REVIEW: Amyloid Properties of Titin
E. I. Yakupova1,2*, I. M. Vikhlyantsev1,2*, M. Y. Lobanov3, O. V. Galzitskaya3, and A. G. Bobylev1,2*
1Institute of Theoretical and Experimental Biophysics, Russian Academy of Sciences, 142290 Pushchino, Moscow Region, Russia; E-mail: yakupova.mira@mail.ru, ivanvikhlyantsev@gmail.com, bobylev1982@gmail.com2Pushchino State Institute of Natural Sciences, 142290 Pushchino, Moscow Region, Russia
3Institute of Protein Research, Russian Academy of Sciences, 142290 Pushchino, Moscow Region, Russia
* To whom correspondence should be addressed.
Received February 27, 2017; Revision received May 23, 2017
This review considers data on structural and functional features of titin, on the role of this protein in determination of mechanical properties of sarcomeres, and on specific features of regulation of the stiffness and elasticity of its molecules, amyloid aggregation of this protein in vitro, and possibilities of formation of intramolecular amyloid structure in vivo. Molecular mechanisms are described of protection of titin against aggregation in muscle cells. Based on the data analysis, it is supposed that titin and the formed by it elastic filaments have features of amyloid.
KEY WORDS: amyloids, amyloidoses, titin, functional amyloidsDOI: 10.1134/S0006297917130077
Abbreviations: Aβ-peptide, amyloid β-peptide; FnIII, fibronectin III-like titin domain; Ig, immunoglobulin-like titin domain.
The story of amyloid studies started in 1639 when Nicolaus Fontanus
described a strongly increased human spleen containing large white
inclusions, which seemed to be amyloid deposits [1]. After 150 years, in 1789, Antoine Portal for the
first time described liver amyloidosis [2]. In
1854, Rudolf Virchow studied inclusions in a “waxy” liver
and found structures that were stained with iodine similarly to starch
grains in plants. These structures Virchow termed “amyloid”
from the Latin “amylum” or Greek “amylon” that
means starch. However, in 1859 Carl Friedreich and August Kekule showed
that these inclusions did not have a carbohydrate component but
contained proteins. These data became a basis for studies on amyloids
as protein derivatives [3, 4].
It is generally thought that amyloids are protein aggregates with a cross-β-structure capable of binding with the dyes Thioflavin T and Congo Red, possessing apple-green birefringence in polarized light, and resistant to action of solvents and proteases [5-8]. By now, more than thirty proteins/peptides are known that form amyloids found in humans in various diseases [8, 9]. Amyloids have been shown to play a central role in the pathogenesis of Alzheimer’s and Parkinson’s diseases, type II diabetes, prion diseases, and systemic amyloidoses [6, 10-12]. Amyloid deposits have been found in the intima and media of vessels under aorta amyloidosis and in striated muscles in myocardites, myositis, and cardiomyopathies [13-15]. In particular, in blood vessels amyloid aggregates have been found formed by serum A protein and its fragments, which are accumulated in the intima and media of arterioles, under endothelium of venules [13]. In aorta amyloidoses, amyloids of medin are found [14]. In the cardiac muscle amyloids are found formed by such proteins as transthyretin, light and heavy immunoglobulin chains, serum amyloid-A, apolipoprotein, apolipoprotein AIV, fibrinogen α-chain, and atrial natriuretic factor, which contribute to development of “amyloid cardiomyopathy” or “cardiac amyloidoses” [15, 16]. Amyloid deposits containing Aβ-peptide are found in skeletal muscle in myositis [8]. However, at present, the paradigm of amyloids as negative formations for the cell, which cause development only of pathologic processes in living systems, is under revision. There are works that have demonstrated that amyloids can also play a positive role in the organism. Thus, in prokaryotes functional amyloids have been found of such proteins as curli in E. coli [17], tafi in Salmonella spp. [18], and chaplins in Streptomyces coelicolor [19]. Amyloids of these proteins were shown to participate in cell adhesion and formation of biological films, and chaplin amyloids are involved in production of aerial hyphae and dissipation of spores [20]. Functional amyloids are also found in eukaryotes: on the surface of spores and fruiting bodies of some fungi amyloid aggregates form dense hydrophobic monolayers [21], in the silk moth Antheraea polyphemus amyloids have a protective function of the chorion (egg envelope) [22]; amyloids of spidroin are components of spider silk [23]. There are also known functional amyloid fibrils in mammals that are formed in melanosomes from proteolytic fragments of protein Pmel17 [24].
