REVIEW: Enteroviruses: Classification, Diseases They Cause, and Approaches to Development of Antiviral Drugs
O. S. Nikonov, E. S. Chernykh, M. B. Garber, and E. Yu. Nikonova*
Institute of Protein Research, Russian Academy of Sciences, 142290 Pushchino, Moscow Region, Russia; E-mail: katya_nik@vega.protres.ru* To whom correspondence should be addressed.
Received June 13, 2017
The genus Enterovirus combines a portion of small (+)ssRNA-containing viruses and is divided into 10 species of true enteroviruses and three species of rhinoviruses. These viruses are causative agents of the widest spectrum of severe and deadly epidemic diseases of higher vertebrates, including humans. Their ubiquitous distribution and high pathogenicity motivate active search to counteract enterovirus infections. There are no sufficiently effective drugs targeted against enteroviral diseases, thus treatment is reduced to supportive and symptomatic measures. This makes it extremely urgent to develop drugs that directly affect enteroviruses and hinder their development and spread in infected organisms. In this review, we cover the classification of enteroviruses, mention the most common enterovirus infections and their clinical manifestations, and consider the current state of development of anti-enteroviral drugs. One of the most promising targets for such antiviral drugs is the viral Internal Ribosome Entry Site (IRES). The classification of these elements of the viral mRNA translation system is also examined.
KEY WORDS: IRES, enteroviruses, Picornaviridae, translation initiation, drug design, taxonomyDOI: 10.1134/S0006297917130041
Abbreviations: IRES, internal ribosomal entry sites; ITAF, IRES trans-acting factors.
Enterovirus is a genus of ubiquitous small (with capsids
15-30 nm in diameter) RNA-containing viruses. These viruses infect
higher vertebrates and cause a wide spectrum of diseases. These
diseases may appear as short-duration sickness or may cause incurable
damage in an infected organism, central nervous system diseases,
paralysis, swelling, and even death. Exactly these viruses posed (and
do so now) a threat of poliomyelitis outbreaks. They are also a cause
of other severe deadly diseases such as aseptic meningitis, enteroviral
encephalitis, and enteroviral vesicular stomatitis. At the same time,
the common cold is also caused by viruses belonging to this genus.
Enteroviruses are readily transmitted from person to person through an
air and/or via a fecal-oral route. Infection through contaminated
objects is also possible. A serious threat is posed by the long
asymptomatic virus shedding, which provides the possibility for sudden
epidemic bursts of enteroviral infections on different continents and
complicates their prediction.
INTERNAL TRANSLATION INITIATION OF VIRAL RNAs IN CELL
After virus penetration into a cell, the RNA molecule released from the capsid triggers a cascade of events that result in formation of mature viral progeny and eventually cell death. These events begin with synthesis of viral proteins, i.e. with the translation of viral RNA by the cellular translation system. Expression of viral genes is often regulated at the level of initiation of mRNA translation. At this step, the 40S ribosomal subunit binds to an mRNA and scans it in the 5′-3′ direction until it reaches the start codon, where the 80S ribosome is to be assembled. Various host proteins and cis-acting RNA molecules participate in this process. A cap structure is present at the 5′-terminus of most eukaryotic mRNAs, which participates in capturing 40S ribosomal subunits. The scanning mechanism implies that the ribosome initiates translation at the first AUG codon. This is the case for most mRNAs. However, the first AUG codon may be ignored if it is in a non-optimal sequence context. In this case, translation is initiated at the next AUG codon. This initiation mechanism is referred to as leaky scanning. It is realized in many viruses, which allows saving coding sequence length.
Picornavirus mRNAs ((+)ssRNA viruses) lack the cap structure. In 1988, it was demonstrated that initiation of translation of such uncapped mRNAs is implemented via a structural feature in mRNA molecule, which allows assembly of the translation apparatus near a start codon. These stable secondary structure elements were called internal ribosomal entry sites (IRESs) [1, 2]. Since then, such cap-independent translation initiation pathway has been extensively studied [3]. This discovery overturned a major dogma in translation initiation stating that the eukaryotic ribosome can bind mRNA exclusively at the 5′-terminus. IRESs are usually situated in the 5′-untranslated region and frequently have a complex secondary and tertiary structure. Since these elements were discovered in picornaviruses, they have also been found in several other viral mRNAs. The mechanism of IRES-dependent translation is presumably exploited by some cellular mRNAs. Translation of these mRNAs continues when cap-dependent translation is repressed, which may happen during endoplasmic reticulum stress, hypoxia, starving for nutrients, mitosis, and cell differentiation [4, 5]. In addition to the above-mentioned picornaviruses, initiation of translation at internal sites is utilized in representatives of Flaviviridae [6], Retroviridae, Dicistroviridae [7], Herpesviridae [8], some insect viruses [9], and plants viruses [10], and also retrotransposons in insects and rodents [11].
However, it appeared that, unlike cap-dependent translation initiation (scanning), there is no common mechanism for functioning of all IRESs. Furthermore, the IRESs are very different: no structural element has been found that is shared by all IRESs. Their sequences also lack significant homology [3, 12]. However, it was shown that the majority of viral IRESs have stable secondary and tertiary structure that facilitates their efficient binding to the 40S subunit. Such binding can be either direct or require participation of additional canonical translation initiation factors along with some other host proteins referred to as ITAF (IRES trans-acting factors). It is possible that some ITAF directly participate in specific interaction of mRNA with the 40S subunit, whereas others stabilize specific functionally active IRES conformations [13-15]. Unlike viral mRNAs, existence of IRES-dependent translation of cellular mRNAs is currently being vigorously discussed [4, 5, 16].
CLASSIFICATION OF PICORNAVIRUS IRESs
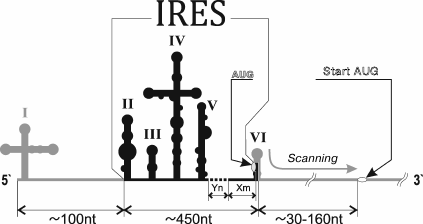
Since the discovery of viral IRESs, difficulties in their classification have arisen due to their dissimilarity. However, extensive studies on viruses, their mRNA, and mechanisms of its translation revealed several common features that may be used to clearly distinguish between the IRES types. The viral IRESs that are now classified according to their sequence and secondary structure are divided into separate families: 1 – intergenic IRESs of dicistroviruses of invertebrates (Dicistroviridae family; for instance, the cricket paralysis virus); 2 – IRES of hepatitis C (HCV) and related viruses of animals (Flaviviridae family), and 3 – IRESs of picornaviruses that are in turn divided into five classes (I-V). Besides, there are polypurine A-rich IRESs (PARS) [17]. An IRES of this type was first discovered in tobamovirus CrTMV [18]. However, we return to reviewing picornavirus IRESs. Picornavirus type I IRESs include IRESs of all representatives of the genus Enterovirus and a single representative of the genus Harkavirus [19]. Length of such IRES is approximately 450 nucleotides (nt). It comprises domains two to six (Fig. 1); it contains Yn-Xm-AUG at the 3′-terminus, where Yn – pyrimidine sequence (n = 8-10 nt), X – a linker between Yn and the AUG triplet (m = 18-20 nt) [20-22]. This motif is considered to be a region of ribosome-binding at the 5′-UTR [23]. It is separated from the start codon with a non-conserved region whose length ranges from <30 nt in rhinoviruses to >150 nt in poliovirus. Conserved functionally important nucleotides are situated at the base of the second domain, in the cruciform fourth domain, and in the fifth domain [12, 19, 24]. Specific binding of the fifth domain to eIF4G and RNA helicase eIF4A promotes binding of the 43S complex to the IRES [12]. Translation initiation on all the studied type I IRESs depends on the presence of ITAF, whose complete list is not yet determined (Fig. 2).
Fig. 1. Scheme of a type I IRES structure.
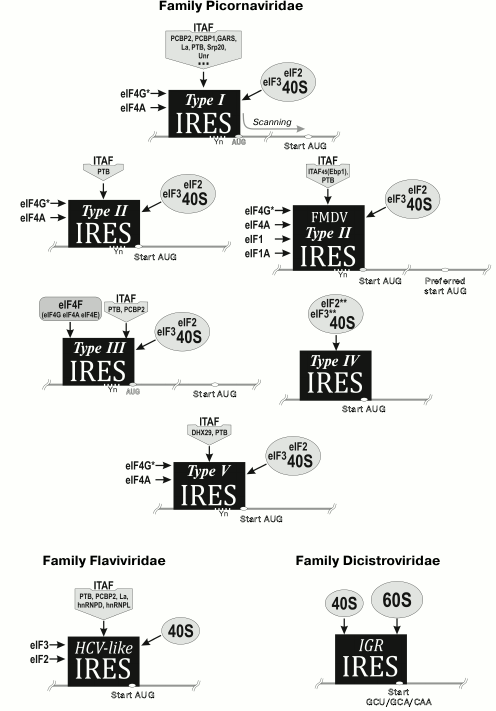
Fig. 2. Main types of classified viral IRESs.
The type II IRES was discovered in representatives of genera Cardiovirus, Aphthovirus, Avisivirus, Cosavirus, Erbovirus, Hunnivirus, Mischivirus, Rosavirus, Parechovirus (Parechovirus A, Parechovirus B), Sicinivirus (http://www.picornaviridae.com). This IRES is approximately 450 nt long and contains a Yn-Xm-AUG motif at the 3′-terminus. However, in this case the AUG codon can be the start codon. IRESs of this type also contain five domains (H, I, J-K, and L), but they do not resemble domains of the type I IRES, excluding domain I, which like the fourth domain of type I IRES contains a C-rich loop and GNRA tetraloop [19]. Initiation of translation on type II IRES requires specific binding of factors eIF4G and eIF4A with domains J-K [25, 26]. At the same time, such IRESs can function without eIF4E and factors participating in ribosome scanning (eIF1 and eIF1A). Hence, type II IRES differs from type I IRES by the lack of ITAF requirement (with one exception: it has been demonstrated that in certain cases, cellular RNA-binding protein PTB (pyrimidine-binding protein) is required) [25, 26] (Fig. 2).
The type III IRES is only found in hepatitis A virus. It is around 410 nt long [27]. IRESs of this type significantly differ from the first two types both by sequence and structural elements. Efficiency of the translation initiation on this IRES is considerably lower compared to the first two IRES types. The hepatitis A IRES requires the host cap-binding protein eIF4E for functioning, though the exact reason for that is not clear [28] (Fig. 2).
The type IV IRES is found in representatives of genera Kobuvirus (Aichivirus C (porcine kobuvirus)), Teschovirus, Sapelovirus, Senecavirus, Tremovirus, Limnipivirus, Megrivirus, Parechovirus (Ferret parechovirus), Pasivirus, Sakobuvirus, and Avihepatovirus. This type of IRES is approximately 330 nt long. It resembles the hepatitis C virus IRES (family Flaviviridae). Viruses with this type of IRES directly bind eIF3 and the 40S subunit facilitating formation of the 48S initiation complex. They do not need factors that are required for binding of the pre-initiator 43S complex to mRNA (eIFs 4A, 4B, 4E, or 4G) or for scanning (eIF1 and eIF1A) [25, 29-31] (Fig. 2).
