despite preserving full protein functionality [84], makes FRP dimers somewhat more stable and inhibits tetramer formation, but FRP crystals from Synechocystis were also obtained when using a protein carrying the N-terminal His-tag (although it is not visible in the crystal structure) [94]. Moreover, very recently a new crystal structure of FRP from Tolypothrix sp. PCC 7601 (=Fremyella diplosiphon) was obtained (PDB 5TZ0 [70]), which was found to be completely equivalent to the dimeric FRP structure from Synechocystis (PDB 4JDX, chains B and D), and the tetrameric organization was not detected in this case either. Even though FRP from Synechocystis and Tolypothrix are identical only to 51%, the structures of their dimers are practically equivalent, only the angle between monomers differs slightly (Fig. 3b). The same observation can be made from the careful alignment of FRP dimers built up of different chains even within the same crystal structure (PDB 4JDX, AC and BD chains): in this case, the angle formed by FRP monomers within the dimer differs even more. This serves as an important indication that the contact of subunits in a FRP dimer is not rigid, but rather allows some conformational flexibility and sliding of the monomers relative to each other.
Fig. 3. Structural organization of an FRP dimer according to X-ray diffraction data. a) The general outline of an FRP dimer from Synechocystis (PDB 4JDX) in two orthogonal projections. Monomers are shown in different colors. b) Superposition of structures of FRP dimers from Tolypothrix (blue, PDB 5TZ0) and Synechocystis (dimers formed by different chains present in the crystal structure are shown in different colors). The angle illustrating the difference in the position of the second monomer relative to the first is marked. c) FRP dimerization area. The key polar contacts are marked by dotted lines, and the residues forming the hydrophobic core are shown in gray.
The interface of Synechocystis FRP dimers is constructed by an antiparallel contact of α-helices α1 and α2; it is approximately 900-1000 Å2 (~10% of the total dimer surface) and is stabilized by a pronounced hydrophobic core (residues L29, L32, L33, V36, A40, I43, I46, L49, W50, L52, L56) and also by the conserved polar contacts D54–R60, H53–S57 and the cation–π interaction R60–W50 on the concave side of the dimer, and, due to it being antiparallel, each of the contacts is present twice in a dimer (Fig. 3c). The same principle of FRP dimerization is preserved in Tolypothrix. It was assumed that the dimerization region is directly involved in the interaction between FRP and OCP, which is consistent with the conservativity of several residues within this region and with the influence of the corresponding mutations on the ability of FRP to accelerate relaxation of OCP [96]. This assumption is not however the only possible one, since it can also be suggested that the studied mutations affect the stability of the FRP interface, and only for this reason they affect FRP functioning. This possibility requires more detailed analysis, especially considering the recently demonstrated ability of FRP to monomerize during binding to OCP [84, 90, 99] (see below). The most enigmatic are the data obtained using the R60K mutant, since this apparently synonymous substitution did not affect FRP folding (the structures of the wild-type FRP (PDB 4JDX) and of the R60K mutant (PDB 4JDQ) are virtually the same), but it made FRP almost incapable of accelerating OCP relaxation [96].
Importantly, a reversible photoprotection system consisting of PBs (the object of quenching), OCP (the inducer of quenching), and FRP (the inhibitor, or the off switch, of quenching) was reconstructed in vitro [68]. For instance, despite certain important assumptions and differences in the processes occurring in vivo and in vitro, it was convincingly demonstrated that the presence of these three components is necessary and sufficient for the operation of the minimum regulated PBs quenching mechanism. OCP2 variants, that were discovered very recently, can quickly and spontaneously relax without the participation of FRP (whose genes may be absent in some OCP2-containing cyanobacteria), and it can be assumed that they are more primitive forms of OCP compared to OCP1 [70], but the more systematic study of this subject is needed.
KEY PROTEIN–PROTEIN INTERACTIONS
Interaction of OCP with the components of phycobilisomes. As mentioned earlier, photoactivation of OCP ultimately leads to dramatic changes in its conformation – separation of protein domains and a significant shift of the carotenoid deep into the N domain. It is believed that such changes in OCP, also caused by some mutations (W288A and Y201A/W288A) or chaotropic agents [83, 84, 100], determine the ability of OCP to interact with proteins within PBs and dissipate excess energy, which is experimentally observed as PBs fluorescence quenching. It is important that binding of the activated form of OCP to PBs does not require illumination, and the intensity of blue-green light determines only the amount of the active form obtained [84, 85, 97], while the original, non-photoactivated orange OCP is unable to bind PBs and quench their fluorescence. Considering complex structure of PBs and the lack of direct structural data on PBs–OCP complexes, the exact site of interaction is unknown, and numerous accumulated indirect data are quite contradictory. The situation is complicated by the enormous difference in the sizes of interacting OCP (~35 kDa) and PBs (several MDa [29, 101]), and the need to use sufficiently high concentrations of phosphate to maintain PBs integrity in vitro [102], and therefore the set of promising approaches to address the problem of the mechanism of the OCP–PBs interaction is very limited. It is not entirely clear how PBs integrity is maintained in vivo.
It was shown that the physical interaction of the photoactivated red form of OCP with PBs is quite strong, since their complex is coprecipitated in a sucrose gradient [68]. As mentioned earlier, OCP is practically incapable of quenching the fluorescence of PCs within PBs rods (Fig. 1), and, conversely, it can quench fluorescence of mutant PBs that lack rods [46, 68]. For this reason, the site of OCP interaction is apparently far from the rods and is located in the PBs core. The true picture might be even more complicated, since mutant PBs which lack rods bound OCP significantly weaker and spontaneously restored their fluorescence in comparison to intact PBs [68]. The results of mass spectrometry studies using specific bifunctional agents that covalently modify lysine residues located at a certain distance from each other confirm a hypothesis about the formation of protein–protein complexes of OCP with the components of the PBs core [92].
The main subject to fluorescence quenching, according to a model elaborated by Zhang et al., was core APC660 trimers, and it was suggested that the OCP binding site should be located between polypeptides ApcB and ApcE [64]. APC660 quenching was proposed in studies by Kirilovsky et al. [68, 89]. In other works [103-105], no fluorescence quenching was detected for APC trimers from PBs, in contrast to fluorescence of the large linker polypeptide ApcE, or LCM (see above), which, like APC, carries a phycocyanobilin chromophore and is one of the major TEs [103-108]. The hypothesis regarding the role of LCM is indirectly confirmed by information obtained using mutants of other core components – genes apcD and apcF – and it supports the view that the OCP-binding site is located near the LCM protein or even on its surface [109], which was also supported by molecular docking in silico [61, 105, 110]. Unfortunately, it was not possible to directly test this assumption by creating a mutant completely lacking the apcE gene, since the product of this gene, LCM, plays an important structural role and is required for PBs assembly and integrity [108]. Nevertheless, removal of the LCM domain containing the chromophore (residues 13-246) resulted in the loss of OCP-dependent PBs fluorescence quenching, which is an important argument in favor of the hypothesis regarding the role of LCM in OCP binding [111]. Other data obtained from the LCM protein isolated from PBs under rather rigorous conditions [109, 112] should be interpreted with caution, since preservation of the completely intact structure of the components of multiprotein complexes during their isolation raises some doubts. On the other hand, attempts to obtain individual PBs components, for example, the LCM protein, in a heterologous expression system in Escherichia coli strains producing phycocyanobilins [107, 113-115] to investigate interactions with OCP appear promising and feasible.
According to another hypothesis, interaction with OCP occurs in the PBs core, near another, low molecular mass linker protein, Lc (with C meaning a region within the core), which is a product of the apcC gene and lacks chromophores. This hypothesis is based on the similarity of the spatial structure of Lc and the C domain of OCP and implies that Lc is displaced/bent away under the action of OCP when it binds to the PBs, and Lc participates in the stabilization of the binding of OCP [47]. PBs variants with a mutation in the Lc protein (ΔApcC) demonstrated reduced OCP-dependent fluorescence quenching compared to control PBs with an intact apcC gene, but increasing phosphate concentration significantly leveled the difference between the mutant and control PBs [63]. In our opinion, unambiguous interpretation of these results is difficult, since the removal of an important component of PBs may have a mixed and poorly predictable effect on its features.
The concept of the OCP region that is responsible for the interaction with PBs has changed over time. According to one of the earlier hypotheses, the C domain of OCP was considered responsible for this interaction [47, 94]. The interaction of OCP through the N domain was also postulated [64], and this hypothesis is supported by the role of the R155 residue in interaction with PBs [69] and the fact that isolated RCP, which is a fragment of OCP corresponding to its proteolyzed N domain [116], is an effective PBs quencher [67]. Very recently, using chemical cross-linking by glutaraldehyde of PBs–OCP complexes followed by mass spectrometry of their proteolysis products, it was found that not the domains, but the interdomain OCP linker provides the greatest number of contacts with PBs core proteins. This is somewhat consistent with the data of Zhang et al. [64], with the deepest domain within PBs being the N domain according to this model, and the C domain of OCP being more accessible [63]. Unfortunately, even this model is based on numerous assumptions and leaves a certain degree of freedom for interpretation. Moreover, the complete absence of cross-linked peptides belonging to the N domain of OCP and to the protein components of the core in this study apparently contradicts the widely known fact that RCP can quench PBs even without containing the C domain or the linker region. It also remains unclear why, given the postulated accessibility of the C-terminal OCP domain in its complex with PBs, Gwizdala et al. could not achieve copurification of PBs and OCP on a metal-affinity column when OCP was immobilized by the C-terminal polyhistidine tag [68].