Recent studies have shown that many proteins under certain conditions can form amyloids in vitro. The concept of an “amylome” appears, which denotes the universe of proteins that are capable of producing amyloid-like aggregates/fibrils [25]. These proteins also include muscle ones: myosin subfragment-1 [26], myosin-binding proteins of the titin family (C-, X-, H-proteins) [27, 28], and titin [29]. Can amyloid aggregation of titin and other muscle proteins occur in vivo, and what role might these aggregates play in the cell? These questions require further investigations. Nevertheless, recent studies allow us to state that on folding of unfolded, titin domains misfolded conformations can be produced, which the authors have named “intramolecular amyloid” [30].
This review presents data on structural and functional features of titin, the role of this protein in determination of mechanical properties of sarcomeres, specific features of regulation of the stiffness and elasticity of its molecules, amyloid aggregation of this protein in vitro, and possibilities of formation of intramolecular amyloid structures in vivo. Molecular mechanisms are described of titin protection against aggregation of its molecules in muscle cells. Based on analysis of the data, it is supposed that titin and the formed by it elastic filaments have features of amyloid.
STRUCTURE—FUNCTIONAL FEATURES OF TITIN AND ITS
DOMAINS
Titin (connectin) is a protein with the highest molecular weight among known proteins: the molecular weight of titin isoforms reaches ~3.0-3.7 MDa in striated and ~2 MDa in smooth muscles [31, 32]. Titin has a multidomain structure, and its gene can encode a protein with the following structure: a protein kinase domain, 152 Ig-domains, 132 FnIII-domains, 31 PEVK domains, 7 Z-repeats, and 33 unidentified domains (based on the UniProt database).
In sarcomeres of cardiac and skeletal muscles, the amount of titin is third after the amounts of actin and myosin. Titin molecules with the length of about 1 µm and diameter of 3-4 nm [33-36] occupy a half of the sarcomere from the M-line to the Z-disc and form the third type of so-called elastic filaments [37]. In the sarcomere A-zone, titin is bound with myosin (thick) filaments [38]. In the sarcomere I-band, some regions of the titin molecule can interact with actin (thin) filaments [39-49]. However, the major part of the titin molecule can pass easily in this zone connecting the ends of myosin filaments with the Z-disc. It is supposed that for each myosin filament, there are six titin molecules [50], with the N-ends overlapped in the Z-line and the C-ends overlapped in the sarcomere M-line [37]. It has been shown that the structure of this giant polypeptide is different in various regions of the sarcomere and contributes to their architecture and functioning. The major part (to 90%) of the titin molecule consists of repeated immunoglobulin-like (Ig) and fibronectin III-like (FnIII) domains with β-folded structure [37]. In addition to these domains, titin contains unique sequences: the kinase domain near the sarcomere M-line, N2A, N2B, and PEVK-elements in the sarcomere I-band, as well as phosphorylated regions in the sarcomere M-, I-, and Z-zones [37, 51-55]. Titin localization in all sarcomere zones, its elastic features, and interactions with many proteins promote the polyfunctionality of this protein. It has been shown that titin: (i) is a carcass for assembly of myosin filaments and sarcomere [56-60]; (ii) contributes to maintaining a highly ordered sarcomere structure and, therefore, to the muscle contractile function [61, 62]; (iii) participates in triggering and regulation of the actin–myosin interaction mediated through the binding with proteins of thin filaments [41, 63] and through changes in the ATPase activity of myosin [36, 64, 65]. It is supposed that titin, in complex with signaling proteins united by titin in the network, acts as a sensor of stretching and tension, participating in intracellular signalization, in particular, in the regulation of expression of genes of muscle proteins and of protein metabolism in the sarcomere [52, 54, 66-77].