Recently, the type V IRES was discovered. It was found in representatives of genera Oscivirus, Kobuvirus (Aichivirus A, Aichivirus B), and Salivirus. This type of IRES is a “hybrid”: its central domain is homologous to the fourth domain of the type I IRESs, whereas the next domain that binds to eIF4G is homologous to domain J of the type II IRESs [32, 33]. The start codon of the viral polyprotein is a part of the Yn-Xm-AUG motif and is situated in the stable hairpin of domain L. Translation initiation on these IRESs requires participation of an ATP-dependent RNA helicase DHX29, PTB, and a set of canonical factors (Fig. 2).
The simplest mechanism of translation initiation is a hallmark of an intergenic 180 nt long IRES of representatives of dicistroviruses (for instance, the intergenic IRES of the cricket paralysis virus). Like the hepatitis C IRES, it forms a complex tRNA-like structure, binds directly to the ribosome (P-site), and triggers initiation without involving eukaryotic translation initiation factors [25, 34] (Fig. 2).
CLASSIFICATION OF ENTEROVIRUSES HAVING TYPE I IRES
The modern classification of enteroviruses was accepted in 2012 and published as an update to the 9th issue of virus taxonomy from the International Committee on Taxonomy of Viruses. Since then, corrections to this classification have been issued [35-39].
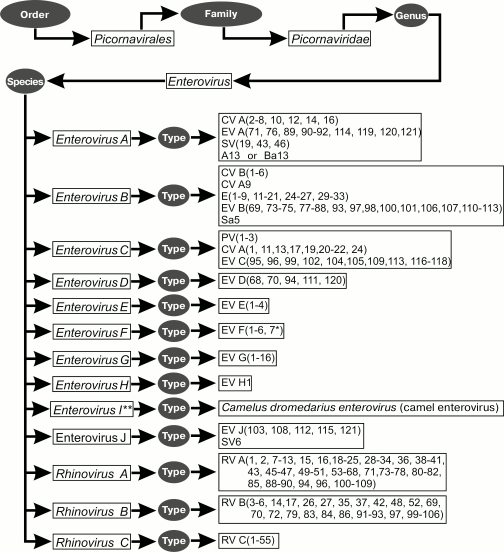
At present, genus Enterovirus belonging to family Picornaviridae includes nine enterovirus species (namely, Enterovirus A, B, C, D, E, F, G, H, and J) and three rhinovirus species (Rhinovirus A, B, and C). A new enterovirus was discovered in camels in 2015, which is apparently the first representative of a new species, Enterovirus I (Fig. 3).
Fig. 3. Taxonomy of enteroviruses.
The species Enterovirus A includes 25 (sero)types: coxsackievirus A2 (CV-A2), CV-A3, CV-A4, CV-A5, CV-A6, CV-A7, CV-A8, CV-A10, CV-A12, CV-A14, CV-A16, enterovirus A71 (EV-A71), EV-A76, EV-A89, EV-A90, EV-A91, EV-A92, EV-A114, EV-A119, EV-A120, EV-A121, simian enteroviruses SV19, SV43, SV46, and baboon enterovirus A13 (BA13).
The species Enterovirus B is one of the most numerous. It consists of 63 (sero)types: coxsackievirus B1, CV-B2 – B6, CV-A9, echovirus 1 (E-1), E-2 – E-7, E-9, E-11 – E-21, E-24 – E-27, E-29 – E-33, enterovirus B69 (EV-B69), EV-B73 – EV-B75, EV-B77 – EV-B88, EV-B93, EV-B97, EV-B98, EV-B100, EV-B101, EV-B106, EV-B107, EV-B110 (from chimpanzee), EV-B111, EV-B112 (from chimpanzee), EV-B113 (from mandrill), and simian enterovirus SA5.
The species Enterovirus C includes 23 (sero)types: poliovirus (PV) 1, PV-2, PV-3, coxsackievirus A1 (CV-A1), CV-A11, CV-A13, CV-A17, CV-A19, CV-A20, CV-A21, CV-A22, CV-A24, EV-C95, EV-C96, EV-C99, EV-C102, EV-C104, EV-C105, EV-C109, EV-C113, EV-C116, EV-C117, and EV-C118.
The species Enterovirus D is relatively uncommon; it includes five (sero)types: EV-D68, EV-D70, EV-D94, EV-D111 (from human and chimpanzee), and EV-D120 (from gorilla).
The species Enterovirus E includes bovine enterovirus group A: from EV-E1 to EV-E4.
The species Enterovirus F includes bovine enterovirus group B (at present, six types are described): from EV-F1 to EV-F6.
The species Enterovirus G consists of 16 (sero)types: from EV-G1 to EV-G16.
The species Enterovirus H includes three monkey viruses isolated in 1950 (SV4, SV28, and SA4) and A-2 plaque virus. However, these four viruses were joined into a single (sero)type enterovirus H1 (EV-H1) due to their strong similarity at the molecular level.
The species Enterovirus J contains six simian enterovirus species: SV6, EV-J103, EV-J108, EV-J112, EV-J115, and EV-J121.
The species Rhinovirus A is the most numerous; it contains 80 (sero)types: rhinovirus (RV) A1, A2, A7-A13, A15, A16, A18, A19-A25, A28-A36, A38-A41, A43, A45-A47, A49-A51, A53-A68, A71, A73-A78, A80-A82, A85, A88-A90, A94, A96, and A100-A109.
The species Rhinovirus B consists of 32 (sero)types: rhinovirus (RV) B3-B6, B14, B17, B26, B27, B35, B37, B42, B48, B52, B69, B70, B72, B79, B83, B84, B86, B91-B93, B97, and B99-B106.
As to viruses belonging to the species Rhinovirus C, despite its difference from Rhinovirus A and Rhinovirus B, until recently there were difficulties and misunderstanding in its classification and discriminating the individual types within the species. Some authors on the basis of numerous tests considered these viruses closely related to Rhinovirus A and called all of them HRV-A2 [40, 41]. Others preferred to refer to these viruses as HRV-C [42-45] or HRV-X [46]. Since 2010, based on significant phylogenetic clustering of the considered enterovirus species, reasonable suggestions appeared for creating a genetics-based system that would allow discriminating its types similarly to (sero)types of other enterovirus species [47, 48]. By now, this species consists of 55 (sero)types (C1-C55).
In addition, the genus includes several yet unclassified enteroviruses: one monkey enterovirus (SV-47) (it was not assigned to a certain species as its genome is not sequenced) and EV-122 and EV-123, which do not match any of the existing species.
Thus, the genus Enterovirus includes many viruses including those highly dangerous for humans. At the same time, these viruses are widespread and highly resistant to the action of physicochemical factors.
DISEASES CAUSED BY VIRUSES BELONGING TO GENUS
Enterovirus
At the beginning, enteroviral infections in humans were classified as acute respiratory diseases caused by intestinal viruses. It was generally accepted to discriminate infections caused by polioviruses (certain (sero)types of Enterovirus C) to a separate group called poliomyelitis. We have reviewed polioviruses and their current position in the viral taxonomy. The disease they cause was known already in ancient Egypt [49, 50].
In 1840, the German orthopedist Jacob von Heine discriminated poliomyelitis as a separate disease. In 1890, the Swedish pediatrician O. Medin suggested infectious nature of this disease based on its epidemic dissemination pattern. Poliomyelitis mainly affects children under 5 years old. Unfortunately, there is no antiviral drug for poliomyelitis treatment, only prevention is possible. This disease has several clinical forms.
The abortive form proceeds with no symptoms of nervous system damage. This form of poliomyelitis is called a minor illness as it passes relatively gently, lasts around one week, and ends by recovery [51].
The nonparalytic form is serous meningitis caused by poliovirus. The disease proceeds significantly more severely than the abortive form with a complete set of meningeal symptoms. However, it also passes favorably and patients make a full recovery [51].
The paralytic form of poliomyelitis is the most dangerous [52]. The above-mentioned forms may convert into the paralytic form upon adverse development of the disease. However, it should be mentioned that this form develops in only 1% of patients. The disease may proceed rapidly, and general paralysis may occur within hours due to damage to the central nervous system. The recovery period may last up to 2 years. It is followed by the aftermath stage with stable paralyses, contractures, and deformations. Irreversible paralysis (usually in legs) occurs in one out of 200 patients. Mortality in this group of patients reaches 10% due to further development of paralysis and its expansion to the respiratory muscles.
In turn, the paralytic form is also divided into several kinds or forms [53].
The spinal form of paralytic poliomyelitis is the most common. It is characterized by lesion of predominantly the lumbar section of the spinal cord. Cervical and other sections are damaged less often. However, lesion of cervical and thoracic sections of the spinal cord is the most severe as it may cause paralysis of the respiratory muscles and thus disturb respiration. The most dangerous in this respect is diaphragmatic paralysis.
The pontine form occurs upon lesion of the bridge of Varolius [54]. It can be isolated or be accompanied by damage to the spinal cord (pontospinal form) or medulla (pontobulbar form). It is characterized by facial muscle paralysis [55]. Most patients make a full recovery, which begins at 10-14 days of the disease.
The bulbar form occurs in 10-15% cases of paralytic poliomyelitis. In this case, bulbar and glossopharyngeal nerves are affected [56]. The disease proceeds rapidly and is characterized by very serious general condition. Fast development of paralysis of corresponding muscle groups is typical. Pharyngeal paralysis (lesion of the palate and the larynx) may develop, leading to disturbance of respiration and upper airway obstruction with saliva, mucus, and sputum. Earlier, this pharyngeal form of poliomyelitis was characterized by high mortality. At present, however, the pharyngeal paralysis may have a good prognosis and with recovery without consequences if it is treated in due time and correctly. In some patients, pharyngeal paralysis is combined with other disorders (spinal, oblongata). Development of collateral laryngeal paralysis (lesion of the larynx and ligaments) is possible. The acute form of this paralysis can cause sudden asphyxiation and cyanosis. The respiratory center may be affected in bulbar poliomyelitis, which results in disturbance in breathing rhythm and frequency and appearance of other breathing pathologies. Breathing disorders are accompanied with vasomotor and vegetative disorders. Early lesion of the vasomotor center may cause death due to sudden decrease in arterial pressure and cardiac arrest. In approximately half of lethal cases evoked by this form of poliomyelitis, an acute interstitial myocarditis is registered.
A rare encephalitic form of poliomyelitis with high mortality has also been described. This form proceeds rapidly, mental confusion developing very fast and transforming into stupor and coma [53].
Poliomyelitis in pregnant patients is considered separately [57]. During the first half of pregnancy, the disease may cause miscarriage, or preterm birth if infection occurred later. However, most women infected with poliomyelitis during pregnancy give birth in due time without obstetric surgery. Typically, the fetus is affected by intoxication and hypoxia rather than by direct transmission of the infection. Respiratory disorders pose an elevated threat, which remains even after the acute phase of the disease is passed, for pregnant woman as well.
Poliomyelitis outbreaks were widespread from the end of the 19th century. At the middle of the 20th century, anti-poliomyelitis vaccines appeared and were widely used. In 1988, the World Health Organization set the task of elimination of poliomyelitis worldwide by the 2000 [58]. Active prevention measures with wide use of vaccines decreased the incidence level by 99% (as of 1988) [59]. Currently, high risk of poliomyelitis outbreaks remains only in Afghanistan and Pakistan. In 2013, a new strategic plan for elimination of poliomyelitis by 2018 was presented at the global vaccine summit in Abu-Dhabi (United Arab Emirates).