Despite certain success in the analysis of the interaction between OCP and PBs in silico, achieved by the methods of molecular dynamics and modeling [61, 63, 64, 105], the exact mechanism of this interaction remains far from being understood and requires direct experimental data from structural methods, for example, from X-ray crystallography or cryoelectron microscopy. With the identification and characterization of stable mutant OCP forms mimicking its activated state (for example, OCP-W288A [84]), this task appears feasible.
However, even in the case of intact OCP, photoactivation results in an extremely strong interaction of OCP with PBs in vitro, especially at high phosphate concentrations, and such binding significantly stabilizes the red form of OCP [68, 78]. Phosphate may significantly stabilize the complexes, and the interaction is probably somewhat weaker in vivo than in vitro, however, it seems that in vivo a factor (or factors) is required that, upon a decrease in illumination intensity, would make the interaction of OCP with PBs reversible and would effectively restore PBs fluorescence and energy flux to the photosystems. As noted, one such factor that substantially accelerates PBs fluorescence recovery in vitro and in vivo is FRP [94].
Interaction of OCP with FRP. Incubation of OCP–PBs complexes in the presence of FRP caused a significant decrease in OCP content in the PBs fraction obtained by centrifugation in a sucrose gradient, which correlates with the recovery of PBs fluorescence that was quenched upon the addition of OCP [68]. Therefore, it was suggested that FRP promotes dissociation of OCP from PBs and thereby creates the prerequisites for its relaxation from the photoactivated red to the orange form. The question how FRP contributes to PBs fluorescence recovery is extremely complex and virtually comes down to clarifying whether competition occurs between FRP and OCP for binding to PBs, or whether FRP binds to available sites on OCP bound to PBs, actively ejecting it from PBs.
A mechanism based on simple competition between FRP and OCP for a certain site on PBs dictates a higher FRP affinity to PBs compared to that of OCPR. Otherwise, a significant concentration of FRP would be required to displace OCP, whereas it is known that FRP is effective even at low FRP/OCP molar ratios, but without further studies of the interactions between FRP and PBs this remains only a matter of discussion. The possibility that there are still undiscovered factors involved in photoprotection regulation in addition to OCP and FRP cannot be ruled out completely. The following scenario is hypothetically possible: FRP interacts only with free OCP, since the rate of PBs fluorescence recovery is much lower than the conversion rate of the red form to the orange form [46], indicating that the rate-limiting stage is precisely the stage of OCP detachment from PBs. This point of view is indirectly supported by the fact that FRP is not required for OCP2 relaxation and spontaneous recovery of PBs fluorescence with a decrease of insolation levels [70]. However, more recent data have been obtained showing that FRP possesses two independent activities, and the mechanisms of FRP interactions with free OCP and PBs-bound OCP may be different. This hypothesis is supported by mutations in both OCP (primarily, of F299 and D220 residues) and FRP (mutation R60L) that greatly reduced the ability of FRP to accelerate the transition from the red to orange form, and have practically no effect on the ability of FRP to detach OCP from PBs [117].
In contrast to complexes of FRP with the PBs-bound OCP, which are still completely uncharacterized, the interaction of FRP with free OCP has been studied in more detail. The genes encoding OCP and FRP are often located close to each other, but they are under the control of different promoters [94], which probably indicates that different, independently changing concentrations of these proteins take place under varying illumination, which is necessary for adjusting the photoprotection mechanism. Initially, it was assumed that FRP can only interact with the photoactivated red form of OCP and that this interaction occurs through the N domain of OCP, with FRP binding destabilizing the red form of OCP [94]. Recent biochemical studies have shown that FRP can also interact with the orange form of OCP [84, 98, 118], but with much less affinity compared to the photoactivated form of OCP, and the apparent dissociation constants are approximately 35 and 2.3 µM, respectively [84]. Evaluation of the parameters of the FRP interaction was possible using the purple mutant form, OCP-W288A, which is a structural and functional analog of the red form of OCP and is also characterized by a larger hydrodynamic radius and, apparently, by separated N and C domains that are distant from each other [83, 84]. Using a combination of biochemical methods, it was found that, during the interaction of OCP and FRP, stable complexes with an apparent 1 : 1 stoichiometry are obtained, meaning that, under the investigated conditions, FRP dimers dissociated during their interaction with OCP [84]. In this case, the affinity and stoichiometry of the FRP interaction with OCP-W288A and the OCP apo form (which lacks the carotenoid molecule that normally stabilizes the compact protein conformation) practically coincided [99]. Considering these data, as well as the similarity in the hydrodynamic behavior of OCP-W288A and the OCP apo form (i.e., increased Stokes radius and pronounced dimerization propensity), it was suggested that FRP can recognize and specifically bind such forms of OCP in which the N- and C-terminal domains are separated from each other, regardless of the presence of a carotenoid molecule. Moreover, an increase in compactness of OCP and FRP complexes was systematically observed, suggesting an adapter or chaperone-like function of FRP towards OCP, so that FRP may bring together the separated OCP domains, facilitating its reverse conversion from the red to the orange form [84, 99]. It can be suggested that, in addition to its determined important role in the photoprotective mechanism, FRP performs a certain role in the process of OCP folding and maturation. In a recent study, Blankenship et al. confirmed the assumption that FRP can act as a bridge that brings together the OCP domains during its functioning [118]. It was also postulated that significant conformational rearrangements of FRP occur during its functioning, which include monomerization and partial unfolding of the C-terminal region of the protein (see Fig. 3a), which is presumably a prerequisite for the transition of OCP from the red to orange form [118].
Until recently, the question of how the area of contact between FRP and OCP is arranged, and what areas are involved in the interaction of these proteins, remained unclear. First, it was necessary to determine which OCP domain contains the FRP binding site(s), and this question was answered by using the “divide and conquer” approach. It turned out that in vitro FRP practically does not interact neither with RCP (the equivalent of the OCP N domain), nor with its apo form, which were obtained in a bacterial expression system, but it binds with a high affinity to a polypeptide corresponding to the OCP C domain, forming complexes of varying stoichiometry [119]. Surprisingly, it turned out that during expression in E. coli cells producing complex carotenoids [83, 120], this protein forms extremely stable dimers capable of binding a carotenoid, conferring a unique violet color to the protein solution [119]. This protein was named COCP (C-terminal OCP-related carotenoid protein), and in the case of Synechocystis, it corresponds to the C domain of OCP (residues 165-317). However, as shown by phylogenetic and bioinformatic analysis, COCP may have homologs expressed independently of OCP or RCP and possessing functions that are not fully understood [121, 122]. The carotenoid molecule was found to stabilize COCP dimers by forming hydrogen bonds to the tyrosine-201 and tryptophan-288 residues. In the absence of a carotenoid or with the W288A substitution, COCP dimers demonstrate a significant propensity to dissociation. Modeling of structures of such dimers based on symmetry dictated by the nature of a symmetric carotenoid, canthaxanthin, which is bound to identical subunits, as well as based on data from Raman spectroscopy, circular dichroism, and secondary structure analysis suggests that subunits within such a dimer are structurally equivalent to the C domain of OCP [119]. Even though the considered data do not exclude the possibility of the FRP interaction with the N domain of OCP or with the interdomain linker, especially in the context of the already formed OCP–FRP complex, they indicate that the main specific FRP binding site is located on the C domain of OCP.