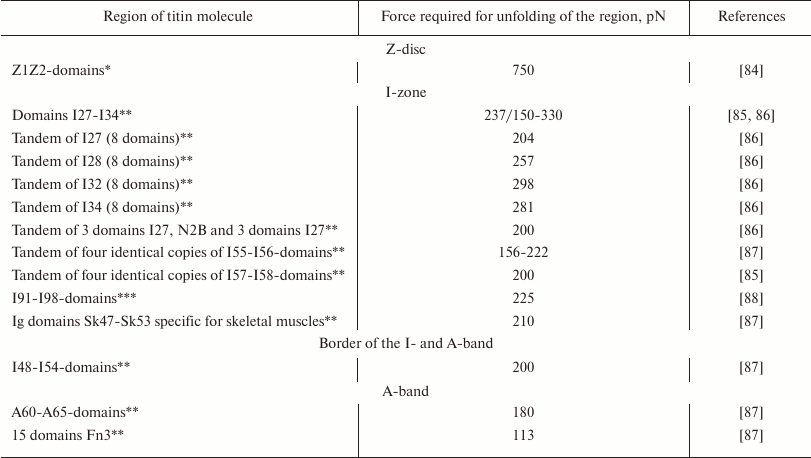
As differentiated from the sarcomere A-zone where titin is firmly bound to myosin filaments, in the I-zone the elastic part of its molecule can develop a passive tension under stretching [78] and restoring force under sarcomere contraction [79-82]. Biophysical studies have shown that this part of the titin molecule behaves as a “nonlinear entropy spring”, which straightens under the influence of a force from 20 to 30 pN and demonstrates elastic resistance under compression with the force of 2.5 pN [83]. Using atomic force microscopy or modeling the molecular dynamics resulted in the higher values of the force required for unfolding of individual titin domains or of their tandem sequences (table).
Force values required for unfolding of individual titin domains or of
tandem sequences of its domains
* The method of molecular dynamic simulations was used.
** Measured using atomic force microscopy.
*** Measured using a modified method of atomic force microscopy termed
“molecular force probe”.
The table shows that the Z1Z2-repeats of titin are the most resistant to stretching. In the stretchable I-band, the Ig domains of titin are unfolding in the range of forces of ~150-330 pN [86, 88]. In the A-part of the titin molecule, which is tightly bound to the myosin filament and does not change its length under natural conditions, the FnIII domains are unfolding at force from 100 to 200 pN [87, 89]. It should be noted that such values of the force required for unfolding the amyloid structure were obtained also for some amyloid fibrils: 115 pN for amyloids of the human prion protein (huPrP90-231) [90], 250 pN for amyloids of glucagon [91] (this value was obtained using atomic force microscopy), 522 pN for amyloids of Aβ-peptide [90] (the values were calculated using molecular dynamics simulations). Obviously, the method of molecular dynamics simulations gives overestimated values of the force, and therefore, comparing these values with others obtained using atomic force microscopy is not correct. The analogy found between titin and amyloids seems to be a consequence of a high content in them of the β-folded structure, which can be unfolded by nearly the same force. Thus, this parameter cannot be used as a basis for comparing mechanical properties of amyloid aggregates and titin molecules. Such comparisons should be based on specific structural features of the protein that determine its mechanical features or the protein itself and of its aggregates. In this connection, titin is interesting because its elasticity and resiliency mediate mechanical features of sarcomeres, cells, and the muscle as a whole.
role of titin in mechanical PROPERTIES of muscle cells. regulation of titin molecule stiffness in sarcomereS. Functional role of IDENTITY OF amino acid sequence of titin domainS
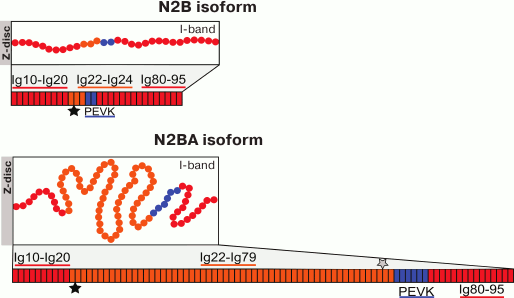
There is now no doubt that titin plays a role in mediating the elasticity of muscle cells, in particular, cardiomyocytes. The coexpression of N2B- and N2BA-isoforms of titin differing in the length of the stretchable I-part and the change in their ratio under the influence of external or internal factors (during ontogenesis, development of pathological processes, hibernation, microgravity (for references see [92]) are considered as one of molecular mechanisms of changes in the stiffness of cardiomyocytes and cardiac muscle as a whole. The titin N2BA-isoform (molecular weight ~3300 kDa) has the longer, more elastic and correspondingly less stiff I-part of the molecule; the N2B-isoform (molecular weight ~3000 kDa) of titin has the shorter, less elastic, and correspondingly stiffer I-part (Fig. 1). The content of the titin N2B-isoform is shown to directly correlate with an increase in passive tension on stretching of cardiac muscle myofibrils [93-96]. The regulation of cardiomyocyte stiffness on the level of ratio of long and short titin isoforms can be easily followed in the ontogenesis of mammals [96-98]. During the early postnatal period, the ratio of titin isoforms changes along with an increase in the pump function of the heart: the fraction of the shorter (stiffer) isoforms of this protein increases [93, 96, 97]. In adult animals, there is a correlation between heart rate and the fraction of the short N2B-isoform of titin in the left ventricle myocardium. In particular, in small animals with heart rate of 140-650 beats/min (rabbit, hamster, mouse, rat), the content of the stiff N2B-titin isoform in the left ventricle is 80-94%. In big animals with heart rate of 60-80 beats/min (sheep, pig, goat, cow), the content of the short titin isoform decreases to 41-76% [99]. It is supposed that the predominance of this titin isoform promotes an increase in the rate of the myocardium active contraction during the early systolic shortening and for the faster relaxation of the heart [99]. Thus, variability in the length of the resilient–elastic region of the titin isoforms in the I-band of the sarcomere is an important component of the molecular mechanism involved in the regulation of mechanical and contractile properties of cardiac muscle. Variability in the length of the extensible region of the titin molecule is characteristic also for its N2A-isoform in skeletal muscles. In particular, alternative splicing leads to generation of the titin N2A-isoform with shorter I-part of the molecule in the m. psoas (molecular weight ~3300 kDa) and longer I-part of the molecule in the m. soleus (molecular weight ~3700 kDa) [100]. The functional significance of such variability in the length of the elastic I-part of the titin N2A-isoform in skeletal muscle isoforms remains unknown.