The first reliable mention of the disease caused by non-polio enteroviral infection occurred in 1856. Exactly in this year, there was an outbreak of pleurodynia in Iceland, which was described later in 1874. The first publication devoted to this illness is dated to 1872, when a Norwegian medical journal first published a communication of Dr. A. Daae to Dr. C. Homann titled “Epidemics of acute muscular rheumatism transmitted through the air in Drangedal”. The Norwegian name for this illness is “Bamble disease” after the place it first appeared [60]. Later, it was referred to as Bornholm disease after the Danish island Bornholm, whereas it is now known as epidemic myalgia. Normally, the disease is caused by coxsackievirus B infection. More rarely it may be caused by coxsackieviruses A and certain (sero)types of echovirus [61]. It evokes myositis of the upper abdominal muscles and pectoral muscles, fever, and headache. Typically, the prognosis is positive – the patient recovers in 7-8 days. However, there are possible severe complications (including, though rarely, aseptic meningitis) up to lethal outcome.
Viral myopericarditis is a combination of myocarditis and pericarditis, which involves inflammation of both the cardiac muscle and the serous layer of pericardium. In infants, typically, myocarditis is developed, whereas pericarditis is more common in children and adults. Incorrect or late treatment may result in death. The frequent cause of myopericarditis is coxsackievirus B [62, 63] infection or coinfection with coxsackieviruses A and B [64]. Echoviruses may also evoke this disorder [65, 66]. The disease may proceed without symptoms or be accompanied with chest pains, vertigo, general weakness, arrhythmia, heart failure, fever, diarrhea, and sore throat. Swelling in the hands and legs may also occur. Sometimes, myopericarditis causes sudden loss of consciousness, which may be associated with abnormal heart rhythms. Breathing difficulties may occur in children. Viral myopericarditis may convert into acute myocardial infarction [67].
Acute hemorrhagic conjunctivitis or enteroviral hemorrhagic conjunctivitis is a highly contagious ophthalmic infection that first appeared in 1969-1970 [68, 69]. It proceeds with visible hyperemia, chemosis, eye irritation, photophobia, eye discharge, and subconjunctival hemorrhage. These symptoms appear along with general symptoms (preauricular adenopathy, headache, increased body temperature, tracheobronchitis, etc.) [70]. Recovery occurs in 7-10 days. Causative agents for acute hemorrhagic conjunctivitis are enterovirus 70 and coxsackievirus A24 [71].
The most frequent manifestation of enteroviral infections is a nonspecific febrile illness. Usually, these infections are well-tolerated and pass within a week. The disease may proceed in two phases [72]. Acute respiratory viral infections (ARVI) are also ascribed to low-hazard enteroviral infections caused by rhinoviruses and known as nasopharyngitis, rhinopharyngitis, rhinovirus infection, rhinonasopharyngitis, epipharyngitis, and the common cold [73]. However, some respiratory enteroviral infections (for instance, those caused by enterovirus 68) may lead to serious consequences resulting in severe complications such as pneumonia [74].
Aseptic meningitis is a viral infectious disease that affects humans of all ages. However, individuals under 30 years old are more susceptible. The most common cause for this disease is non-polio enteroviruses [75], namely coxsackieviruses A and B, echoviruses, and enteroviruses 69 and 73 [76, 77]. During infection, the meninges are affected. Patients suffer from headache, fever, muscle aches, stomach aches, and stiff neck. Other possible symptoms are light sensitivity, rush, nausea, diarrhea, sore throat, and cough. As a rule, this illness has good prognosis and passes without consequences in 7-10 days. However, especially in newborns, the infection may develop to symptoms of encephalitis with focal neurologic signs and cramps. In this case, prognosis may be very poor up to lethal outcome caused by heart failure or liver damage [78]. Such infectious damage of the central nervous system in children may be associated also with enterovirus A71. In this case, the disease proceeds in more severe form and may evoke paresis and brainstem encephalitis [79].
Herpangina is an acute infectious disease caused mainly by coxsackieviruses A, which affects 3-10 years old children. This disease may be caused also by enterovirus A71 [80-82]. Distinctive symptom of herpangina is appearance of vesicles with serous contents on the soft palate, tonsils, and back of throat. They are small and resemble herpetic damage. Usually, vesicles open rapidly, dry up with a crust formation, and then heal. Upon bacterial coinfection, they may suppurate or ulcerate. The disease proceeds with general symptoms such as fever, headache, rhinitis, hypersalivation, severe sore throat, hyperemia, and pain during palpation of regional lymph nodes. The disease usually passes in a few days.
Enteroviral vesicular stomatitis (hand, foot, and mouth disease, HFMD) is an acute disease caused by coxsackieviruses A, B, and enterovirus A71. It predominantly occurs in children under 10 years old. However, it may also affect adults [83, 84].
Incubation period lasts approximately 3-6 days. During the prodromal period (12 to 36 h), patients experience such symptoms as cough, sore throat, general illness, and loss of appetite. After that, vesicular rash appears on hands, legs, and oral cavity. If the disease goes on auspiciously, it ends in 5-7 days. However, sometimes (especially if infection is caused by enterovirus A71) it may lead to severe neurological complications such as encephalitis, meningitis, and paralyses like those caused by poliovirus. This form is highly severe and features high mortality. From 2008 to 2012, over seven million cases were registered in China. It was fatal in 2457 cases [85].
Enteroviral encephalitis amounts to approximately 5% of cases of enteroviral infections [86]. The main cause of this severe neurological disease is coxsackieviruses A and B, echoviruses [87], and enterovirus A71 [86]. The disease involves inflammation of the brain. It is accompanied by fever, vomiting, headache, and weakness. Disorders of consciousness, cramps, behavior disorder, and paresis may occur. Severe disease may result in coma. Acute cerebellar ataxia, drop attacks, and hemichorea may occur in children. Several clinical types of enteroviral encephalitis are discriminated according to localization of inflammation: brainstem, cerebellar, hemispheric. The cerebellar form is the most auspicious, which ends with full recovery [88]. However, enteroviral encephalitis is a deadly disease [89]. Encephalitis caused by enterovirus A71 infection usually has brainstem clinical features and high mortality [86, 90].
Polio-like illnesses, acute flaccid paralysis and acute paralytic poliomyelitis of non-polio etiology, are diseases having symptoms similar to those of poliomyelitis but caused by other viruses, namely enteroviruses 68-71, coxsackieviruses, and echoviruses [91, 92]. These diseases affect predominantly children. The most severe forms are commonly caused by enterovirus A71 [93, 94]. Lesion of the central nervous system occurring during development of severe forms of the diseases, similarly to poliomyelitis, may evoke very serious consequences including fatal outcome [95].
Enteroviral infections are dangerous not only for humans. Many animal species are susceptible to these viruses, especially higher mammals. Enteroviral infections of animals may worsen life of pets and even cause significant damage to entire branches of agriculture associated with livestock farming. Cases of severe gastroenteritis caused by enteroviral infections leading to 50% mortality in young stock were registered in poultry farm birds [96]. Livestock farming suffers from outbreaks of enteroviral infections causing high mortality in farm animals (for instance, pigs) [97]. In addition, entire populations of rare or endangered species become victims of these infectious diseases. Even dolphins are susceptible to these infections [98]. Thus, counteraction to spread of these diseases is of great importance.
Early diagnosis of enteroviral infection followed by antiviral therapy may prevent occurrence of severe complications in patients. However, now there are no highly efficient and widely used anti-enteroviral preparations. Therefore, treatment of enteroviral infections is limited to a complex of procedures for relieving the general condition of patients, counteracting concomitant bacterial infections, and minimizing possible complications. Viral infection as such must be dealt with by the immune system of the patient. Therefore, there is a great need for development of highly efficient antiviral agents for treatment of enteroviral infections.
APPROACHES TO DEVELOPING ANTIVIRAL DRUGS
The life cycle of enteroviruses includes virus adsorption, release of genetic material from the envelope, RNA translation, maturation of viral proteins, replication of viral RNA, and virus assembly. Any of these stages can be a target for antiviral agents.
The enterovirus envelope consists of four viral proteins (VP1-VP4). VP1 is one of the most frequently used targets in counteracting enteroviral infections. A great number of chemical compounds have antiviral properties in vitro through interaction with VP1 and prevention of virus adsorption or release of viral RNA from the envelope. One of the most successful experiences in designing antiviral preparation that interacted with the viral envelope was pleconaril [99]. It inhibited replication of several enteroviral (sero)types by 50% (though it did not affect EV-A71) [100]. It was demonstrated that intake of this drug alleviated disease passage [101]. Although the preparation has numerous negative side effects and it did not pass clinical trials yet, it served as a basis for developing other more efficient and less toxic antiviral drugs [102]. Preparations based on other drugs that interact with viral capsid, but have not passed clinical trials due to side effects, are also being developed [103].
Viral proteases are also targets in drug design. Proteins 2A and 3C are proteases of picornaviruses that play an important role in the processing of the viral polyprotein. In addition, they affect host cap-dependent protein synthesis by cleaving elongation factor eIF4GI/II [104], disturb nuclear transport [105], and impair cellular splicing and transcription [106]. Successful propagation of many viruses depends on correct processing of the viral polyprotein. For instance, polyprotein EV-A71 is cleaved by viral proteases, giving rise to four structural envelope proteins (VP1-VP4) and seven nonstructural proteins (2Apro, 2B, 2C, 3A, 3B, 3Cpro, and 3Dpol), which are required for virus replication [107]. During translation, 2A protease cleaves its own N-terminus from C-terminus of VP1, thus separating capsid protein precursor from precursor of replicative proteins. However, 3C protease in considered the main viral protease, as it is responsible for cleaving the other linkers joining proteins in the viral polyprotein [108, 109].
It was shown that alkylating agents (iodoacetamide and N-ethylmaleimide) reduce activity of 2A protease [110]. Caspase inhibitors also may block 2A protease of rhinoviruses and coxsackievirus 2A both in vitro and in vivo [111]. Some antiviral agents affecting 3C protease were obtained based on the substrate of this protease. Many of them are peptides comprising 3-5 amino acid residues (a.a.), aldehyde groups of which are used as electrophilic anchors [112]. Some peptide inhibitors of the 3C protease were modified so that they could form irreversible covalent bonds with the protease [113]. Such agents possess high antiviral activity toward rhinoviruses CV-A21, CV-B3, and EV-A70, and echovirus 11.
It is known that some alkaloids have antiviral properties. So, lycorine (an alkaloid of the family Amaryllidaceae) having a broad spectrum of biological activities inhibits development of polioviruses and EV-A71 by affecting, in particular, 2A protease [114].
Rupintrivir was initially designed as an inhibitor of rhinoviral 3C protease. Later it demonstrated antiviral activity toward other representatives of the family Picornaviridae. Derivatives of this preparation were also able to inhibit enteroviruses EV-A71 and CV-A16 [115].