During a systematic comparison of the ability of FRP to interact with various forms and fragments of OCP, a hypothesis was put forward that the FRP binding site at the C-terminus of OCP always exists, but in some OCP states it may be blocked by a certain structural element of OCP preventing strong FRP binding typical of the photoactivated OCP form [90]. A remarkable feature of the OCP structure in its orange form is the presence of the N-terminal segment (N-terminal extension, NTE) that contains a short α1 helix, adhering externally to the β-sheet of the OCP’s C domain (see Fig. 2). This segment apparently acts like a latch or a fuse in OCPO, but disconnects and unfolds during photoactivation of OCP [62, 91] (see the corresponding section above). This element is also absent in certain studied OCP forms, which might explain their ability to tightly bind to OCP even without requiring domain separation. Indeed, OCPR, OCP-W288A, Apo-OCP, COCP, and Apo-COCP demonstrate the ability to interact with FRP, and even if OCPO, RCP, and Apo-RCP are capable of binding FRP, they do it with a much lesser affinity [90, 119]. These data lead us to the suggestion that the FRP binding site overlaps or coincides with the NTE binding site within the β-sheet of the OCP’s C domain, and the NTE segment itself prevents OCP from premature binding of FRP in the absence of photoactivation and, presumably, releases FRP at the very end of a photocycle, upon the OCP transition from the red to orange form. In full agreement with this suggestion, it turned out that deletion of the α-helix in the NTE structure of the ΔNTE protein results in it binding to FRP with the highest affinity among all OCP forms studied so far, without requiring photoactivation and separation of the domains [90]. The apparent stoichiometry of the ΔNTE–FRP complexes, determined by a combination of methods (analytical gel-filtration, native gel-electrophoresis, and chemical crosslinking with glutaraldehyde) was close to 1 : 1, in agreement with the conclusions drawn for OCP-W288A and Apo-OCP, indicating monomerization of FRP during its interaction with OCP [84, 99]. The apparent submicromolar dissociation constant of the ΔNTE–FRP complex appears to be due to the absence of the NTE element in OCP–ΔNTE, while normally it competes with FRP for a tentative common binding site, and/or since in this case the domains are not separated, and other hypothetical FRP binding sites might be easily accessible, with FRP functioning as an adapter or a bridge that brings together the OCP domains [84, 90, 98, 118]. It was suggested that competition between FRP and NTE may be due to a certain degree of similarity between both the primary structure and the spatial arrangement of the N-terminal regions in FRP (peptide 12AETQSAHALFR22 in the Synechocystis FRP) and OCP (peptide 3FTIDSARGIFP13 in the Synechocystis OCP) [90]. Indeed, similar to these peptides, the surface of the C domain of OCP in the NTE (and FRP)-binding region is enriched in hydrophobic amino acid residues (A222, F264, L291, F299, F300, A302), which may point to the nature of protein–protein interactions. It is not surprising that mutations at this site of OCP cause disruptions in FRP binding and functioning [96, 117]. Moreover, inspection of this region demonstrates why OCP photoactivation and disruption of hydrogen bonds between the keto group of the carotenoid and W288/Y201 results in the NTE detachment and further conformational rearrangements in OCP [62, 71, 91]. It should be noted that the W288 residue is located on the inner side of the same β-sheet as the hydrophobic NTE binding site; therefore, it is easy to assume changes in its hydrophobicity/polarity, and, accordingly, in binding affinity of NTE, when the hydrogen bond with W288 is disrupted. At the same time, it is not entirely clear why OCP sequences, in particular, those of the C domains, are much more evolutionary conserved than the FRP sequences. This may reflect the process of evolution of protein–protein interactions and indicate some differences in the mechanism of FRP binding in different cyanobacterial species, or it may be due to the lesser specificity of the FRP binding site in OCP. The third possibility is that only identical/similar regions in FRPs take part in the universal mechanism of their action on OCPs. In any case, it will be interesting to learn from future studies whether FRPs from certain organisms can bind and accelerate the transition of the red form to the orange form of OCP obtained from other sources. This should elucidate the key areas in the FRP structure that are important for its functioning, and those that determine species-specificity.
Despite the attractiveness of the hypothesis about the existence of more than one contact site between FRP and OCP and the data of native mass-spectrometry indicating the possibility of formation of the N domain–FRP–C domain transient complexes [98, 118, 123], new investigations will undoubtedly be required to accurately ascertain the mechanism of the FRP binding to OCP and its functioning. Experimental data indicating FRP monomerization are extremely interesting and may explain many observations, and they are also in agreement with the results of kinetic modeling of the processes occurring during the functioning of the photoprotection system in vitro and in vivo [97], but they will require more detailed verification in the future, in particular using the methods of structural biology. Unfortunately, extensive attempts to crystallize the FRP complexes with different forms of OCP resulted in the growth of beautiful, colored, but not diffracting crystals (Sluchanko et al., unpublished data).
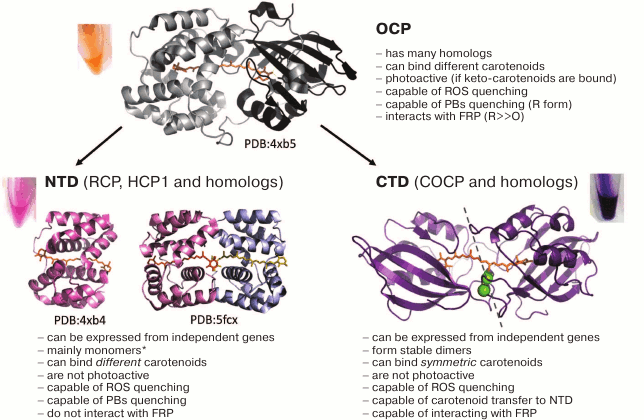
Interaction of isolated OCP domains and the unique mechanism of carotenoid transfer. As mentioned earlier, a notable feature of OCP is the modular principle of its structural and functional organization – the protein consists of two relatively rigid domains connected by an extended linker, which encapsulate a carotenoid molecule [67] (Fig. 4). Successful sequencing of numerous cyanobacterial genomes in recent years has determined progress in understanding the diversity of the structure and many important mechanisms at the molecular level. With respect to OCP, it turned out that many cyanobacterial genomes contain, in addition to full-length OCP genes, also genes corresponding to the individual N and C domains of the protein, and in certain representatives, several versions of such homologs are present. In 2016, it was shown that homologs of the N-terminal OCP domain are carotenoid-binding proteins called HCP (helical carotenoid proteins) [121]. Their spatial structure is similar to that of RCP (the product of OCP proteolysis [116]). Despite their similar structure and common ability to bind carotenoids of a different nature, which causes their reddish-purple color (Fig. 4), the physiological role of HCPs is not fully understood – their ability to quench PBs and ROS fluorescence varied greatly for the analyzed HCPs and was poorly predictable [121, 124]. For example, despite their similarity to RCP and the presence of a bound carotenoid in all HCP1-4 from Anabaena, the HCP1 variant (all1123) is an inefficient quencher for both ROS and PBs, while HCP2 (all3221) and HCP3 (alr4783) demonstrate the ability to quench only ROS, and HCP4 (all4941) – to quench PBs [124, 125]. It can be assumed that the difference in the properties of different homologs of the N-terminal domain is related to the type of bound carotenoids, to their different exposure to the solvent, and to the oligomeric state of HCP. For instance, monomeric state is apparently typical of RCP and HCP [67, 119], even with increased protein concentration upon crystallization (PDB 4XB4), whereas HCP1 was crystallized as a dimer with a rather large contact area of the monomers, each of which binds a carotenoid molecule (PDB 5FCX) (see Fig. 4). Perhaps a wide repertory of various carotenoid-binding OCP homologs is necessary for distributing functions (the so-called subfunctionalization) and helps cyanobacteria to acquire additional adaptations.
Fig. 4. Structure and properties of OCP from Synechocystis 6803 and homologs of its N and C domains. The models of crystal structures of OCP, RCP, and HCP1 (PDB identifiers are given) are presented, as well as a hybrid model of a COCP dimer built based on biochemical and biophysical data and optimized by molecular dynamics [119]. The major properties of the proteins are listed, and the typical colors of samples obtained in the bacterial expression system are shown [83, 120]. The asterisk (*) indicates that the monomeric state may not be typical of all HCPs, since at least one of them (HCP1) was crystallized in a dimeric form (PDB 5FCX). In the COCP model, the dashed line indicates the arbitrary symmetry plane of the dimer, and the position of F278 residues (numbering is equivalent to OCP from Synechocystis), where the homologs of the C domain may contain a cysteine, is shown in green (see also Fig. 5).
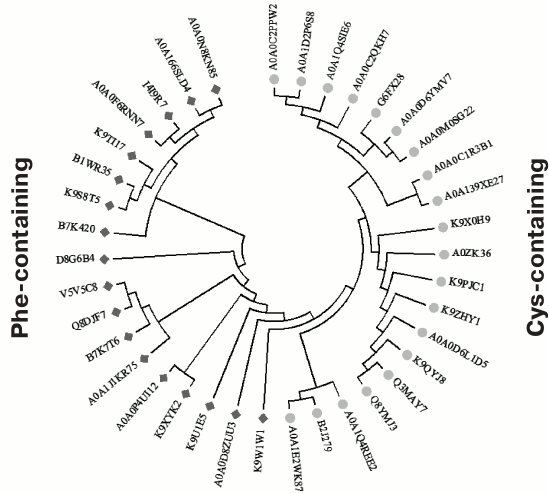
Fig. 5. Phylogenetic tree constructed based on the alignment of 38 sequences of various homologs of the OCP’s C domain from various cyanobacteria (UniProt identifiers are given), which demonstrates the presence of two alternative groups containing either a phenylalanine residue or a cysteine residue in position equivalent to F278 in the sequence of the full-length OCP from Synechocystis. The dimers presumably formed by the Cys-containing variants of the sequence can form disulfide bridges upon oxidation, since the cysteine residues from the two monomers are at the optimal distance for this (<10 Å; see Fig. 4). Phylogenetic analysis was carried out by the method of maximum likelihood in the MEGA7 program [128].