Fig. 1. Schematic picture of the domain structure of titin cardiac isoforms in the sarcomere I-band. The sequence of the titin domain structure is presented according to the UniProtKB database – Q8WZ42 (TITIN_HUMAN). Both isoforms contain identical sequences: the proximal sequence Ig10-Ig20 and the distal sequence Ig80-95. As differentiated from the shorter and stiffer N2B-isoform, the N2BA-isoform of titin has the longer middle region consisting of immunoglobulin-like domains Ig22-Ig79, the majority of which is a variable (differently spliced in N2BA-isovariants) region (Ig25-79). The PEVK sequence in the N2B-isoform is shorter. Both isoforms contain the unique N2-B sequence consisting of three Ig-like domains and a unique sequence containing 572 a.a. that is located after Ig22. In the N2BA-isoform there is also the N2A sequence consisting of four Ig-like domains and a region of unique sequence containing 106 a.a. (located before the PEVK region); in the scheme, their localization is shown by asterisks (N2B – black, N2A – gray).
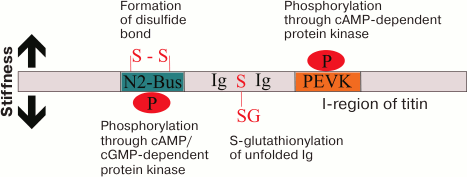
Stiffness and elasticity of titin molecules are regulated not only through changes in their length in the sarcomere I-zone, but also through posttranslational modifications [101] (Fig. 2). It has been shown that phosphorylation of the N2-B region by cGMP- or cAMP-dependent protein kinases decreases the stiffness of the titin molecule, whereas phosphorylation of the PEVK-sequence (enriched with residues of proline, glutamic acid, valine, and lysine) by cAMP-dependent protein kinase increases its stiffness [102, 103]. Stiffness of titin molecules can be induced by oxidative stress emerging in myocardium infarction, obesity, or diabetes mellitus, which makes worse the diastolic function of the left ventricle [104]. This is associated with generation of disulfide bonds in the titin N2-B sequence, which enhances the stiffness of the molecule. It has been also shown that the enhanced stiffness of titin can be compensated due to reversible S-glutathionylation of cysteines in the unfolded (due to increased load on the sarcomere) Ig-domains [104].
Fig. 2. Posttranslational mechanisms of changes in the stiffness of titin. Generation of disulfide bonds in the N2B region and phosphorylation of PEVK by cAMP-dependent protein kinase contributes to the stiffness of the molecule. On the contrary, phosphorylation through cAMP/cGMP-dependent protein kinase of the N2-B-sequence and S-glutathionylation of unfolded Ig-domains of titin reduces stiffness of titin molecules.