The following group includes antiviral drugs affecting viral proteins involved in replication of viral RNA, or cell systems that are used by viruses for RNA replication. Replication of viral RNA occurs with the participation of replicative complex comprising various viral proteins: 2B, 2C, 2BC, 3A, 3B, 3AB, 3CD, and 3D. Some of these were tested as targets for antiviral agents. The preparations obtained featured a narrow activity spectrum and also had side effects [116]. For example, 5-(3,4-dichlorophenyl)methylhydantoin (a hydantoin derivative) inhibits replication of EV-A71 RNA. The exact mechanism of this effect is to be studied, but, apparently, the process of viral RNA replication is disturbed due to interaction of the preparation with the capsid protein VP3, thus blocking activity of 2C protein, which also directly interacts with VP3 [117]. Compound BPR-3P012 (6-bromo-2-[1-(2,5-dimethylphenyl)-5-methyl-1H-pyrazol-4-yl]quinoline-4-carboxylic acid) interacts with viral RNA-dependent RNA polymerase (3D) and inhibits translation of EV-A71 RNA [118]. Already known drugs frequently possess antiviral activity by affecting replication of viral RNA. For instance, isoflavone formononetin, which is commonly obtained from red clover [119], or antifungal broad-spectrum preparation itraconazole [120]. Besides, experiments are carried out for discovery of antiviral preparations based on noncoding regulatory microRNAs, which are also used for vaccine preparation [121].
IRESs OF VIRAL RNAs AS A PHARMACEUTICAL TARGET
A promising target for antiviral drugs is the internal ribosome entry site (IRES) on viral mRNA. A region of the 5′-UTR of viral mRNA, on which the preinitiation complex assembles, plays a pivotal role in regulation of its translation [122]. Viral IRESs differ from cellular IRESs by the presence of highly ordered secondary structures, a set of factors used for the translation initiation, and requirement for IRES trans-acting factors (ITAF). Therefore, the process of translation initiation of viral mRNA is a promising target for pharmacological action. Besides, it was demonstrated that mutations in IRES affecting interaction of ITAF with viral mRNA also affect viral tissue tropism [123, 124]. IRESs of some viruses may act as chaperones influencing development of viral infection not only during the initiation of translation of viral mRNA [125]. As soon as viral IRESs were discovered, attempts were made to use them for therapy [126-129]. The main efforts were focused on developing a compound that would be able to modify IRES structure to make it ineligible for initiation of protein synthesis, or disturbing its interaction with the ribosome, translation initiation factors, and ITAF [126-128, 130-133].
Approaches associated with designing or searching for antiviral agents whose action is directed against IRESs are being extensively developed. The following compounds are considered as such preparations or a basis for their development: complementary oligonucleotides [131], peptide nucleic acids [130], locked nucleic acids [130], morpholines [134, 135], short RNA hairpins [133, 136, 137], small interfering RNAs [133, 136, 137], RNA aptamers, ribozymes [138, 139], DNAzymes [140, 141], peptides [142, 143], and low molecular weight inhibitors [141-148].
Historically, the first agents directed against IRESs were complementary oligonucleotides. The majority of early attempts were made to prevent hepatitis C virus gene expression [149-151].
Two approaches were applied in these works. One approach used complementary DNA oligonucleotides as a target for RNase H cleavage. The second approach consisted in designing DNA oligonucleotides that would block interaction between IRES and the ribosome. Unfortunately, the latter approach has several typical disadvantages associated with efficiency of transport of DNA oligonucleotides, their intracellular stability, and in certain cases negative side effects. To improve stability and affinity of complementary DNA oligonucleotides, their modified analogs were developed, which consisted of peptide nucleic acids and locked nucleic acids [151-153]. However, it did not solve the problem of delivery, intracellular transport, and toxicity of these compounds.
During development of this approach, attention of researchers was drawn to morpholines (single-stranded DNA-like cell-penetrating complementary agents able to decrease levels of gene expression by blocking complementary RNA sequences). They represent a third generation of complementary oligonucleotides and feature acceptable toxicity and resistance to nucleases [154]. Morpholino–RNA duplexes are significantly more stable than similar DNA–RNA duplexes. Morpholines sterically block target RNA. They are widely used for modulating expression of genes of certain organisms (such as frogs and zebrafish) [154]. A set of peptide-conjugated phosphorodiamidate morpholino oligomers (PPMO) was designed that were complementary to conserved type I IRES regions of RNA viruses (rhinovirus B14, coxsackievirus B2, and poliovirus type 2) [155]. These compounds are soluble in water and resistant to the action of nucleases. They efficiently penetrate cells and inhibit virus replication through forming a duplex with complementary viral mRNA. In cell culture, they reduce virus titer by several orders of magnitude. Application of PPMO increases survivability in mice infected by poliovirus, coxsackievirus B3, Ebola virus, and influenza virus [155, 156].
Octa-guanidine conjugated to morpholines (Vivo-morpholinos, vPMOs) are also single-stranded DNA-like complementary agents. It was shown that these compounds reduced RNA replication and expression of a capsid protein of EV-A71 virus. Besides, these compounds inhibited development of poliovirus and coxsackievirus A16 [157].
Single-stranded mRNA regions potentially available for binding are preferable targets when designing a complementary antiviral agent using the above-mentioned technologies. These regions are usually located within apical loops of various hairpins, in bulges, and in other elements of RNA secondary structures [151, 152, 158].
Another kind of antiviral drugs is RNA hairpins or small interfering RNAs (siRNAs) [159]. Upon transfection of poliovirus-infected murine fibroblasts with siRNAs, strong inhibition of poliovirus replication occurs [160]. Though siRNAs might be used as a basis for development of medications, they suffer the same cell-transport problems as the above-mentioned drugs. Furthermore, these RNA agents carry net negative charge and they are less stable, which impedes their delivery into the cell. To overcome these problems, liposomes and polymeric nanoparticles are used as drug delivery vehicles [161]. Besides, siRNAs suffer a significant disadvantage: they can activate protein kinase K, which inhibits translation of cellular proteins due to phosphorylation of the α-subunit of translation initiation factor 2 [13, 162]. Therefore, approaches based on use of DNA oligonucleotide agents are currently considered more promising as these agents feature higher intracellular stability and increased affinity toward target viral RNA.
The next kind of antiviral drugs are ribozymes, DNAzymes, or ribozyme-conjugated RNA aptamers. DNAzymes are catalytic DNAs that can cleave the phosphodiester bond in an RNA molecule [163]. They can be obtained more easily than synthetic ribozymes, and they are more stable. Unlike siRNAs, DNAzymes do not activate protein kinase K [164]. As ribozymes and DNAzymes can specifically inhibit viral IRESs, they represent a reasonable basis for developing therapeutic preparations directed against viruses that use IRES-dependent translation initiation [164-167].
Peptide-inhibitors and small molecules are extensively used in medicine [168, 169]. These peptides usually consist of 5-40 a.a. They mimic functionally active regions of intact proteins that serve as the basis for their design. Due to their small size, they can specifically bind target RNA, disrupting functional complexes that were formed already [168, 170]. Several such peptides were designed to block an IRES of hepatitis C virus [133, 142, 144]. They are based on an RNA-recognition motif of La autoantigen and prevent binding of La to the IRES of hepatitis C virus [142]. However, La is also an ITAF for numerous viral and cellular IRESs [4, 5, 171, 172]. Therefore, such peptides are not specific to hepatitis C virus.
To solve the problem of intracellular peptide resistance against proteolysis, unnatural amino acids are introduced into them (the corresponding compounds are referred to as peptidomimetics). Liposomes and polymeric nanoparticles are used for delivery of these peptides to cells. Also, a peptide can be linked to a protein domain that assists its transmembrane transport. Alternatively, they may be synthesized in cells upon transduction with viral vectors during gene therapy [168].
Currently, small molecules are preferred drugs [169]. There are new approaches directed toward generation of libraries of small molecules with desired properties [169]. Using combinatorial chemistry, large libraries are generated of closely related structural analogs that are further tested in biological screening. Numerous attempts are made to of small molecules able to deactivate viral IRESs [173-176]. As a result, some potential low molecular weight antiviral agents were obtained [173, 176]. For instance, it was shown that the 9-aminoacridine derivative quinacrine inhibits translation of poliovirus in a cell-free system and in infected HeLa cells, which makes it prospective for further studies [174]. Only a few molecules become subject to clinical trials. However, despite these failures, such approach is still considered one of the most prospective.
Summarizing the above-said, it should be noted that despite several promising direct-action antiviral preparations including the ones already approved for medical application [177], there is no worldwide certified and commonly recognized antiviral drug for treatment of diseases caused by enteroviruses. Physicians are forced to counteract consequences of disease rather than its cause. Thus, valuable time is lost, which increases risks of irreversible damage in patients. Currently, the only very efficient remedy against enteroviral infections is prevention. Joint global efforts in this direction may provide great outcome like that reached in fighting poliomyelitis [59]. For this reason, vaccines against other dangerous representatives of the genus Enterovirus are being developed extensively [85]. Nevertheless, despite 99% reduction in infection cases because of global efforts toward elimination of poliomyelitis, this disease persists. At the same time, one should clearly recognize that if there is still a single poliovirus-infected individual, unvaccinated people worldwide are at risk of infection.
It should be mentioned that development of antiviral drugs is associated with significant difficulties, not only scientific ones, but also organizational and financial difficulties on a global scale. To be efficient, prevention must be ubiquitous and constant, which is still a problem. Besides, vaccine usually is highly specific, whereas enteroviruses are very diverse. Furthermore, under certain conditions vaccination may lead to negative events, such as vaccine-borne virus infection as occurs during overall successful fighting against poliomyelitis. A main point is that prevention does not help those who are already infected. They may only rely on their own immune system and symptomatic treatment. Therefore, the urgent need for designing drugs that directly affect enteroviruses causing numerous dangerous diseases is clear. Drugs with wide spectrum of specificity are especially required, which would allow suppressing enterovirus epidemics at the beginning, before they spread. Development of such preparations is being carried out worldwide.
Acknowledgments
We express our gratitude to A. P. Korepanov for careful reading and valuable critical remarks.
This work was supported by the Russian Science Foundation (project No. 15-14-00028).
REFERENCES
1.Pelletier, J., and Sonenberg, N. (1988) Internal
initiation of translation of eukaryotic mRNA directed by a sequence
derived from poliovirus RNA, Nature, 334, 320-325.
2.Jang, S. K., Krausslich, H. G., Nicklin, M. J.,
Duke, G. M., Palmenberg, A. C., and Wimmer, E. (1988) A segment of the
5′-nontranslated region of encephalomyocarditis virus RNA directs
internal entry of ribosomes during in vitro translation, J.
Virol., 62, 2636-2643.
3.Skulachev, M. V. (2005) Internal translation
initiation: diversity of mechanisms and possible role in cell life,
Usp. Biol. Khim., 45, 123-172.
4.Komar, A. A., and Hatzoglou, M. (2005) Internal
ribosome entry sites in cellular mRNAs: mystery of their existence,
J. Biol. Chem., 280, 23425-23428.
5.Komar, A. A., and Hatzoglou, M. (2011) Cellular
IRES-mediated translation: the war of ITAFs in pathophysiological
states, Cell Cycle, 10, 229-240.
6.Niepmann, M. (2009) Internal translation initiation
of picornaviruses and hepatitis C virus, Biochim. Biophys. Acta,
1789, 529-541.
7.Balvay, L., Soto-Rifo, R., Ricci, E. P., Decimo,
D., and Ohlmann, T. (2009) Structural and functional diversity of viral
IRESes, Biochim. Biophys. Acta, 1789, 542-557.
8.Tahiri-Alaoui, A., Smith, L. P., and Baigent, S.