In 2017, it was shown that homologs of the C-terminal domain of OCP can also exist independently and bind a carotenoid [110, 119, 122]. This was shown for the first time with an artificially obtained C-terminal OCP domain from Synechocystis (which does not contain independent genes encoding homologs of the C-terminal domain) [93, 119], and only later this was demonstrated for other homologs of the C domain of OCP [110, 122]. Such proteins turned out to be carotenoid-binding and capable of quenching reactive oxygen species (ROS), but incapable of quenching PBs fluorescence [119], which further confirms a hypothesis about the location of the PBs binding site not on the C domain of OCP, but rather on the N domain and/or the linker region, and it is also fully consistent with the modular principle of OCP structure [67]. The specialization of OCP domains, albeit not fully understood, suggests the evolutionary origin of such a two-domain protein. Once arisen independently, the domains could undergo fusion after nature stopped using their noncovalently bound heterodimers as less effective assemblies [119, 121, 122]. However, this hypothesis should be considered with caution, since the most primitive cyanobacterium that currently exist, Gloeobacter, lacking thylakoids [126], possesses a full-length OCP (UniProt Q7NPK5) and uses the corresponding mechanism of PBs fluorescence quenching [127].
In an attempt to recapitulate this situation in vitro for OCP domains from Synechocystis synthesized separately in carotenoid-producing cells of E. coli, it turned out that the interaction of the domains led to the formation of unstable OCP-like complexes that demonstrated photoactivation and were capable of quenching PBs, but after photoactivation were practically unable to return into an OCPO-like state [119]. First, these data experimentally demonstrated a possible evolutionary scenario of the OCP emergence [121], and second, they were indicative of a carotenoid transfer from one protein to another during the recombination of the apo form of the N domain and the carotenoid-bound COCP. During this process, purple COCP solution (the carotenoid is located between C domains within a COCP dimer) (see Fig. 4) was gradually turning orange, which is characteristic of OCPO, in which the carotenoid is located between the N and C domains. This process was even more effective if the apo form of the N domain was replaced by the apo form of the full-length OCP [93, 119]. Following the direct transfer of the carotenoid from COCP to Apo-OCP, a much more stable OCPO form was formed that was capable of multiple rounds of photoactivation and PBs quenching [93]. This discovery suggests that, in addition to the antioxidant function in the cell, COCP homologs carry out the function of storing and transporting carotenoids and promote the maturation of OCP and HCP proteins, transferring carotenoids to their apo forms.
It is interesting to note that with such a unique mechanism of transferring a carotenoid from protein to protein, disassembly of rather stable COCP dimers coordinating a carotenoid occurred, and the W288A mutation within COCP, that destabilized the bond to carotenoid and protein dimers, significantly increased the rate of carotenoid transfer [93]. The detected tendency of Apo-COCP to self-associate into dimers even in the absence of a carotenoid and the corresponding concentration dependences that were similar for Apo-COCP and full-length Apo-OCP or OCPR and OCP-W288A suggested that C domains of OCP are responsible for the previously discovered tendency of the photoactivated OCP form (and of other forms with separated domains) to form dimers [83, 84]. OCPO, having a compact form, exhibited a much less pronounced dimerization propensity, which apparently proceeded in a different way, with the formation of another intersubunit interface [93]. Based on these observations, as well as data from small-angle X-ray scattering, low-resolution models were obtained for OCP and Apo-OCP (OCPR) dimers and monomers in solution [93]. It is important to note that in these studies, as well as others [98, 118], OCPO dimers were efficiently formed only at very high protein concentrations. These data to a certain degree contradict the mechanism postulated earlier, according to which OCP exists in the form of dimers [70, 72], and photoactivation causes their monomerization [64]. Apparently, photoactivation of OCP that eventually causes separation of its domains, in contrast, enhances dimerization propensity of the protein, mainly due to the formation of an interface between the C domains [93]. It should be noted that such an interface between C domains, also present in the model of COCP dimer binding a carotenoid, implies proximity of the loops containing a remarkable phenylalanine residue in position 278 (Synechocystis OCP numbering), in which many homologs of the C domain contain a cysteine residue (for example, the all4940 homolog from Anabaena) (Fig. 5) [110]. Very recently, it has been shown that dimers of such homologs are able to form covalent disulfide bridges during oxidation and that such a modification may affect carotenoid transfer from COCP in vitro [110].
Thus, dimer–monomer transitions are extremely important for understanding the mechanism of OCP and FRP functioning in vitro, but their role in the processes occurring in vivo remains to be determined.
Despite the relatively simple construction of the photosynthetic apparatus in cyanobacteria, compared to that of higher plants, it took decades of research and analysis to determine the mechanism of their photoprotection and to realize that it is associated with the unique orange carotenoid protein and its ability to be photoactivated and to interact with light-harvesting antennae. The function of OCP was first demonstrated only about 10 years ago. Now it is difficult to imagine how the area of photosynthesis and photoprotection research would develop if this event occurred much earlier (since OCP was first isolated and described 36 years ago). Improvement of numerous methodological approaches in biochemistry, biophysics, and structural biology, that occurred over the past decade, greatly facilitates understanding of the details of the discovered photoprotection mechanism associated with OCP functioning. In this regard, tremendous progress in recent years has been made in characterizing the OCP photocycle (on a scale from picoseconds to seconds and minutes), in studying its ability to quench PBs fluorescence and interact with FRP. Nevertheless, despite titanic efforts of several laboratories around the world, several fundamental issues remain subject of heated debate and are still far from being understood.
Strictly speaking, the area of the OCP binding within PBs is still unknown. Not to mention that some indirect data on this topic are in a somewhat contradiction with each other. It is also completely inexplicable how a relatively conservative protein, OCP, whose sequence is extremely similar in different cyanobacterial representatives, can interact with phycobilisomes of different architectures and protein compositions from different cyanobacteria [129]. There are certain disagreements in the literature regarding the OCP–PBs complex: whether it is specific and stable, or vice versa – less specific and more dynamic. Significant progress in the field of structural methods, primarily in cryoelectron microscopy, may be very timely and may contribute to clarifying the structure of tentative OCP–PBs complexes.
There is still no clear understanding of why the OCP sequence is much more conservative than the FRP sequence, if it is admitted that these two components of the photoprotection system interact according to a general principle in cyanobacteria. The site and the mechanism of the interaction between OCP and FRP are defined only approximately – even the oligomeric state of OCP and FRP proteins in vitro and in vivo is debated in the literature and is far from consensus. Main results related to studying the interaction between OCP and FRP were obtained in the last 2-3 years. FRP was discovered only seven years ago, and it was structurally studied only four years ago. Structural biology approaches are also promising in this respect.
OCP is an amazing example of how light can regulate dramatic, but reversible changes in protein conformation. During photoactivation of OCP, not merely does the local structure of its active site or of a single domain change: according to modern views, the protein is completely rearranged, which is accompanied by an alteration of its surface and exposure of previously masked zones (interface of the N and C domains, the NTE segment, and its binding site on the C domain, disordering of the linker, etc.). Scenarios for using such light-controlled conformational changes in OCP in applied research or in designing various sensors, or for the needs of optogenetics, appear promising. In might be possible, for example, through the OCP interaction with a certain (artificial or modified natural) partner that might facilitate transduction of such a light-induced signal. In any case, we do not doubt the future rapid development of this field of research and expect many new and interesting discoveries explaining the features of the photoprotection mechanism in cyanobacteria and key protein–protein interactions underlying it.
Acknowledgments
We thank N. B. Gusev, I. N. Stadnichuk, D. A. Los, P. M. Krasilnikov, and N. P. Yurina for valuable consultations.
This study was supported by the Russian Foundation for Basic Research (grant No. 15-04-01930a) and by Moscow Government within the framework of a scientific project No. 15-34-70007 “mol_a_mos”.
REFERENCES
1.Guerrero, F., Zurita, J. L., Roncel, M.,
Kirilovsky, D., and Ortega, J. M. (2014) The role of the high potential
form of the cytochrome b559: study of Thermosynechococcus
elongatus mutants, Biochim. Biophys. Acta, 1837,
908-919.
2.Herbert, S. K., Martin, R. E., and Fork, D. C.
(1995) Light adaptation of cyclic electron transport through
photosystem I in the cyanobacterium Synechococcus sp. PCC 7942,
Photosynth. Res., 46, 277-285.
3.Mullineaux, C. W. (2014) Co-existence of
photosynthetic and respiratory activities in cyanobacterial thylakoid
membranes, Biochim. Biophys. Acta, 1837, 503-511.
4.Mullineaux, C. W. (2014) Electron transport and
light-harvesting switches in cyanobacteria, Front. Plant Sci.,
5, 7.
5.Trubitsin, B. V., Ptushenko, V. V., Koksharova, O.
A., Mamedov, M. D., Vitukhnovskaya, L. A., Grigor’ev, I. A.,
Semenov, A. Y., and Tikhonov, A. N. (2005) EPR study of electron
transport in the cyanobacterium Synechocystis sp. PCC 6803:
oxygen-dependent interrelations between photosynthetic and respiratory
electron transport chains, Biochim. Biophys. Acta, 1708,
238-249.
6.Peschek, G. A., Obinger, C., and Renger, G. (2011)
Bioenergetic Processes of Cyanobacteria, Springer.
7.Allakhverdiev, S. I., Kinoshita, M., Inaba, M.,
Suzuki, I., and Murata, N. (2001) Unsaturated fatty acids in membrane
lipids protect the photosynthetic machinery against salt-induced damage
in Synechococcus, Plant Physiol., 125,
1842-1853.