Mechanical properties of titin can be regulated more finely through variations in the identity of the amino acid sequence of its domains. This hypothesis was proposed based on data about the different aggregability in vitro of titin domains with different degree of amino acid sequence identity [105] and on our calculations of the Ig-domains of the variable region of the I-band of the N2B- and N2BA-isoforms of titin (Fig. 1). Data obtained using the dynamic light scattering method revealed that the aggregation rate in vitro of titin domains with higher identity of the amino acid sequence was higher than that of titin domains with lower identity of the amino acid sequence [105]. Using the BLAST program, we found that the domain of a variable region expressed only in the N2BA-isoform has higher identity of amino acid sequence (~32%, calculated for 26 domains) than that of other domains of the I-band of the N2B- and N2BA-titin isoforms (~23-25%, calculated for 30 domains). Up to now, there is no exact idea about the number of molecules of this giant protein and about their arrangement in the sarcomere I-band. However, considering data on titin aggregation in vitro [29, 30, 106, 107], including its amyloid aggregation [29], the binding in vivo of closely localized domains of one or several molecules of this protein cannot be neglected. In this case, it may be supposed that the domains of the variable region of the N2BA-isoform will be more prone to aggregation than other domains. This will lead to increase in the stiffness of the aggregated region of the N2BA isoform of titin, which can directly influence changes in the mechanical and contractile properties of muscle cells.
AGGREGATION PROPERTIES OF TITIN in vitro AND in
vivo. MECHANISMS OF CELL PROTECTION AGAINST TITIN
AGGREGATION
It is unknown whether the above-mentioned aggregation of titin can occur in vivo. However, studies on the in vitro aggregation of short amino acid sequences of some titin domains performed in 2005 [105] and other proteins [106] led to the conclusion that the capability of aggregating increases if the identity of the amino acid sequence of the domains is more than 30-40% [105]. These studies were preconditioned by the earlier works, which revealed the ability of neighboring identical domains of titin to form in vitro misfolded structures [107]. Recent studies published in Nature Communications in 2015 showed that, independently of the amino acid identity, misfolded conformations are produced during the folding of unfolded titin domains [30]. Molecular simulations allowed the authors to suppose that a significant part of these misfolded conformations can be an intramolecular amyloid [30]. Note that the higher identity in the amino acid sequence was favorable for production of more stable forms [30]. The authors of this study supposed that multidomain proteins including titin during evolution could undergo changes reducing the identity of the amino acid sequence of their domains for preventing or decreasing the probability of formation in vivo of resistant protein aggregates, including amyloid aggregates [30].
Nevertheless, the possibility of titin molecules to form aggregates in vivo in the sarcomere I-band cannot be excluded. It has been shown that during sarcomere extension, the titin domains can unfold, uncovering latent hydrophobic regions, and this can lead to aggregation of the protein and disturbance of its functions [108, 109]. The aggregation of unfolded Ig-domains in the I-band can result in an abnormally high stiffness of titin molecules and, consequently, of myocytes [110]. Interaction of this part of titin molecule with small heat shock proteins (sHsps) capable of suppressing aggregation of many proteins is one of the mechanisms preventing Ig-domain aggregation in the sarcomere. In particular, it has been shown that Hsp27 and αB-crystallin do not interact with distal Ig-domains localized near the sarcomere A-band, but they bind to Ig-domains of the titin molecule extensible part, which is localized between the Z-disc and PEVK [110]. It should be noted that these Ig-domains, as discriminated from PEVK and N2-B-sequences, have a higher tendency for aggregating under conditions of partial denaturation [110]. How can these differences be explained? It is known that proteins with unordered structure are resistant to aggregation due to different factors: a high total charge and a low hydrophobicity, a small number of amyloidogenic regions, and a high content of proline residues [105, 111]. The PEVK region of titin is enriched with proline and has a comparatively high total charge [34]. The N2-B-sequence also contains many proline residues and higher total charge than the typical Ig-domain of titin [110]. Thus, the presence in the titin molecule of PEVK and N2-B-sequences decreases the probability of in vivo aggregation of this part of the protein. However, under conditions of muscle overextension, which occurs during intensive physical exercises and in pathologies (e.g. at ischemia when the death of myocytes can lead to overextension of the adjacent cells), the probability of unfolding and, consequently, of aggregation of titin Ig-domains increases [86]. Although the unfolded Ig-domains of titin are folding repeatedly in vitro in the absence of small heat shock proteins [108], this process can be difficult in vivo under a sufficiently tight localization of proteins in the sarcomere even if the above-mentioned proteins are present. The higher identity of the amino acid sequence of the variable region of the N2BA-isoform increases the probability of titin aggregation in vivo, which undoubtedly will lead to a significant increase in the stiffness of titin filaments. In turn, this will have negative consequences for mechanical properties and contractility of myocytes and the muscle as a whole. Increased proteolysis of titin in the sarcomere can protect against these changes. However, this pathway does not exclude accumulation of titin fragments in cells and formation of amyloid aggregates by them in the cytosol. In this case, cell autophagy is the last stage of the organism’s protection against uncontrolled aggregation of cytoskeletal proteins of the sarcomere [110, 112, 113].