(2009) Identification of an intercistronic internal ribosome entry site
in a Marek’s disease virus immediate-early gene, J.
Virol., 83, 5846-5853.
9.Wilson, J. E. (2000) Naturally occurring
dicistronic cricket paralysis virus RNA is regulated by two internal
ribosome entry sites, Mol. Cell Biol., 20, 4990-4999.
10.Wong, S. M., Koh, D. C., and Liu, D. (2008)
Identification of plant virus IRES, Methods Mol. Biol.,
451, 125-133.
11.Ronfort, C., De Breyne, S., Sandrin, V., Darlix,
J. L., and Ohlmann, T. (2004) Characterization of two distinct RNA
domains that regulate translation of the Drosophila gypsy
retroelement, RNA, 10, 504-515.
12.De Breyne, S., Yu, Y., Unbehaun, A., Pestova, T.
V., and Hellen, C. (2009) Direct functional interaction of initiation
factor eIF4G with type 1 internal ribosomal entry sites, Proc. Natl.
Acad. Sci. USA, 106, 9197-9202.
13.Jackson, R. J., Hellen, C. U., and Pestova, T. V.
(2010) The mechanism of eukaryotic translation initiation and
principles of its regulation, Nat. Rev. Mol. Cell Biol.,
11, 113-127.
14.Hellen, C. U. (2009) IRES-induced conformational
changes in the ribosome and the mechanism of translation initiation by
internal ribosomal entry, Biochim. Biophys. Acta, 1789,
558-570.
15.Pisarev, A. V., Shirokikh, N. E., and Hellen, C.
U. T. (2005) Translation initiation by factor-independent binding of
eukaryotic ribosomes to internal ribosomal entry sites, C. R.
Biol., 328, 589-605.
16.Shatsky, I. N., Dmitriev, S. E., Terenin, I. M.,
and Andreev, D. E. (2010) Cap- and IRES-independent scanning mechanism
of translation initiation as an alternative to the concept of cellular
IRESs, Mol. Cells, 30, 285-293.
17.Dorokhov, Y. L., Skulachev, M. V., Ivanov, P. A.,
Zvereva, S. D., Tjulkina, L. G., Merits, A., Gleba, Y. Y., Hohn, T.,
and Atabekov, J. G. (2002) Polypurine (A)-rich sequences promote
cross-kingdom conservation of internal ribosome entry, Proc. Natl.
Acad. Sci. USA, 99, 5301-5306.
18.Dorokhov, Yu. L., Ivanov, P. A., Novikov, V. K.,
Agranovsky, A. A., Morozov, S. Yu., Efimov, V. A., Casper, R., and
Atabekov, J. G. (1994) Complete nucleotide sequence and genome
organization of a tobamovirus infecting Cruciferae plants, FEBS
Lett., 350, 5-8.
19.Belsham, G. J., and Jackson, R. J. (2000)
Translation initiation on picornavirus RNA, in Translational Control
of Gene Expression (Sonenberg, N., Hershey, J. W. B., and Mathews,
M. B., eds.) Cold Spring Harbor Laboratory Press, Cold Spring Harbor,
NY, pp. 869-900.
20.Jang, S. K., Pestova, T. V., Hellen, C. U. T.,
Witherell, G. W., and Wimmer, E. (1990) Cap-independent translation of
picornavirus RNAs: structure and function of the internal ribosomal
entry site, Enzyme, 44, 292-309.
21.Willcocks, M. M. (2011) Structural features of
the Seneca Valley virus internal ribosome entry site element; a
picornavirus with a pestivirus-like IRES, J. Virol., 85,
4452-4461.
22.Pestova, T. V., Hellen, C. U., and Wimmer, E.
(1991) Translation of poliovirus RNA: role of an essential
cis-acting oligopyrimidine element within the
5′-nontranslated region and involvement of a cellular
57-kilodalton protein, J. Virol., 65, 6194-6204.
23.Pestova, T. V., Hellen, C. U., and Wimmer, E.
(1994) A conserved AUG triplet in the 5′-nontranslated region of
poliovirus can function as an initiation codon in vitro and
in vivo, Virology, 204, 729-737.
24.Bailey, J. M., and Tapprich, W. E. (2007)
Structure of the 5′-nontranslated region of the coxsackievirus b3
genome: chemical modification and comparative sequence analysis, J.
Virol., 81, 650-668.
25.Pestova, T. V., Hellen, C. U., and Shatsky, I. N.
(1996) Canonical eukaryotic initiation factors determine initiation of
translation by internal ribosomal entry, Mol. Cell. Biol.,
16, 6859-6869.
26.Pilipenko, E. V., Pestova, T. V., Kolupaeva, V.
G., Khitrina, E. V., Poperechnaya, A. N., Agol, V. I., and Hellen, C.
U. (2000) A cell cycle-dependent protein serves as a template-specific
translation initiation factor, Genes Dev., 14,
2028-2045.
27.Brown, E. A., Zajac, A. J., and Lemon, S. M.
(1994) In vitro characterization of an internal ribosomal
entry site (IRES) present within the 5′-nontranslated region of
hepatitis A virus RNA: comparison with the IRES of encephalomyocarditis
virus, J. Virol., 68, 1066-1074.
28.Ali, I. K., McKendrick, L., Morley, S. J., and
Jackson, R. J. (2001) Activity of the hepatitis A virus IRES requires
association between the cap-binding translation initiation factor
(eIF4E) and eIF4G, J. Virol., 75, 7854-7863.
29.De Breyne, S., Yu, Y., Pestova, T. V., and
Hellen, C. U. (2008) Factor requirements for translation initiation on
the simian picornavirus internal ribosomal entry site, RNA,
14, 367-380.
30.Pestova, T. V., Borukhov, S. I., and Hellen, C.
U. (1998) Eukaryotic ribosomes require 872 initiation factors 1
and 1A to locate initiation codons, Nature, 394,
854-859.
31.Pestova, T. V., De Breyne, S., Pisarev, A. V.,
Abaeva, I. S., and Hellen, C. U. T. (2008) eIF2-dependent and
eIF2-independent modes of initiation on the CSFV IRES: a common 875
role of domain II, EMBO J., 27, 1060-1072.
32.Kafasla, P., Morgner, N., Robinson, C. V., and
Jackson, R. J. (2010) Polypyrimidine tract-binding protein
stimulates the poliovirus IRES by modulating eIF4G binding, EMBO
J., 29, 3710-3722.
33.Kaminski, A., Hunt, S. L., Gibbs, C. L., and
Jackson, R. J. (1994) Internal initiation of mRNA translation in
eukaryote, in Genetic Engineering (Setlow, J. K., ed.)
Vol. 16, Plenum Press, New York, pp. 115-155.
34.Wilson, J. E., Pestova, T. V., Hellen, C. U. T.,
and Sarnow, P. (2000) Initiation of protein synthesis from the A
site of the ribosome, Cell, 102, 511-520.
35.King, A. M. Q., Adams, M. J., Carstens, E. B.,
and Lefkowitz, E. J. (2012) Virus Taxonomy: Classification and
Nomenclature of Viruses: Ninth Report of the International Committee on
Taxonomy of Viruses, Elsevier Academic Press, San Diego.
36.Adams, M. J., King, A. M. Q., and Carstens, E. B.
(2013) Ratification vote on taxonomic proposals to the International
Committee on Taxonomy of Viruses, Arch. Virol., 158,
2023-2030.
37.Adams, M. J., Lefkowitz, E. J., King, A. M. Q.,
and Carstens, E. B. (2014) Ratification vote on taxonomic proposals to
the International Committee on Taxonomy of Viruses, Arch.
Virol., 159, 2831-2841.
38.Adams, M. J., Lefkowitz, E. J., King, A. M. Q.,
Bamford, D. H., Breitbart, M., Davison, A. J., Ghabrial, S. A.,
Gorbalenya, A. E., Knowles, N. J., Krell, P., Lavigne, R.,
Prangishvili, D., Sanfaçon, H., Siddell, S. G., Simmonds, P.,
and Carstens, E. B. (2015) Ratification vote on taxonomic proposals to
the International Committee on Taxonomy of Viruses, Arch.
Virol., 160, 1837-1850.
39.Adams, M. J., Lefkowitz, E. J., King, A. M. Q.,
Harrach, B., Harrison, R. L., Knowles, N. J., Kropinski, A. M.,
Krupovic, M., Kuhn, J. H., Mushegian, A. R., Nibert, M., Sabanadzovic,
S., Sanfacon, H., Siddell, S. G., Simmonds, P., Varsani, A., Zerbini,
F. M., Gorbalenya, A. E., and Davison, A. J. (2016) Ratification vote
on taxonomic proposals to the International Committee on Taxonomy of
Viruses, Arch. Virol., 161, 2921-2949.
40.Arden, K. E., McErlean, P., Nissen, M. D.,
Sloots, T. P., and Mackay, I. M. (2006) Frequent detection of human
rhinoviruses, paramyxoviruses, coronaviruses, and bocavirus during
acute respiratory tract infections, J. Med. Virol., 78,
1232-1240.
41.McErlean, P., Shackelton, L. A., Lambert, S. B.,
Nissen, M. D., Sloots, T. P., and Mackay, I. M. (2007) Characterization
of a newly identified human rhinovirus, HRV-QPM, discovered in infants
with bronchiolitis, J. Clin. Virol., 39, 67-75.
42.Lamson, D., Renwick, N., Kapoor, V., Liu, Z.,
Palacios, G., Ju, J., Dean, A., St George, K., Briese, T., and Lipkin,
W. I. (2006) MassTag polymerase-chain-reaction detection of respiratory
pathogens, including a new rhinovirus genotype, that caused
influenza-like illness in New York State during 2004-2005, J.
Infect. Dis., 194, 1398-1402.
43.Lau, S. K., Yip, C. C., Tsoi, H. W., Lee, R. A.,
So, L. Y., Lau, Y. L., Chan, K. H., Woo, P. C., and Yuen, K. Y. (2007)
Clinical features and complete genome characterization of a distinct
human rhinovirus genetic cluster, probably representing a previously
undetected HRV species, HRV-C, associated with acute respiratory
illness in children, J. Clin. Microbiol., 45,
3655-3664.
44.Lee, W.-M., Kiesner, C., Pappas, T., Lee, I.,
Grindle, K., Jartti, T., Jakiela, B., Lemanske, R. F., Jr., Shult, P.
A., and Gern, J. E. (2007) A diverse group of previously unrecognized
human rhinoviruses are common causes of respiratory illnesses in
infants, PLoS One, 2, e966.
45.McErlean, P., Shackelton, L. A., Andrews, E.,
Webster, D. R., Lambert, S. B., Nissen, M. D., Sloots, T. P., and
Mackay, I. M. (2008) Distinguishing molecular features and clinical
characteristics of a putative new rhinovirus species, Human
rhinovirus C (HRV C), PLoS One, 3, e1847.
46.Kistler, A., Avila, P. C., Rouskin, S., Wang, D.,
Ward, T., Yagi, S., Schnurr, D., Ganem, D., DeRisi, J. L., and Boushey,
H. A. (2007) Pan-viral screening of respiratory tract infections in
adults with and without asthma reveals unexpected human coronavirus and
human rhinovirus diversity, J. Infect. Dis., 196,
817-825.
47.McIntyre, C. L., Knowles, N. J., and Simmonds, P.