8.Mironov, K. S., Sidorov, R. A., Trofimova, M. S.,
Bedbenov, V. S., Tsydendambaev, V. D., Allakhverdiev, S. I., and Los,
D. A. (2012) Light-dependent cold-induced fatty acid unsaturation,
changes in membrane fluidity, and alterations in gene expression in
Synechocystis, Biochim. Biophys. Acta,
1817, 1352-1359.
9.Maksimov, E. G., Mironov, K. S., Trofimova, M. S.,
Nechaeva, N. L., Todorenko, D. A., Klementiev, K. E., Tsoraev, G. V.,
Tyutyaev, E. V., Zorina, A. A., Feduraev, P. V., Allakhverdiev, S. I.,
Paschenko, V. Z., and Los, D. A. (2017) Membrane fluidity controls
redox-regulated cold stress responses in cyanobacteria, Photosynth.
Res., 133, 215-223.
10.He, J.-A., Hu, Y.-Z., and Jiang, L.-J. (1997)
Photodynamic action of phycobiliproteins: in situ generation of
reactive oxygen species, Biochim. Biophys. Acta,
1320, 165-174.
11.Pascal, A. A., Liu, Z., Broess, K., Van Oort, B.,
Van Amerongen, H., Wang, C., Horton, P., Robert, B., Chang, W., and
Ruban, A. (2005) Molecular basis of photoprotection and control of
photosynthetic light-harvesting, Nature, 436,
134-137.
12.Stirbet, A., and Govindjee (2011) On the relation
between the Kautsky effect (chlorophyll a fluorescence induction) and
photosystem II: basics and applications of the OJIP fluorescence
transient, J. Photochem. Photobiol. B, 104,
236-257.
13.Conrad, W., and Mullineaux, J. F. A. (1988)
Fluorescence induction transients indicate dissociation of photosystem
II from the phycobilisome during the state-2 transition in the
cyanobacterium Synechococcus 6301, Biochim. Biophys.
Acta, 934, 96-107.
14.Mullineaux, C. W., and Emlyn-Jones, D. (2005)
State transitions: an example of acclimation to low-light stress, J.
Exp. Bot., 56, 389-393.
15.Kirilovsky, D. (2015) Modulating energy arriving
at photochemical reaction centers: orange carotenoid protein-related
photoprotection and state transitions, Photosynth. Res.,
126, 3-17.
16.Liu, H., Zhang, H., Niedzwiedzki, D. M., Prado,
M., He, G., Gross, M. L., and Blankenship, R. E. (2013) Phycobilisomes
supply excitations to both photosystems in a megacomplex in
cyanobacteria, Science, 342, 1104-1107.
17.De Marsac, N. T., and Cohen-Bazire, G. (1977)
Molecular composition of cyanobacterial phycobilisomes, Proc. Natl.
Acad. Sci. USA, 74, 1635-1639.
18.Gantt, E. (1981) Phycobilisomes, Annu. Rev.
Plant Physiol., 32, 327-347.
19.Glazer, A. N. (1985) Light harvesting by
phycobilisomes, Annu. Rev. Biophys. Biophys. Chem.,
14, 47-77.
20.Samsonoff, W. A., and MacColl, R. (2001)
Biliproteins and phycobilisomes from cyanobacteria and red algae at the
extremes of habitat, Arch. Microbiol., 176,
400-405.
21.Liu, L.-N., Chen, X.-L., Zhang, Y.-Z., and Zhou,
B.-C. (2005) Characterization, structure and function of linker
polypeptides in phycobilisomes of cyanobacteria and red algae: an
overview, Biochim. Biophys. Acta, 1708,
133-142.
22.Zhang, J., Ma, J., Liu, D., Qin, S., Sun, S.,
Zhao, J., and Sui, S. F. (2017) Structure of phycobilisome from the red
alga Griffithsia pacifica, Nature, 551, 57-63.
23.Bryant, D. A., Guglielmi, G., De Marsac, N. T.,
Castets, A.-M., and Cohen-Bazire, G. (1979) The structure of
cyanobacterial phycobilisomes: a model, Arch. Microbiol.,
123, 113-127.
24.Mimuro, M., and Gantt, E. (1986) A high molecular
weight terminal pigment (“anchor polypeptide”) and a minor
blue polypeptide from phycobilisomes of the cyanobacterium
Nostoc sp. (MAC): isolation and characterization, Photosynth.
Res., 10, 201-208.
25.Maksimov, E. G., Kuzminov, F. I., Konyuhov, I.
V., Elanskaya, I. V., and Paschenko, V. Z. (2011) Photosystem 2
effective fluorescence cross-section of cyanobacterium
Synechocystis sp. PCC6803 and its mutants, J. Photochem.
Photobiol. B, 104, 285-291.
26.Adir, N. (2005) Elucidation of the molecular
structures of components of the phycobilisome: reconstructing a giant,
Photosynth. Res., 85, 15-32.
27.Betz, M. (1997) One century of protein
crystallography: the phycobiliproteins, Biol. Chem.,
378, 167-176.
28.McGregor, A., Klartag, M., David, L., and Adir,
N. (2008) Allophycocyanin trimer stability and functionality are
primarily due to polar enhanced hydrophobicity of the phycocyanobilin
binding pocket, J. Mol. Biol., 384, 406-421.
29.Sonani, R. R., Gupta, G. D., Madamwar, D., and
Kumar, V. (2015) Crystal structure of allophycocyanin from marine
cyanobacterium Phormidium sp. A09DM, PLoS One,
10, e0124580.
30.Zuber, H. (1986) Structure of light-harvesting
antenna complexes of photosynthetic bacteria, cyanobacteria and red
algae, Trends Biochem. Sci., 11, 414-419.
31.MacColl, R. (1998) Cyanobacterial phycobilisomes,
J. Struct. Biol., 124, 311-334.
32.Maksimov, E. G., Gostev, T. S., Kuz’minov,
F. I., Sluchanko, N. N., Stadnichuk, I. N., Pashchenko, V. Z., and
Rubin, A. B. (2010) Hybrid systems of quantum dots mixed with the
photosensitive protein phycoerythrin, Nanotechnol. Russ.,
5, 531-537.
33.Karpulevich, A. A., Maksimov, E. G., Sluchanko,
N. N., Vasiliev, A. N., and Paschenko, V. Z. (2016) Highly efficient
energy transfer from quantum dot to allophycocyanin in hybrid
structures, J. Photochem. Photobiol. B, 160,
96-101.
34.Holzwarth, A. R. (1991) Structure–function
relationships and energy transfer in phycobiliprotein antennae,
Physiol. Plant., 83, 518-528.
35.Zlenko, D. V., Krasilnikov, P. M., and
Stadnichuk, I. N. (2016) Structural modeling of the phycobilisome core
and its association with the photosystems, Photosynth. Res.,
130, 347-356.
36.MacColl, R. (2004) Allophycocyanin and energy
transfer, Biochim. Biophys. Acta, 1657, 73-81.
37.Schmitt, F. J., Maksimov, E. G., Suedmeyer, H.,
Jeyasangar, V., Theiss, C., Paschenko, V. Z., Eichler, H. J., and
Renger, G. (2011) Time resolved temperature switchable excitation
energy transfer processes between CdSe/ZnS nanocrystals and
phycobiliprotein antenna from Acaryochloris marina, Photon.
Nanostruct. Fundament. Appl., 9, 190-195.
38.Maksimov, E. G., Tsoraev, G. V., Paschenko, V.
Z., and Rubin, A. B. (2012) Nature of anomalous temperature dependence
of allophycocyanin fluorescence lifetime, Rep. Acad. Sci.,
443, 377.
39.Maksimov, E. G., Schmitt, F. J., Hatti, P.,
Klementiev, K. E., Paschenko, V. Z., Renger, G., and Rubin, A. B.
(2013) Anomalous temperature dependence of the fluorescence lifetime of
phycobiliproteins, Las. Phys. Lett., 10,
055602.
40.Mao, H.-B., Li, G.-F., Li, D.-H., Wu, Q.-Y.,
Gong, Y.-D., Zhang, X.-F., and Zhao, N.-M. (2003) Effects of glycerol
and high temperatures on structure and function of phycobilisomes in
Synechocystis sp. PCC 6803, FEBS Lett.,
553, 68-72.
41.Ajlani, G., and Vernotte, C. (1998) Construction
and characterization of a phycobiliprotein-less mutant of
Synechocystis sp. PCC 6803, Plant Mol. Biol.,
37, 577-580.
42.Manodori, A., and Melis, A. (1986) Cyanobacterial
acclimation to photosystem I or photosystem II light, Plant
Physiol., 82, 185-189.
43.El Bissati, K., Delphin, E., Murata, N., Etienne,
A. L., and Kirilovsky, D. (2000) Photosystem II fluorescence quenching
in the cyanobacterium Synechocystis PCC 6803: involvement of two
different mechanisms, Biochim. Biophys. Acta,
1457, 229-242.
44.Rakhimberdieva, M. G., Stadnichuk, I. N.,
Elanskaya, I. V., and Karapetyan, N. V. (2004) Carotenoid-induced
quenching of the phycobilisome fluorescence in photosystem II-deficient
mutant of Synechocystis sp, FEBS Lett.,
574, 85-88.