Can titin aggregates accumulate in vivo? Our studies in vitro have shown that such possibility exists. In particular, it has been shown that under conditions close to physiological, smooth-muscle titin can form amyloid aggregates within short time intervals (tens of minutes) [29]. Circular dichroism did not reveal structural rearrangements of the type α-helix transition into β-structure, which are characteristic for other amyloidogenic proteins [114, 115]. A pronounced cytotoxic effect of titin amyloid aggregations was detected on smooth muscle cells of bovine aorta culture, and this effect was accompanied by disorganization of the actin cytoskeleton [29]. These data not only demonstrated that titin amyloid aggregation can occur in vivo, but also suggest that this protein should be involved in the development of muscle amyloidoses.
POSSIBLE FUNCTIONAL ROLE OF TITIN AGGREGATION IN
SARCOMERES
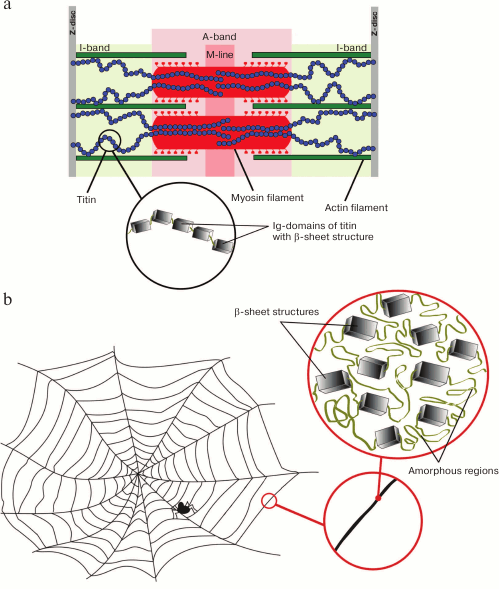
It is generally believed that amyloids play a negative role in living cells. Due to their stiffness, amyloid fibrils can mechanically tear the cell membrane; therefore, the accumulation of amyloid aggregates, which are resistant to proteolytic degradation, leads to cell death [116, 117] and development of a pathologic process [118]. However, it should be noted that a high stiffness is also inherent in functional amyloids, e.g. amyloids of spidroin, a protein participating in formation of solid elastic threads of the spider silk. Functional amyloids of spidroin and the titin molecule are similar in morphology (Fig. 3). The silk protein is enriched with a β-folded structure (up to 40-50% of the total volume of a silk fibril [119]), whereas the remaining part is filled with less ordered, possibly amorphous structures [120]. In addition to the β-folded structure, the titin molecule has about 50% amorphous structures that has been shown by circular dichroism [29] and short α-helical regions, e.g. in the PEVK sequence [40] – in the kinase domain [121].
Fig. 3. Morphological similarity of titin molecule (a) and functional amyloids of spidroin – the protein of spider silk (b).
Thin elastic threads of titin molecules form the intracellular cytoskeletal extensible carcass, which determines mechanical properties of muscle tissue. Perhaps the aggregation of titin molecules in the sarcomere I-band, including “formation of intramolecular amyloid structures”, can play a functional role – to contribute to increasing muscle stiffness. In turn, this can play a protective role counteracting overextension of sarcomeres having unfavorable consequences for the muscle. Changes in the mechanosensory role of titin in the case of aggregation of its molecule also cannot be excluded.
In conclusion, based on analysis of data on the properties of titin, we pay special attention to the following facts: titin forms the intracellular cytoskeletal elastic carcass that determines mechanical properties of sarcomeres and muscle as a whole; titin forms amyloid aggregates in vitro; on folding of unfolded domains in the titin molecule, amyloid-like structures can be produced; titin has a morphological similarity with functional amyloids of spidroin. Based on these data, can we state that titin is a molecular amyloid? Up to now, we cannot answer this question in the affirmative. However, it is clear that during evolution one of the most unique structures of living nature has been created that combines features of amyloid and elastic protein participating not only in formation of sarcomere and maintenance of its structure, but also in the fine regulation of the actin–myosin interaction and intracellular signalization.
Acknowledgments
This work was supported by the program “Molecular and Cell Biology” of the Russian Academy of Sciences Presidium (project No. 01201353567), and by Russian Science Foundation (project No.14-14-00879).