(2013) Proposals for the classification of human rhinovirus species A,
B and C into genotypically assigned types, J. Gen. Virol.,
94, 1791-1806.
48.Simmonds, P., McIntyre, C. L., Savolainen-Kopra,
C., Tapparel, C., Mackay, I. M., and Hovi, T. (2010) Proposals for the
classification of human rhinovirus species C into
genotypically-assigned types, J. Gen. Virol., 91,
2409-2419.
49.Galassi, F. M., Habicht, M. E., and Ruhli, F. J.
(2017) Poliomyelitis in Ancient Egypt? Neurol. Sci., 38,
375.
50.Horstmann, D. M., and Yale, J. (1985) The
poliomyelitis story: a scientific hegira, Biol. Med., 58,
79-90.
51.De Jesus, N. H. (2007) Epidemics to eradication:
the modern history of poliomyelitis, Virol. J., 4,
70.
52.Ritchie, W., and Russell, B. (1949) Paralytic
poliomyelitis, Med. J., 1, 465-471.
53.Kidd, D., Williams, A. J., and Howard, R. S.
(1996) Poliomyelitis, Postgrad. Med. J., 72,
641-647.
54.Reznik, B. I., and Kurakina, L. T. (1961) A
pontine form of poliomyelitis and isolated facial neuritis, Sov.
Med., 25, 87-91.
55.Matzke, H. A., and Baker, A. B. (1952)
Poliomyelitis. V. The pons, AMA Neurol. Psychiatry,
68, 1-15.
56.Noran, H. H. (1968) Poliomyelitis. The bulbar
type, Minn. Med., 51, 1249-1252.
57.Schaefer, J., and Edward, B. (1949) Poliomyelitis
in pregnancy, Calif. Med., 70, 16-18.
58.Global Polio Eradication Initiative. Wild
Poliovirus Weekly Update. Sept 8, 2009. Available at http://www.polioeradication.org/casecount.asp.
59.WHO Poliomyelitis. Online at: http://www.who.int/topics/poliomyelitis/en/.
Accessed 11 Aug 2016.
60.Norway, T. M. (1967) The occurrence of Bamble
disease (epidemic pleurodynia), Vogelsang Med. Hist., 11,
86-90.
61.Leendertse, M., Van Vugt, M., Benschop, K. S.,
Van Dijk, K., Minnaar, R. P., Van Eijk, H. W., Hodiamont, C. J., and
Wolthers, K. C. (2013) Pleurodynia caused by an echovirus 1 brought
back from the tropics, J. Clin. Virol., 58, 490-493.
62.Gaaloul, I., Riabi, S., Harrath, R., Hunter, T.,
Hamda, K. B., Ghzala, A. B., Huber, S., and Aouni, M. (2014)
Coxsackievirus B detection in cases of myocarditis, myopericarditis,
pericarditis and dilated cardiomyopathy in hospitalized patients,
Mol. Med. Rep., 10, 2811-2818.
63.Novikov, Iu. I., Stulova, M. A., and Lavrova, I.
K. (1984) Myocarditis caused by coxsackie B viruses in adults, Ter.
Arkh., 56, 37-43.
64.Lee, W. S., Lee, K. J., Kwon, J. E., Oh, M. S.,
Kim, J. E., Cho, E. J., and Kim, C. J. (2012) Acute viral
myopericarditis presenting as a transient effusive-constrictive
pericarditis caused by coinfection with coxsackieviruses A4 and B3,
Korean J. Intern. Med., 27, 216-220.
65.Shanmugam, J., Raveendranath, M., and
Balakrishnan, K. G. (1986) Isolation of ECHO virus type-22 from a child
with acute myopericarditis – a case report, Indian Heart
J., 38, 79-80.
66.Fukuhara, T., Kinoshita, M., Bito, K., Sawamura,
M., Motomura, M., Kawakita, S., and Kawanishi, K. (1983)
Myopericarditis associated with ECHO virus type 3 infection – a
case report, Jpn. Circ. J., 47, 1274-1280.
67.Liapounova, N. A., Mouquet, F., and Ennezat, P.
V. (2011) Acute myocardial infarction spurred by myopericarditis in a
young female patient: coxsackie B2 to blame, Acta Cardiol.,
66, 79-81.
68.Chatterjee, S., Quarcoopome, C. O., and Apenteng,
A. (1970) Unusual type of epidemic conjunctivitis in Ghana, Br. J.
Ophthalmol., 54, 628-630.
69.Lim, K. H., and Yin-Murphy, M. (1971) An epidemic
of conjunctivitis in Singapore in 1970, Singapore Med. J.,
12, 247-249.
70.Wright, P. W., Strauss, G. H., and Langford, M.
P. (1992) Acute hemorrhagic conjunctivitis, Am. Fam. Physician,
45, 173-178.
71.Langford, M. P., Anders, E. A., and Burch, M. A.
(2015) Acute hemorrhagic conjunctivitis: anti-coxsackievirus A24
variant secretory immunoglobulin A in acute and convalescent tear,
Clin. Ophthalmol., 10, 1665-1663.
72.Kogon, A., Spigland, I., Frothingham, T. E.,
Elveback, L., Williams, C., and Hall, C. E. (1969) The virus watch
program: a continuing surveillance of viral infections in metropolitan
New York families. VII. Observations on viral excretion, seroimmunity,
intrafamilial spread and illness association in coxsackie and echovirus
infections, Am. J. Epidemiol., 89, 51-61.
73.Simasek, M., and Blandino, D. A. (2007) Treatment
of the common cold, Am. Fam. Physician, 75, 515-520.
74.Jacobson, L. M., Redd, J. T., Schneider, E., Lu,
X., Chern, S. W., Oberste, M. S., Erdman, D. D., Fischer, G. E.,
Armstrong, G. L., Kodani, M., Montoya, J., Magri, J. M., and Cheek, J.
E. (2012) Outbreak of lower respiratory tract illness associated with
human enterovirus 68 among American Indian children, Pediatr.
Infect. Dis. J., 31, 309-312.
75.Rotbart, H. A. (1995) Enteroviral infections of
the central nervous system, Clin. Infect. Dis., 20,
971-981.
76.Lee, B. E., and Davies, H. D. (2007) Aseptic
meningitis, Curr. Opin. Infect. Dis., 20, 272-277.
77.Cui, A., Yu, D., Zhu, Z., Meng, L., Li, H., Liu,
J., Liu, G., Mao, N., and Xu, W. (2010) An outbreak of aseptic
meningitis caused by coxsackievirus A9 in Gansu, the People’s
Republic of China, Virol. J., 7, 72.
78.Irani, D. N. (2008) Aseptic meningitis and viral
myelitis, Neurol. Clin., 26, 635.
79.Huang, C. C., Liu, C. C., Chang, Y. C., Chen, C.
Y., Wang, S. T., and Yeh, T. F. (1999) Neurologic complications in
children with enterovirus 71 infection, N. Engl. J. Med.,
341, 936-942.
80.Lukashev, A. N., Koroleva, G. A., Lashkevich, V.
A., and Mikhailov, M. I. (2009) Enterovirus 71: epidemiology and
diagnostics, J. Microbiol. Epidemiol. Immunobiol., 3,
110-116.
81.Jubelt, B., and Lipton, H. L. (2014)
Enterovirus/picornavirus infections, Handb. Clin. Neurol.,
123, 379-416.
82.Chang, L.-Y., King, Ch.-Ch., Hsu, K.-H., Ning,
H.-Ch., Tsao, K.-Ch., Li, Ch.-Ch., Huang, Yh.-Ch., Shih, S.-R., Chiou,
S.-T., Chen, P.-Y., Chang, H.-J., and Lin, T. Y. (2002) Risk factors of
enterovirus 71 infection and associated hand, foot, and mouth
disease/herpangina in children during an epidemic in Taiwan,
Pediatrics, 109, e88.
83.Laga, A. C., Shroba, S. M., and Hanna, J. (2016)
A typical hand, foot and mouth disease in adults associated with
coxsackievirus A6: a clinicopathologic study, J. Cutan. Pathol.,
43, 940-945.
84.Chiu, W. Y., Lo, Y. H., and Yeh, T. C. (2016)
Coxsackievirus associated hand, foot and mouth disease in an adult,
QJM, 109, 823-824.
85.Lee, K. Y. (2016) Enterovirus 71 infection and
neurological complications, Korean J. Pediatr., 59,
395-401.
86.Fowlkes, A. L., Honarmand, S., Glaser, C., Yagi,
S., Schnurr, D., Oberste, M. S., Anderson, L., Pallansch, M. A., and
Khetsuriani, N. J. (2008) Enterovirus-associated encephalitis in the
California encephalitis project, 1998-2005, Infect. Dis.,
198, 1685-1691.
87.Zhang, L., Yan, J., Ojcius, D. M., Lv, H., Miao,
Z., Chen, Y., Zhang, Y., and Yan, J. (2013) Novel and predominant
pathogen responsible for the enterovirus-associated encephalitis in
eastern China, PLoS One, 8, e85023.
88.Gusev, E. A., Burd, G. S., and Konovalov, A. N.
(2000) Neurology and Neurosurgery [in Russian], Meditsina,
Moscow, p. 656.
89.Zuckerman, M. A., Sheaff, M., Martin, J. E., and
Gabriel, C. M. (1993) Fatal case of echovirus type 9 encephalitis,
J. Clin. Pathol., 46, 865-866.
90.Wang, S. M., and Liu, C. C. (2009) Enterovirus
71: epidemiology, pathogenesis and management, Expert Rev.
Anti-Infect. Ther., 7, 735-742.
91.Skripachenkom, N. V., Sorokina, M. N., Ivanova,
V. V., and Komantsev, V. N. (1999) Acute flaccid paralyses in children
under modern conditions, Ross. Vestn. Perinatol. Pediatr.,
3, 31-35.
92.Tang, J., Yoshida, H., Ding, Z., Tao, Z., Zhang,
J., Tian, B., Zhao, Z., and Zhang, L. (2014) Molecular epidemiology and
recombination of human enteroviruses from AFP surveillance in Yunnan,
China from 2006 to 2010, Sci. Rep., 14, 6058.
93.Ong, K. C., and Wong, K. T. (2015) Understanding
enterovirus 71 neuropathogenesis and its impact on other neurotropic
enteroviruses, Brain Pathol., 25, 614-624.
94.Perez-Velez, C. M., Anderson, M. S., Robinson, C.
C., McFarland, E. J., Nix, W. A., Pallansch, M. A., Oberste, M. S., and
Glode, M. P. (2007) Outbreak of neurologic enterovirus type 71 disease:
a diagnostic challenge, Clin. Infect. Dis., 45,
950-957.
95.Landry, M. L., Fonseca, S. N., Cohen, S., and
Bogue, C. W. (1995) Fatal enterovirus type 71 infection: rapid
detection and diagnostic pitfalls, Pediatr. Infect. Dis. J.,
14, 1095-100.
96.Aliev, A. S., and Alieva, A. K. (2009) Poultry
gastrointestinal diseases of viral etiology, Poult. Chicken
Products, 4, 50-54.
97.Mitchell, D., Corner, A. H., Bannister, G. L.,
and Greig, A. S. (1961) Studies on pathogenic porcine enteroviruses: 1.
Preliminary investigations, Can. J. Compar. Med. Vet. Sci.,
25, 85-93.