45.Rakhimberdieva, M. G., Bolychevtseva, Y. V.,
Elanskaya, I. V., and Karapetyan, N. V. (2007) Protein–protein
interactions in carotenoid triggered quenching of phycobilisome
fluorescence in Synechocystis sp. PCC 6803, FEBS Lett.,
581, 2429-2433.
46.Wilson, A., Ajlani, G., Verbavatz, J.-M., Vass,
I., Kerfeld, C. A., and Kirilovsky, D. (2006) A soluble carotenoid
protein involved in phycobilisome-related energy dissipation in
cyanobacteria, Plant Cell, 18, 992-1007.
47.Wilson, A., Punginelli, C., Gall, A., Bonetti,
C., Alexandre, M., Routaboul, J.-M., Kerfeld, C. A., Van Grondelle, R.,
Robert, B., Kennis, J. T. M., and Kirilovsky, D. (2008) A photoactive
carotenoid protein acting as light intensity sensor, Proc. Natl.
Acad. Sci. USA, 105, 12075-12080.
48.Punginelli, C., Wilson, A., Routaboul, J. M., and
Kirilovsky, D. (2009) Influence of zeaxanthin and echinenone binding on
the activity of the orange carotenoid protein, Biochim. Biophys.
Acta, 1787, 280-288.
49.Wilson, A., Kinney, J. N., Zwart, P. H.,
Punginelli, C., D’Haene, S., Perreau, F., Klein, M. G.,
Kirilovsky, D., and Kerfeld, C. A. (2010) Structural determinants
underlying photoprotection in the photoactive orange carotenoid protein
of cyanobacteria, J. Biol. Chem., 285,
18364-18375.
50.Rakhimberdieva, M. G., Vavilin, D. V., Vermaas,
W. F., Elanskaya, I. V., and Karapetyan, N. V. (2007)
Phycobilin/chlorophyll excitation equilibration upon carotenoid-induced
non-photochemical fluorescence quenching in phycobilisomes of the
cyanobacterium Synechocystis sp. PCC 6803, Biochim. Biophys.
Acta, 1767, 757-765.
51.Gorbunov, M. Y., Kuzminov, F. I., Fadeev, V. V.,
Kim, J. D., and Falkowski, P. G. (2011) A kinetic model of
non-photochemical quenching in cyanobacteria, Biochim. Biophys.
Acta, 1807, 1591-1599.
52.Berera, R., Van Stokkum, I. H., Gwizdala, M.,
Wilson, A., Kirilovsky, D., and Van Grondelle, R. (2012) The
photophysics of the orange carotenoid protein, a light-powered
molecular switch, J. Phys. Chem. B, 116,
2568-2574.
53.Tian, L., Gwizdala, M., Van Stokkum, I. H.,
Koehorst, R. B., Kirilovsky, D., and Van Amerongen, H. (2012)
Picosecond kinetics of light harvesting and photoprotective quenching
in wild-type and mutant phycobilisomes isolated from the cyanobacterium
Synechocystis PCC 6803, Biophys. J., 102,
1692-1700.
54.Maksimov, E. G., Schmitt, F. J., Shirshin, E. A.,
Svirin, M. D., Elanskaya, I. V., Friedrich, T., Fadeev, V. V.,
Paschenko, V. Z., and Rubin, A. B. (2014) The time course of
non-photochemical quenching in phycobilisomes of Synechocystis
sp. PCC6803 as revealed by picosecond time-resolved fluorimetry,
Biochim. Biophys. Acta, 1837, 1540-1547.
55.Krasilnikov, P. M., Zlenko, D. V., and
Stadnichuk, I. N. (2015) The efficiency of non-photochemical
fluorescence quenching of phycobilisomes by the orange carotenoid
protein, Biophysics, 60, 752-758.
56.Maksimov, E. G., Klementiev, K. E., Shirshin, E.
A., Tsoraev, G. V., Elanskaya, I. V., and Paschenko, V. Z. (2015)
Features of temporal behavior of fluorescence recovery in
Synechocystis sp. PCC6803, Photosynth. Res.,
125, 167-178.
57.Polivka, T., Kerfeld, C. A., Pascher, T., and
Sundstrom, V. (2005) Spectroscopic properties of the carotenoid
3′-hydroxyechinenone in the orange carotenoid protein from the
cyanobacterium Arthrospira maxima, Biochemistry,
44, 3994-4003.
58.Chabera, P., Durchan, M., Shih, P. M., Kerfeld,
C. A., and Polivka, T. (2011) Excited-state properties of the 16 kDa
red carotenoid protein from Arthrospira maxima, Biochim.
Biophys. Acta, 1807, 30-35.
59.Slouf, V., Kuznetsova, V., Fuciman, M., De
Carbon, C. B., Wilson, A., Kirilovsky, D., and Polivka, T. (2017)
Ultrafast spectroscopy tracks carotenoid configurations in the orange
and red carotenoid proteins from cyanobacteria, Photosynth.
Res., 131, 105-117.
60.Polivka, T., Chabera, P., and Kerfeld, C. A.
(2013) Carotenoid–protein interaction alters the S1 energy of
hydroxyechinenone in the orange carotenoid protein, Biochim.
Biophys. Acta, 1827, 248-254.
61.Zlenko, D. V., Krasilnikov, P. M., and
Stadnichuk, I. N. (2016) Role of inter-domain cavity in the attachment
of the orange carotenoid protein to the phycobilisome core and to the
fluorescence recovery protein, J. Biomol. Struct. Dyn.,
34, 486-496.
62.Liu, H., Zhang, H., King, J. D., Wolf, N. R.,
Prado, M., Gross, M. L., and Blankenship, R. E. (2014) Mass
spectrometry footprinting reveals the structural rearrangements of
cyanobacterial orange carotenoid protein upon light activation,
Biochim. Biophys. Acta, 1837, 1955-1963.
63.Harris, D., Tal, O., Jallet, D., Wilson, A.,
Kirilovsky, D., and Adir, N. (2016) Orange carotenoid protein burrows
into the phycobilisome to provide photoprotection, Proc. Natl. Acad.
Sci. USA, 113, E1655-E1662.
64.Zhang, H., Liu, H., Niedzwiedzki, D. M., Prado,
M., Jiang, J., Gross, M. L., and Blankenship, R. E. (2014) Molecular
mechanism of photoactivation and structural location of the
cyanobacterial orange carotenoid protein, Biochemistry,
53, 13-19.
65.Wilson, A., Punginelli, C., Couturier, M.,
Perreau, F., and Kirilovsky, D. (2011) Essential role of two tyrosines
and two tryptophans on the photoprotection activity of the orange
carotenoid protein, Biochim. Biophys. Acta, 1807,
293-301.
66.Holt, T. K., and Krogmann, D. W. (1981) A
carotenoid-protein from cyanobacteria, Biochim. Biophys. Acta,
637, 408-414.
67.Leverenz, R. L., Jallet, D., Li, M. D., Mathies,
R. A., Kirilovsky, D., and Kerfeld, C. A. (2014) Structural and
functional modularity of the orange carotenoid protein: distinct roles
for the N- and C-terminal domains in cyanobacterial photoprotection,
Plant Cell, 26, 426-437.
68.Gwizdala, M., Wilson, A., and Kirilovsky, D.
(2011) In vitro reconstitution of the cyanobacterial
photoprotective mechanism mediated by the orange carotenoid protein in
Synechocystis PCC 6803, Plant Cell, 23,
2631-2643.
69.Wilson, A., Gwizdala, M., Mezzetti, A.,
Alexandre, M., Kerfeld, C. A., and Kirilovsky, D. (2012) The essential
role of the N-terminal domain of the orange carotenoid protein in
cyanobacterial photoprotection: importance of a positive charge for
phycobilisome binding, Plant Cell, 24,
1972-1983.
70.Bao, H., Melnicki, M. R., Pawlowski, E. G.,
Sutter, M., Agostoni, M., Lechno-Yossef, S., Cai, F., Montgomery, B.
L., and Kerfeld, C. A. (2017) Additional families of orange carotenoid
proteins in the photoprotective system of cyanobacteria, Nat.
Plants, 3, 17089.
71.Maksimov, E. G., Shirshin, E. A., Sluchanko, N.
N., Zlenko, D. V., Parshina, E. Y., Tsoraev, G. V., Klementiev, K. E.,
Budylin, G. S., Schmitt, F. J., Friedrich, T., Fadeev, V. V.,
Paschenko, V. Z., and Rubin, A. B. (2015) The signaling state of orange
carotenoid protein, Biophys. J., 109, 595-607.
72.Kerfeld, C. A., Sawaya, M. R., Brahmandam, V.,
Cascio, D., Ho, K. K., Trevithick-Sutton, C. C., Krogmann, D. W., and
Yeates, T. O. (2003) The crystal structure of a cyanobacterial
water-soluble carotenoid binding protein, Structure,
11, 55-65.
73.Kerfeld, C. A. (2004) Structure and function of
the water-soluble carotenoid-binding proteins of cyanobacteria,
Photosynth. Res., 81, 215-225.