REFERENCES
1.Fonteyn, N. (1639) Responsionum et Curationum
Medicinalium, Amstelodami, Amsterdam.
2.Kyle, R. (2001) Amyloidosis: a convoluted story,
Br. J. Haematol., 114, 529-538.
3.Sipe, J. D., and Cohen, A. S. (2000) Review:
History of amyloid fibril, J. Struct. Biol., 130,
88-89.
4.Kelly, J. J. (1987) Amyloidosis. Topics in the
Neurosciences, Top. Neurosci., 5, 105-127.
5.Langkilde, A. E., and Vestergaard, B. (2009)
Methods for structural characterization of prefibrillar intermediates
and amyloid fibrils, FEBS Lett., 583, 2600-2609.
6.Dobson, C. M. (2004) Principles of protein folding,
misfolding and aggregation, Semin. Cell Dev. Biol., 15,
3-16.
7.Ross, C. A., and Poirier, M. A. (2004) Protein
aggregation and neurodegenerative disease, Nat. Med., 10,
10-17.
8.Uversky, V. N., and Fink, A. L. (2004)
Conformational constraints for amyloid fibrillation: the importance of
being unfolded, Biochim. Biophys. Acta, 1698,
131-153.
9.Chiti, F., and Dobson, C. M. (2006) Protein
misfolding, functional amyloid, and human disease, Annu. Rev.
Biochem., 75, 333-366.
10.Knowles, T. P., Vendruscolo, M., and Dobson, C.
M. (2014) The amyloid state and its association with protein misfolding
diseases, Nat. Rev. Mol. Cell Biol., 15, 384-396.
11.Dobson, C. M. (2004) Experimental investigation
of protein folding and misfolding, Methods, 34, 4-14.
12.Buxbaum, J. N., and Linke, R. P. (2000) A
molecular history of the amyloidoses, J. Mol. Biol., 421,
142-159.
13.Rosenthal, C. J., Franklin, E. C., Frangione, B.,
and Greenspan, J. (1976) Isolation and partial characterization of SAA
– an amyloid-related protein from human serum, J.
Immunol., 116, 1415-1418.
14.Haggqvist, B., Naslund, J., Sletten, K.,
Westermark, G. T., Mucchiano, G., Tjernberg, L. O., Nordstedt, C.,
Engstrom, U., and Westermark, P. (1999) Medin: an integral fragment of
aortic smooth muscle cell-produced lactadherin forms the most common
human amyloid, Proc. Natl. Acad. Sci. USA, 96,
8669-8674.
15.Barsukov, A., Shustov, S., Shkodkin, I.,
Vorob’ev, S., and Pronina, E. (2005) Hypertrophic cardiomyopathy
and heart amyloidosis, Vrach. Delo, 10, 42-46.
16.Guan, J., Mishra, S., Falk, R. H., and Liao, R.
(2012) Current perspectives on cardiac amyloidosis, Am. J. Physiol.
Heart Circ. Physiol., 302, 544-552.
17.Olsen, A., Jonsson, A., and Normark, S. (1989)
Fibronectin binding mediated by a novel class of surface organelles on
Escherichia coli, Nature, 338, 652-655.
18.Rcmling, U., Bian, Z., Hammar, M., Sierralta, W.
D., and Normark, S. (1998) Curli fibers are highly conserved between
Salmonella typhimurium and Escherichia coli with respect
to open structure and regulation, J. Bacteriol., 180,
722-731.
19.Claessen, D., Rink, R., De Jong, W., Siebring,
J., De Vreughd, P., Boersma, F. G. H., Dijkhuizen, L., and Wcsten, H.
A. B. (2003) A novel class of secreted hydrophobic proteins is involved
in aerial hyphae formation in Streptomyces coelicolor by forming
amyloid-like fibrils, Genes Dev., 17, 1714-1726.
20.Otzen, D., and Nielsen, P. H. (2008) We find them
here, we find them there: functional bacterial amyloid, Cell Mol.
Life Sci., 65, 910-927.
21.Woesten, H. A. B., and De Vocht, M. L. (2000)
Hydrophobins, the fungal coat unraveled, Biochim. Biophys. Acta,
1469, 79-86.
22.Iconomidou, V. A., Chryssikos, G. D., Gionis, V.,
Galanis, A. S., Cordopatis, P., Hoenger, A., and Hamodrakas, S. J.
(2006) Amyloid fibril formation propensity is inherent into the
hexapeptide tandemly repeating sequence of the central domain of silk
moth chorion proteins of the A-family, J. Struct. Biol.,
156, 480-488.