98.Nollens, H. H., Rivera, R., Palacios, G.,
Wellehan, J. F., Saliki, J. T., Caseltine, S. L., Smith, C. R., Jensen,
E. D., Hui, J., Lipkin, W. I., Yochem, P. K., Wells, R. S., St. Leger,
J., and Venn-Watson, S. (2009) Short communication: New recognition of
enterovirus infections in bottlenose dolphins (Tursiops
truncatus), Vet. Microbiol., 139, 170-175.
99.Abzug, M. J., Michaels, M. G., Wald, E., Jacobs,
R. F., Romero, J. R., Sanchez, P. J., Wilson, G., Krogstad, P., Storch,
G. A., Lawrence, R., Shelton, M., Palmer, A., Robinson, J., Dennehy,
P., Sood, S. K., Cloud, G., Jester, P., Acosta, E. P., Whitley, R., and
Kimberlin, D. (2016) Controlled trial of pleconaril for the treatment
of neonates with enterovirus sepsis. National institute of allergy and
infectious diseases collaborative antiviral study group, J.
Pediatric Infect. Dis. Soc., 5, 53-62.
100.Pevear, D. C., Tull, T. M., Seipel, M. E., and
Groarke, J. M. (1999) Activity of pleconaril against enteroviruses,
Antimicrob. Agents Chemother., 43, 2109-2115.
101.Hayden, F. G., Herrington, D. T., Coats, T. L.,
Kim, K., Cooper, E. C., Villano, S. A., Liu, S., Hudson, S., Pevear, D.
C., Collett, M., and McKinlay, M. (2003) Efficacy and safety of oral
pleconaril for treatment of colds due to picornaviruses in adults:
results of 2 double-blind, randomized, placebo-controlled trials,
Clin. Infect. Dis., 36, 1523-1532.
102.Shia, K. S., Li, W. T., Chang, C. M., Hsu, M.
C., Chern, J. H., Leong, M. K., Tseng, S. N., Lee, C. C., Lee, Y. C.,
Chen, S. J., Peng, K. C., Tseng, H. Y., Chang, Y. L., Tai, C. L., and
Shih, S. R. (2002) Design, synthesis, and structure-activity
relationship of pyridyl imidazolidinones: a novel class of potent and
selective human enterovirus 71 inhibitors, J. Med. Chem.,
45, 1644-1655.
103.Laconi, S., Madeddu, M. A., and Pompei, R.
(2011) Study of the biological activity of novel synthetic compounds
with antiviral properties against human rhinoviruses, Molecules,
16, 3479-3487.
104.Gradi, A., Svitkin, Y. V., Imataka, H., and
Sonenberg, N. (1998) Proteolysis of human eukaryotic translation
initiation factor eIF4GII, but not eIF4GI, coincides with the shutoff
of host protein synthesis after poliovirus infection, Proc. Natl.
Acad. Sci. USA, 95, 11089-11094.
105.Park, N., Katikaneni, P., Skern, T., and
Gustin, K. E. J. (2008) Differential targeting of nuclear pore complex
proteins in poliovirus-infected cells, Virology, 82,
1647-1655.
106.Almstead, L. L., and Sarnow, P. (2007)
Inhibition of U snRNP assembly by a virus-encoded proteinase, Genes
Dev., 21, 1086-1089.
107.Zhou, H., Sun, Y., Guo, Y., and Lou, Z. (2013)
Structural perspective on the formation of ribonucleoprotein complex in
negative-sense single stranded RNA viruses, Trends Microbiol.,
21, 475-484.
108.Tan, J., George, S., Kusov, Y., Perbandt, M.,
Anemuller, S., Mesters, J. R., Norder, H., Coutard, B., Lacroix, C.,
Leyssen, P., Neyts, J., and Hilgenfeld, R. (2013) 3C protease of
enterovirus 68: structure-based design of Michael acceptor inhibitors
and their broad-spectrum antiviral effects against picornaviruses,
J. Virol., 87, 4339-4351.
109.Racaniello, V. R. (2007) Picornaviridae: The
Viruses and Their Replication (Knipe, D. M., et al., eds.) 5th
Edn., Fields Virology, Lippincott Williams & Wilkins, Philadelphia,
PA, pp. 796-839.
110.Konig, H., and Rosenwirth, B. (1988)
Purification and partial characterization of poliovirus protease 2A by
means of a functional assay, J. Virol., 62,
1243-1250.
111.Deszcz, L., Cencic, R., Sousa, C., Kuechler,
E., and Skern, T. (2006) An antiviral peptide inhibitor that is active
against picornavirus 2A proteinases but not cellular caspases, J.
Virol., 80, 9619-9627.
112.De Palma, A. M., Vliegen, I., De Clercq, E.,
and Neyts, J. (2008) Selective inhibitors of picornavirus replication,
Med. Res. Rev., 28, 823-884.
113.Dragovich, P. S., Webber, S. E., Babine, R. E.,
Fuhrman, S. A., Patick, A. K., Matthews, D. A., Lee C. A., Reich, S.
H., Prins, T. J., Marakovits, J. T., Littlefield, E. S., Zhou, R.,
Tikhe, J., Ford, C. E., Wallace, M. B., Meador, J. W., 3rd, Ferre, R.
A., Brown, E. L., Binford, S. L., Harr, J. E., DeLisle, D. M., and
Worland, S. T. (1998) Structure-based design, synthesis, and biological
evaluation of irreversible human rhinovirus 3C protease inhibitors. 1.
Michael acceptor structure-activity studies, J. Med. Chem.,
41, 2806-2018.
114.Guo, Y., Wang, Y., Cao, L., Wang, P., Qing, J.,
Zheng, Q., Shang, L., Yin, Z., and Sun, Y. (2016) A conserved
inhibitory mechanism of a lycorine derivative against enterovirus and
hepatitis C virus, Antimicrob. Agents Chemother., 60,
913-924.
115.Lu, G., Qi, J., Chen, Z., Xu, X., Gao, F., Lin,
D., Qian, W., Liu, H., Jiang, H., Yan, J., and Gao, G. F. (2011)
Enterovirus 71 and coxsackievirus A16 3C proteases: binding to
rupintrivir and their substrates and anti-hand, foot, and mouth disease
virus drug design, J. Virol., 85, 10319-10331.
116.Chen, T. C., Weng, K. F., Chang, S. C., Lin, J.
Y., Huang, P. N., and Shih, S. R. (2008) Development of antiviral
agents for enteroviruses, J. Antimicrob. Chemother., 62,
1169-1173.
117.Tijsma, A., Thibaut, H. J., Franco, D.,
Dallmeier, K., and Neyts, J. (2016) Hydantoin: the mechanism of its
in vitro anti-enterovirus activity revisited, Antiviral
Res., 133, 106-109.
118.Velu, A. B., Chen, G. W., Hsieh, P. T., Horng,
J. T., Hsu, J. T., Hsieh, H. P., Chen, T. C., Weng, K. F., and Shih, S.
R. (2014) BPR-3P0128 inhibits RNA-dependent RNA polymerase elongation
and VPg uridylylation activities of enterovirus 71, Antiviral
Res., 112, 18-25.
119.Wang, H., Zhang, D., Ge, M., Li, Z., Jiang, J.,
and Li, Y. (2015) Formononetin inhibits enterovirus 71 replication by
regulating COX-2/PGE₂ expression, Virol. J., 12,
35.
120.Strating, J. R., Van der Linden, L., Albulescu,
L., Bigay, J., Arita, M., Delang, L., Leyssen, P., Van der Schaar, H.
M., Lanke, K. H., Thibaut, H. J., Ulferts, R., Drin, G., Schlinck, N.,
Wubbolts, R. W., Sever, N., Head, S. A., Liu, J. O., Beachy, P. A., De
Matteis, M. A., Shair, M. D., Olkkonen, V. M., Neyts, J., and Van
Kuppeveld, F. J. (2015) Itraconazole inhibits enterovirus replication
by targeting the oxysterol-binding protein, Cell Rep.,
10, 600-615.
121.Tsin, I. Y., and Laa, P. C. (2016) Development
of novel miRNA-based vaccines and antivirals against enterovirus 71,
Curr. Pharm. Des., 22, 6694-6700.
122.Lee, K. M., Chen, C. J., and Shih, S. R. (2017)
Regulation mechanisms of viral IRES-driven translation, Trends
Microbiol., pii: S0966-842X(17)30022-7.
123.Pilipenko, E. V., Viktorova, E. G., Guest, S.
T., Agol, V. I., and Roos, R. P. (2001) Cell specific proteins regulate
viral RNA translation and virus induced disease, EMBO J.,
20, 6899-6908.
124.Guest, S., Pilipenko, E., Sharma, K., Chumakov,
K., and Roos, R. (2004) Molecular mechanisms of attenuation of the
Sabin strain of poliovirus type 3, J. Virol., 78,
11097-11107.
125.Romero-Lopez, C., Barroso-Deljesus, A., and
Berzal-Herranz, A. (2017) The chaperone-like activity of the hepatitis
C virus IRES and CRE elements regulates genome dimerization, Sci.
Rep., 24, 43415.
126.Wakita, T., and Wands, J. R. (1994) Specific
inhibition of hepatitis C virus expression by antisense
oligodeoxynucleotides. In vitro model for selection of target
sequence, J. Biol. Chem., 269, 14205-14210.
127.Hanecak, R., Brown-Driver, V., Fox, M. C.,
Azad, R. F., Furusako, S., Nozaki, C., Ford, C., Sasmor, H., and
Anderson, K. P. (1996) Antisense oligonucleotide inhibition of
hepatitis C virus gene expression in transformed hepatocytes, J.
Virol., 70, 5203-5212.
128.Yang, D., Wilson, J. E., Anderson, D. R.,
Bohunek, L., Cordeiro, C., Kandolf, R., and MacManus, B. M. (1997)
In vitro mutational and inhibitory analysis of the
cis-acting translational elements within the 5′
untranslated region of coxsackievirus B3: potential targets for
antiviral action of antisense oligomers, Virology, 228,
63-73.
129.Brown, M. C., and Gromeier, M. (2015) Cytotoxic
and immunogenic mechanisms of recombinant oncolytic poliovirus,
Curr. Opin. Virol., 13, 81-85.
130.Nulf, C. J., and Corey, D. (2004) Intracellular
inhibition of hepatitis C virus (HCV) internal ribosomal entry site
(IRES)-dependent translation by peptide nucleic acids (PNAs) and locked
nucleic acids (LNAs), Nucleic Acids Res., 32,
3792-3798.
131.Martinand-Mari, C., Lebleu, B., and Robbins, I.
(2003) Oligonucleotide-based strategies to inhibit human hepatitis C
virus, Oligonucleotides, 13, 539-548.
132.Dasgupta, A., Das, S., Izumi, R., Venkatesan,
A., and Barat, B. (2004) Targeting internal ribosome entry site
(IRES)-mediated translation to block hepatitis C and other RNA viruses,
FEMS Microbiol. Lett., 234, 189-199.
133.Dibrov, S. M., Parsons, J., Carnevali, M.,
Zhou, S., Rynearson, K. D., Ding, K., Garcia Sega, E., Brunn, N. D.,
Boerneke, M. A., Castaldi, M. P., and Hermann, T. (2014) Hepatitis C
virus translation inhibitors targeting the internal ribosomal entry
site, J. Med. Chem., 57, 1694-1707.