74.Kirilovsky, D. (2010) The photoactive orange
carotenoid protein and photoprotection in cyanobacteria, Adv. Exp.
Med. Biol., 675, 139-159.
75.Kirilovsky, D., and Kerfeld, C. A. (2012) The
orange carotenoid protein in photoprotection of photosystem II in
cyanobacteria, Biochim. Biophys. Acta, 1817,
158-66.
76.Stadnichuk, I. N., Krasilnikov, P. M., Zlenko, D.
V., Freidzon, A. Y., Yanyushin, M. F., and Rubin, A. B. (2015)
Electronic coupling of the phycobilisome with the orange carotenoid
protein and fluorescence quenching, Photosynth. Res.,
124, 315-335.
77.Gupta, S., Guttman, M., Leverenz, R. L.,
Zhumadilova, K., Pawlowski, E. G., Petzold, C. J., Lee, K. K., Ralston,
C. Y., and Kerfeld, C. A. (2015) Local and global structural drivers
for the photoactivation of the orange carotenoid protein, Proc.
Natl. Acad. Sci. USA, 112, E5567-E5574.
78.Maksimov, E. G., Sluchanko, N. N., Slonimskiy, Y.
B., Slutskaya, E. A., Stepanov, A. M., Argentova-Stevens, A. V.,
Shirshin, E. A., Tsoraev, G. V., Klementiev, K. E., Slatinskaya, O. V.,
Lukashev, E. P., Friedrich, T., Paschenko, V. Z., and Rubin, A. B.
(2017) The photocycle of orange carotenoid protein conceals distinct
intermediates and asynchronous changes in the carotenoid and protein
components, Sci. Rep., 7, 15548.
79.Shakhnovich, E. I., and Finkelstein, A. V. (1989)
Theory of cooperative transitions in protein molecules. I. Why
denaturation of globular protein is a first-order phase transition,
Biopolymers, 28, 1667-1680.
80.Semisotnov, G. V., Rodionova, N. A., Razgulyaev,
O. I., Uversky, V. N., Gripas’, A. F., and Gilmanshin, R. I.
(1991) Study of the “molten globule” intermediate state in
protein folding by a hydrophobic fluorescent probe, Biopolymers,
31, 119-128.
81.Frauenfelder, H., Fenimore, P. W., Chen, G., and
McMahon, B. H. (2006) Protein folding is slaved to solvent motions,
Proc. Natl. Acad. Sci. USA, 103, 15469-15472.
82.Hagen, S. J. (2010) Solvent viscosity and
friction in protein folding dynamics, Curr. Prot. Pept. Sci.,
11, 385-395.
83.Maksimov, E. G., Moldenhauer, M., Shirshin, E.
A., Parshina, E. A., Sluchanko, N. N., Klementiev, K. E., Tsoraev, G.
V., Tavraz, N. N., Willoweit, M., Schmitt, F.-J., Breitenbach, J.,
Sandmann, G., Paschenko, V. Z., Friedrich, T., and Rubin, A. B. (2016)
A comparative study of three signaling forms of the orange carotenoid
protein, Photosynth. Res., 130, 389-401.
84.Sluchanko, N. N., Klementiev, K. E., Shirshin, E.
A., Tsoraev, G. V., Friedrich, T., and Maksimov, E. G. (2017) The
purple Trp288Ala mutant of Synechocystis OCP persistently
quenches phycobilisome fluorescence and tightly interacts with FRP,
Biochim. Biophys. Acta, 1858, 1-11.
85.Leverenz, R. L., Sutter, M., Wilson, A., Gupta,
S., Thurotte, A., De Carbon, C. B., Petzold, C. J., Ralston, C.,
Perreau, F., and Kirilovsky, D. (2015) A 12 Å carotenoid
translocation in a photoswitch associated with cyanobacterial
photoprotection, Science, 348, 1463-1466.
86.Maksimov, E. G., Sluchanko, N. N., Mironov, K.
S., Shirshin, E. A., Klementiev, K. E., Tsoraev, G. V., Moldenhauer,
M., Friedrich, T., Los, D. A., Allakhverdiev, S. I., Paschenko, V. Z.,
and Rubin, A. B. (2017) Fluorescent labeling preserving OCP
photoactivity reveals its reorganization during the photocycle,
Biophys. J., 112, 46-56.
87.Kish, E., Pinto, M. M., Kirilovsky, D., Spezia,
R., and Robert, B. (2015) Echinenone vibrational properties: from
solvents to the orange carotenoid protein, Biochim. Biophys.
Acta, 1847, 1044-1054.
88.Bandara, S., Ren, Z., Lu, L., Zeng, X., Shin, H.,
Zhao, K.-H., and Yang, X. (2017) Photoactivation mechanism of a
carotenoid-based photoreceptor, Proc. Natl. Acad. Sci. USA,
114, 6286-6291.
89.Thurotte, A., Lopez-Igual, R., Wilson, A.,
Comolet, L., Bourcier de Carbon, C., Xiao, F., and Kirilovsky, D.
(2015) Regulation of orange carotenoid protein activity in
cyanobacterial photoprotection, Plant Physiol.,
169, 737-747.
90.Sluchanko, N. N., Slonimskiy, Y. B., Moldenhauer,
M., Friedrich, T., and Maksimov, E. G. (2017) Deletion of the short
N-terminal extension in OCP reveals the main site for FRP binding,
FEBS Lett., 591, 1667-1676.
91.Liu, H., Zhang, H., Orf, G. S., Lu, Y., Jiang,
J., King, J. D., Wolf, N. R., Gross, M. L., and Blankenship, R. E.
(2016) Dramatic domain rearrangements of the cyanobacterial orange
carotenoid protein upon photoactivation, Biochemistry,
55, 1003-1009.
92.Zhang, H., Liu, H., Lu, Y., Wolf, N. R., Gross,
M. L., and Blankenship, R. E. (2016) Native mass spectrometry and ion
mobility characterize the orange carotenoid protein functional domains,
Biochim. Biophys. Acta, 1857, 734-739.
93.Maksimov, E. G., Sluchanko, N. N., Slonimskiy, Y.
B., Mironov, K. S., Klementiev, K. E., Moldenhauer, M., Friedrich, T.,
Los, D. A., Paschenko, V. Z., and Rubin, A. B. (2017) The unique
protein-to-protein carotenoid transfer mechanism, Biophys. J.,
113, 402-413.
94.Boulay, C., Wilson, A., D’Haene, S., and
Kirilovsky, D. (2010) Identification of a protein required for recovery
of full antenna capacity in OCP-related photoprotective mechanism in
cyanobacteria, Proc. Natl. Acad. Sci. USA, 107,
11620-11625.
95.Gwizdala, M., Wilson, A., Omairi-Nasser, A., and
Kirilovsky, D. (2013) Characterization of the Synechocystis PCC
6803 fluorescence recovery protein involved in photoprotection,
Biochim. Biophys. Acta, 1827, 348-354.
96.Sutter, M., Wilson, A., Leverenz, R. L.,
Lopez-Igual, R., Thurotte, A., Salmeen, A. E., Kirilovsky, D., and
Kerfeld, C. A. (2013) Crystal structure of the FRP and identification
of the active site for modulation of OCP-mediated photoprotection in
cyanobacteria, Proc. Natl. Acad. Sci. USA, 110,
10022-10027.
97.Shirshin, E. A., Nikonova, E. E., Kuzminov, F.
I., Sluchanko, N. N., Elanskaya, I. V., Gorbunov, M. Y., Fadeev, V. V.,
Friedrich, T., and Maksimov, E. G. (2017) Biophysical modeling of in
vitro and in vivo processes underlying regulated
photoprotective mechanism in cyanobacteria, Photosynth. Res.,
133, 261-271.
98.Lu, Y., Liu, H., Saer, R. G., Zhang, H., Meyer,
C. M., Li, V. L., Shi, L., King, J. D., Gross, M. L., and Blankenship,
R. E. (2017) Native mass spectrometry analysis of oligomerization
states of fluorescence recovery protein and orange carotenoid protein:
two proteins involved in the cyanobacterial photoprotection cycle,
Biochemistry, 56, 160-166.
99.Moldenhauer, M., Sluchanko, N. N., Tavraz, N. N.,
Junghans, C., Buhrke, D., Willoweit, M., Chiappisi, L., Schmitt, F.-J.,
Vukojevic, V., Shirshin, E. A., Ponomarev, V. Y., Paschenko, V. Z.,
Gradzielski, M., Maksimov, E. G., and Friedrich, T. (2017) Interaction
of the signaling state analog and the apoprotein form of the orange
carotenoid protein with the fluorescence recovery protein,
Photosynth. Res., doi: 10.1007/s11120-017-0346-2.
100.King, J. D., Liu, H., He, G., Orf, G. S., and
Blankenship, R. E. (2014) Chemical activation of the cyanobacterial
orange carotenoid protein, FEBS Lett., 588,
4561-5465.
101.Dagnino-Leone, J., Figueroa, M., Mella, C.,
Vorphal, M. A., Kerff, F., Vasquez, A. J., Bunster, M., and
Martinez-Oyanedel, J. (2017) Structural models of the different trimers
present in the core of phycobilisomes from Gracilaria
chilensis based on crystal structures and sequences, PLoS
One, 12, e0177540.
102.Gantt, E., and Lipschultz, C. A. (1972)
Phycobilisomes of Porphyridium cruentum, 1. Isolation, J.
Cell Biol., 54, 313-324.
103.Stadnichuk, I. N., Yanoshin, M. F.,
Zharmahumedov, S. K., Maksimov, E. G., Muronec, E. M., and Paschenko,
V. Z. (2011) Quenching of phycobilisome quenching by orange
carotenoid-protein, Rep. Acad. Sci., 439, 270-273.
104.Stadnichuk, I. N., Yanyushin, M. F., Maksimov,
E. G., Lukashev, E. P., Zharmukhamedov, S. K., Elanskaya, I. V., and
Paschenko, V. Z. (2012) Site of non-photochemical quenching of the
phycobilisome by orange carotenoid protein in the cyanobacterium
Synechocystis sp. PCC 6803, Biochim. Biophys. Acta,
1817, 1436-1445.
105.Stadnichuk, I. N., Yanyushin, M. F., Bernat,
G., Zlenko, D. V., Krasilnikov, P. M., Lukashev, E. P., Maksimov, E.
G., and Paschenko, V. Z. (2013) Fluorescence quenching of the
phycobilisome terminal emitter LCM from the cyanobacterium
Synechocystis sp. PCC 6803 detected in vivo and in
vitro, J. Photochem. Photobiol. B, 125,
137-145.
106.Gao, X., Wei, T.-D., Zhang, N., Xie, B.-B., Su,
H.-N., Zhang, X.-Y., Chen, X.-L., Zhou, B.-C., Wang, Z.-X., Wu, J.-W.,
and Zhang, Y.-Z. (2012) Molecular insights into the terminal energy
acceptor in cyanobacterial phycobilisome, Mol. Microbiol.,
85, 907-915.
107.Tang, K., Ding, W.-L., Hoppner, A., Zhao, C.,
Zhang, L., Hontani, Y., Kennis, J. T. M., Gartner, W., Scheer, H.,
Zhou, M., and Zhao, K.-H. (2015) The terminal phycobilisome emitter,
LCM: a light-harvesting pigment with a phytochrome chromophore,
Proc. Natl. Acad. Sci. USA, 112, 15880-15885.
108.Houmard, J., Capuano, V., Colombano, M. V.,
Coursin, T., and Tandeau de Marsac, N. (1990) Molecular
characterization of the terminal energy acceptor of cyanobacterial
phycobilisomes, Proc. Natl. Acad. Sci. USA, 87,
2152-2156.
109.Stadnichuk, I. N., Yanyushin, M. F., Maksimov,
E. G., Lukashev, E. P., Zharmukhamedov, S. K., Elanskaya, I. V., and
Paschenko, V. Z. (2012) Site of non-photochemical quenching of the
phycobilisome by orange carotenoid protein in the cyanobacterium
Synechocystis sp. PCC 6803, Biochim. Biophys. Acta,
1817, 1436-1445.
110.Muzzopappa, F., Wilson, A., Yogarajah, V., Cot,
S., Perreau, F., Montigny, C., Bourcier de Carbon, C., and Kirilovsky,
D. (2017) Paralogs of the C-terminal domain of the cyanobacterial
orange carotenoid protein are carotenoid donors to helical carotenoid
proteins, Plant Physiol., 175, 1283-1303.
111.Elanskaya, I. V., Kononova, I. A., Lukashev, E.
P., Bolychevtseva, Y. V., Yanushin, M. F., and Stadnichuk, I. N. (2016)
Functions of chromophore-containing domain in the large linker
LCM-polypeptide of phycobilisome, Dokl. Biochem. Biophys.,
471, 403-406.
112.Mimuro, M., and Gantt, E. (1986) A high
molecular weight terminal pigment (“anchor polypeptide”)
and a minor blue polypeptide from phycobilisomes of the cyanobacterium
Nostoc sp. (MAC): isolation and characterization, Photosynth.
Res., 10, 201-208.
113.Gambetta, G. A., and Lagarias, J. C. (2001)
Genetic engineering of phytochrome biosynthesis in bacteria, Proc.
Natl. Acad. Sci. USA, 98, 10566-10571.
114.Miao, D., Ding, W. L., Zhao, B. Q., Lu, L., Xu,
Q. Z., Scheer, H., and Zhao, K. H. (2016) Adapting photosynthesis to
the near-infrared: non-covalent binding of phycocyanobilin provides an
extreme spectral red-shift to phycobilisome core–membrane linker
from Synechococcus sp. PCC7335, Biochim. Biophys. Acta,
1857, 688-694.
115.Ma, Q., Zhou, N., and Zhou, M. (2016) Energy
transfer between fusion biliproteins co-expressed with phycobiliprotein
in Escherichia coli, Protein Expr. Purif.,
126, 84-88.
116.Wu, Y. P., and Krogmann, D. W. (1997) The
orange carotenoid protein of Synechocystis PCC 6803, Biochim.
Biophys. Acta, 1322, 1-7.
117.Thurotte, A., Bourcier de Carbon, C., Wilson,
A., Talbot, L., Cot, S., Lopez-Igual, R., and Kirilovsky, D. (2017) The
cyanobacterial fluorescence recovery protein has two distinct
activities: orange carotenoid protein amino acids involved in FRP
interaction, Biochim. Biophys. Acta, 1858,
308-317.
118.Lu, Y., Liu, H., Saer, R., Li, V. L., Zhang,
H., Shi, L., Goodson, C., Gross, M. L., and Blankenship, R. E. (2017) A
molecular mechanism for nonphotochemical quenching in cyanobacteria,
Biochemistry, 56, 2812-2823.
119.Moldenhauer, M., Sluchanko, N. N., Buhrke, D.,
Zlenko, D. V., Tavraz, N. N., Schmitt, F.-J., Hildebrandt, P.,
Maksimov, E. G., and Friedrich, T. (2017) Assembly of photoactive
orange carotenoid protein from its domains unravels a carotenoid
shuttle mechanism, Photosynth. Res., 133, 327-341.
120.De Carbon, C. B., Thurotte, A., Wilson, A.,
Perreau, F., and Kirilovsky, D. (2015) Biosynthesis of soluble
carotenoid holoproteins in Escherichia coli, Sci.
Rep., 5, 9085.
121.Melnicki, M. R., Leverenz, R. L., Sutter, M.,
Lopez-Igual, R., Wilson, A., Pawlowski, E. G., Perreau, F., Kirilovsky,
D., and Kerfeld, C. A. (2016) Structure, diversity, and evolution of a
new family of soluble carotenoid binding proteins in cyanobacteria,
Mol. Plant, 9, 1379-1394.
122.Lechno-Yossef, S., Melnicki, M. R., Bao, H.,
Montgomery, B. L., and Kerfeld, C. A. (2017) Synthetic OCP heterodimers
are photoactive and recapitulate the fusion of two primitive
carotenoproteins in the evolution of cyanobacterial photoprotection,
Plant J. Cell Mol. Biol., 91, 646-656.
123.Liu, H., Lu, Y., Wolf, B., Saer, R., King, J.
D., and Blankenship, R. E. (2017) Photoactivation and relaxation
studies on the cyanobacterial orange carotenoid protein in the presence
of copper ion, Photosynth. Res., doi:
10.1007/s11120-017-0363-1.
124.Kerfeld, C. A., Melnicki, M. R., Sutter, M.,
and Dominguez-Martin, M. A. (2017) Structure, function and evolution of
the cyanobacterial orange carotenoid protein and its homologs, New
Phytol., 215, 937-951.
125.Lopez-Igual, R., Wilson, A., Leverenz, R. L.,
Melnicki, M. R., Bourcier de Carbon, C., Sutter, M., Turmo, A.,
Perreau, F., Kerfeld, C. A., and Kirilovsky, D. (2016) Different
functions of the paralogs to the N-terminal domain of the orange
carotenoid protein in the cyanobacterium Anabaena sp. PCC 7120,
Plant Physiol., 171, 1852-1866.
126.Mares, J., Hrouzek, P., Kana, R., Ventura, S.,
Strunecky, O., and Komarek, J. (2013) The primitive thylakoid-less
cyanobacterium Gloeobacter is a common rock-dwelling organism,
PLoS One, 8, e66323.
127.Bernat, G., Schreiber, U., Sendtko, E.,
Stadnichuk, I. N., Rexroth, S., Rogner, M., and Koenig, F. (2012)
Unique properties vs. common themes: the atypical cyanobacterium
Gloeobacter violaceus PCC 7421 is capable of state transitions
and blue-light-induced fluorescence quenching, Plant Cell
Physiol., 53, 528-542.
128.Kumar, S., Stecher, G., and Tamura, K. (2016)
MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger
datasets, Mol. Biol. Evol., 33, 1870-1874.
129.Watanabe, M., and Ikeuchi, M. (2013)
Phycobilisome: architecture of a light-harvesting supercomplex,
Photosynth. Res., 116, 265-276.