134.McCaffrey, A. P., Meuse, L., Karimi, M.,
Contag, C. H., and Kay, M. A. (2003) A potent and specific morpholino
antisense inhibitor of hepatitis C translation in mice,
Hepatology, 38, 503-508.
135.Stone, J. K., Rijnbrand, R., Stein, D. A., Ma,
Y., Yang, Y., Iversen, P. L., and Andino, R. (2008) A morpholino
oligomer targeting highly conserved internal ribosome entry site
sequence is able to inhibit multiple species of picornavirus,
Antimicrob. Agents Chemother., 52, 1970-1981.
136.Kanda, T., Steele, R., Ray, R., and Ray, R. B.
(2007) Small interfering RNA targeted to hepatitis C virus
5′-nontranslated region exerts potent antiviral effect, J.
Virol., 81, 669-676.
137.Ma, H., Dallas, A., Ilves, H., Shorenstein, J.,
MacLachlan, I., Klumpp, K., and Johnston, B. H. (2014) Formulated
minimal-length synthetic small hairpin RNAs are potent inhibitors of
hepatitis C virus in mice with humanized livers,
Gastroenterology, 146, 63-65.
138.Mao, X., Li, X., Mao, X., Huang, Z., Zhang, C.,
Zhang, W., Wu, J., and Li, G. (2014) Inhibition of hepatitis C virus by
an M1GS ribozyme derived from the catalytic RNA subunit of
Escherichia coli RNase P, Virol. J., 11, 86.
139.Levesque, M. V., Levesque, D., Briere, F. P.,
and Perreault, J.-P. (2010) Investigating a new generation of ribozymes
in order to target HCV, PLoS ONE, 5, e9627.
140.Sugiyama, R., Hayafune, M., Habu, Y., Yamamoto,
N., and Takaku, H. (2011) HIV-1 RT-dependent DNAzyme expression
inhibits HIV-1 replication without the emergence of escape viruses,
Nucleic Acids Res., 39, 589-598.
141.Silverman, S. K. (2016) Catalytic DNA: scocpe,
applications, and biochemistry of deoxyribozymes, Trends Biochem.
Sci., 41, 595-609.
142.Pudi, R., Ramamurthy, S. S., and Das, S. (2005)
A peptide derived from RNA recognition motif 2 of human La protein
binds to hepatitis C virus internal ribosome entry site, prevents
ribosomal assembly, and inhibits internal initiation of translation,
J. Virol., 79, 9842-9853.
143.Fontanes, V., Raychaudhuri, S., and Dasgupta,
A. (2009) A cell-permeable peptide inhibits hepatitis C virus
replication by sequestering IRES transacting factors, Virology,
394, 82-90.
144.De Clercq, E., and Li, G. (2016) Approved
antiviral drugs over the past 50 years, Clinic. Microbiol. Rev.,
29, 695-747.
145.Novac, O., Guenier, A. S., and Pelletier, J.
(2004) Inhibitors of protein synthesis identified by a high throughput
multiplexed translation screen, Nucleic Acids Res., 32,
902-915.
146.Li, Z., Khaliq, M., Zhou, Z., Post, C. B.,
Kuhn, R. J., and Cushman, M. (2008) Design, synthesis, and biological
evaluation of antiviral agents targeting flavivirus envelope proteins,
J. Med. Chem., 51, 4660-4671.
147.Wang, J., Du, J., Wu, Z., and Jin, Q. (2013)
Quinacrine impairs enterovirus 71 RNA replication by preventing binding
of polypyrimidine-tract binding protein with internal ribosome entry
sites, PLoS One, 8, e52954.
148.Tong, J., Wang, Y., and Lu, Y. (2012) New
developments in small molecular compounds for anti-hepatitis C virus
(HCV) therapy, J. Zhejiang University. Science. B, 13,
56-82.
149.Wakita, T., and Wands, J. R. (1994) Specific
inhibition of hepatitis C virus expression by antisense
oligodeoxynucleotides. In vitro model for selection of target
sequence, J. Biol. Chem., 269, 14205-14210.
150.Hanecak, R., Brown-Driver, V., Fox, M. C.,
Azad, R. F., Furusako, S., Nozaki, C., Ford, C., Sasmor, H., and
Anderson, K. P. (1996) Antisense oligonucleotide inhibition of
hepatitis C virus gene expression in transformed hepatocytes, J.
Virol., 70, 5203-5212.
151.Martinand-Mari, C., Lebleu, B., and Robbins, I.
(2003) Oligonucleotide-based strategies to inhibit human hepatitis C
virus, Oligonucleotides, 13, 539-548.
152.Nulf, C. J., and Corey, D. (2004) Intracellular
inhibition of hepatitis C virus (HCV) internal ribosomal entry site
(IRES)-dependent translation by peptide nucleic acids (PNAs) and locked
nucleic acids (LNAs), Nucleic Acids Res., 32,
3792-3798.
153.Mutso, M., Nikonov, A., Pihlak, A., Zusinaite,
E., Viru, L., Selyutina, A., Reintamm, T., Kelve, M., Saarma, M.,
Karelson, M., and Merits, A. (2015) RNA interference-guided targeting
of hepatitis C virus replication with antisense locked nucleic
acid-based oligonucleotides containing 8-oxo-dG modifications, PLoS
One, 10, e0128686.
154.Karkare, S., and Bhatnagar, D. (2006) Promising
nucleic acid analogs and mimics: characteristic features and
applications of PNA, LNA, and morpholino, Appl. Microbiol.
Biotechnol., 71, 575-586.
155.Stone, J. K., Rijnbrand, R., Stein, D. A., Ma,
Y., Yang, Y., Iversen, P. L., and Andino, R. (2008) A morpholino
oligomer targeting highly conserved internal ribosome entry site
sequence is able to inhibit multiple species of picornavirus,
Antimicrob. Agents Chemother., 52, 1970-1981.
156.Stein, D. A. (2008) Inhibition of RNA virus
infections with peptide-conjugated morpholino oligomers, Curr.
Pharm. Des., 14, 2619-2634.
157.Tan, C. W., Chan, Y. F., Quah, Y. W., and Poh,
C. L. (2014) Inhibition of enterovirus 71 infection by antisense
octaguanidinium dendrimer-conjugated morpholino oligomers, Antiviral
Res., 107, 35-41.
158.Dasgupta, A., Das, S., Izumi, R., Venkatesan,
A., and Barat, B. (2004) Targeting internal ribosome entry site
(IRES)-mediated translation to block hepatitis C and other RNA viruses,
FEMS Microbiol. Lett., 234, 189-199.
159.Holoch, D., and Moazed, D. (2015) RNA-mediated
epigenetic regulation of gene expression, Nat. Rev. Genet.,
16, 71-84.
160.Gitlin, L., Karelsky, S., and Andino, R. (2002)
Short interfering RNA confers intracellular antiviral immunity in human
cells, Nature, 418, 430-434.
161.Torrecilla, J., Del Pozo-Rodriguez, A.,
Apaolaza, P. S., Solinis, M. A., and Rodriguez-Gascon, A. (2015) Solid
lipid nanoparticles as non-viral vector for the treatment of chronic
hepatitis C by RNA interference, Int. J. Pharm., 479,
181-188.
162.Sledz, C. A., Holko, M., De Veer, M. J.,
Silverman, R. H., and Williams, B. R. (2003) Activation of the
interferon system by short-interfering RNAs, Nat. Cell. Biol.,
5, 834-839.
163.Silverman, S. K., and Baum, D. A. (2009) Use of
deoxyribozymes in RNA research, Methods Enzymol., 469,
95-117.
164.Roy, S., Gupta, N., Subramanian, N., Monda, L.
T., Banerjea, A. C., and Das, S. (2008) Sequence-specific cleavage of
hepatitis C virus RNA by DNAzymes: inhibition of viral RNA translation
and replication, J. Gen. Virol., 89, 1579-1586.
165.Macejak, D. G., Jensen, K. L., Jamison, S. F.,
Domenico, K., Roberts, E. C., Chaudhary, N., Von Carlowitz, I., Bellon,
L., Tong, M. J., Conrad, A., Pavco, P. A., and Blatt, L. M. (2000)
Inhibition of hepatitis C virus (HCV)-RNA-dependent translation and
replication of a chimeric HCV poliovirus using synthetic stabilized
ribozymes, Hepatology, 31, 769-776.
166.Romero-Lopez, C., Berzal-Herranz, B., Gomez,
J., and Berzal-Herranz, A. (2012) An engineered inhibitor RNA that
efficiently interferes with hepatitis C virus translation and
replication, Antiviral Res., 94, 131-138.
167.Kumar, D., Chaudhury, I., Kar, P., and Das, R.
H. (2009) Site-specific cleavage of HCV genomic RNA and its cloned core
and NS5B genes by DNAzyme, J. Gastroenterol. Hepatol.,
24, 872-878.
168.Yuan, J., Stein, D. A., Lim, T., Qui, D.,
Coughlin, S., Liu, Z., Wang, Y., Blouch, R., Moulton, H. M., Iversen,
P. L., and Yang, D. (2006) Inhibition of coxsackievirus B3 in cell
cultures and in mice by peptide-conjugated morpholino oligomers
targeting the internal ribosome entry site, J. Virol.,
80, 11510-11519.
169.Abet, V., Mariani, A., Truscott, F. R.,
Britton, S., and Rodriguez, R. (2014) Biased and unbiased strategies to
identify biologically active small molecules, Bioorg. Med.
Chem., 22, 4474-4489.
170.Dietrich, U., Durr, R., and Koch, J. (2013)
Peptides as drugs: from screening to application, Curr. Pharm.
Biotechnol., 14, 501-512.
171.Costa-Mattioli, M., Svitkin, Y., and Sonenberg,
N. (2004) La autoantigen is necessary for optimal function of the
poliovirus and hepatitis C virus internal ribosome entry site in
vivo and in vitro, Mol. Cell Biol., 24,
6861-6870.
172.Wurth, L., and Gebauer, F. (2015) RNA-binding
proteins, multifaceted translational regulators in cancer, Biochim.
Biophys. Acta, 1849, 881-886.
173.Novac, O., Guenier, A. S., and Pelletier, J.
(2004) Inhibitors of protein synthesis identified by a high throughput
multiplexed translation screen, Nucleic Acids Res., 32,
902-915.
174.Gasparian, A. V., Neznanov, N., Jha, S.,
Galkin, O., Moran, J. J., Gudkov, A. V., Gurova, A. V., and Komar, A.
A. (2010) Inhibition of encephalomyocarditis virus and poliovirus
replication by quinacrine: implications for the design and discovery of
novel antiviral drugs, J. Virol., 84, 9390-9397.
175.Wang, J., Du, J., Wu, Z., and Jin, Q. (2013)
Quinacrine impairs enterovirus 71 RNA replication by preventing binding
of polypyrimidine-tract binding protein with internal ribosome entry
sites, PLoS One, 8, e52954.
176.Rynearson, K. D., Charrette, B., Gabriel, C.,
Moreno, J., Boerneke, M. A., Dibrov, S. M., and Hermann, T. (2014)
2-Aminobenzoxazole ligands of the hepatitis C virus internal ribosome
entry site, Bioorg. Med. Chem. Lett., 24, 3521-3525.
177.Direct effect antiviral preparations registered
with WHO: https://www.whocc.no/atc_ddd_index/?code=J05A
(official web site).