23.Slotta, U., Hess, S., Spiess, K., Stromer, T.,
Serpell, L., and Scheibel, T. (2007) Spider silk and amyloid fibrils: a
structural comparison, Macromol. Biosci., 7, 183-188.
24.Fowler, D. M., Koulov, A. V., Alory-Jost, C.,
Marks, M. S., Balch, W. E., and Kelly, J. W. (2006) Functional amyloid
formation within mammalian tissue, PLoS Biol., 4, e6.
25.Goldschmidt, L., Teng, P. K., Riek, R., and
Eisenberg, D. (2010) Identifying the amylome, proteins capable of
forming amyloid-like fibrils, Proc. Natl. Acad. Sci. USA,
107, 3487-3492.
26.Komatsu, H., Shinotani, N., Kimori, Y., Tokuoka,
J., Kaseda, K., Nakagawa, H., and Kodama, T. (2006) Aggregation of
partially unfolded myosin subfragment-1 into spherical oligomers with
amyloid-like dye-binding properties, J. Biochem., 139,
989-996.
27.Marsagishvili, L. G., Shpagina, M. D.,
Emel’yanenko, V. I., and Podlubnaya, Z. A. (2005) Sarcomeric
proteins of the titin family form amyloids, Biophysics,
50, 704-709.
28.Marsagishvili, L. G., Shpagina, M. D., Shatalin,
Y. V., Shubina, V. S., Naumov, A. A., Potselueva, M. M., and
Podlubnaya, Z. A. (2006) Study of the cytotoxicity of protein X amyloid
fibrils, Biophysics, 51, 705-709.
29.Bobylev, A. G., Galzitskaya, O. V., Fadeev, R.
S., Bobyleva, L. G., Yurshenas, D. A., Molochkov, N. V., Dovidchenko,
N. V., Selivanova, O. M., Penkov, N. V., Podlubnaya, Z. A., and
Vikhlyantsev, I. M. (2016) Smooth muscle titin forms in vitro
amyloid aggregates, Biosci. Rep., 36, e00334.
30.Borgia, A., Kemplen, K. R., Borgia, M. B.,
Soranno, A., Shammas, S., Wunderlich, B., Nettels, D., Best, R. B.,
Clarke, J., and Schuler, B. (2015) Transient misfolding dominates
multidomain protein folding, Nat. Commun., 6, 8861.
31.Labeit, S., Lahmers, S., Burkart, C., Fong, C.,
McNabb, M., Witt, S., Witt, C., Labeit, D., and Granzier, H. (2006)
Expression of distinct classes of titin isoforms in striated and smooth
muscles by alternative splicing, and their conserved interaction with
filamins, J. Mol. Biol., 362, 664-681.
32.Meyer, L. C., and Wright, N. T. (2013) Structure
of giant muscle proteins, Front. Physiol., 4, 368.
33.Suzuki, J., Kimura, S., and Maruyama, K. (1994)
Electron microscopic filament lengths of connection and its fragments,
J. Biochem., 116, 406-410.
34.Tskhovrebova, L., and Trinick, J. (1997) Direct
visualization of extensibility in isolated titin molecules, J. Mol.
Biol., 265, 100-106.
35.Vikhlyantsev, I. M. (2011) Polymorphism of
Striated Muscle Titin in Norm, Adaptation and Pathology: doctoral
dissertation [in Russian], Pushchino, Institute of Theoretical and
Experimental Biophysics, Russian Academy of Sciences, p. 235.
36.Vikhlyantsev, I. M., Okuneva, A. D., Shpagina, M.
D., Shumilina, Y. V., Molochkov, N. V., Salmov, N. N., and Podlubnaya,
Z. A. (2011) Changes in isoform composition, structure and functional
properties of titin from Mongolian gerbil (Meriones
unguiculatus) after space flight, Biochemistry (Moscow),
76, 1312-1320.
37.Gregorio, C. C., Granzier, H., Sorimachi, H., and
Labeit, S. (1999) Muscle assembly: a titanic achievement? Curr.
Opin. Cell Biol., 11, 18-25.
38.Houmeida, A., Holt, J., Tskhovrebova, L., and
Trinick, J. (1995) Studies of the interaction between titin and myosin,
J. Cell Biol., 131, 1471-1481.
39.Trombitas, K., and Pollack, G. H. (1993) Elastic
properties of the titin filament in the Z-line region of vertebrate
striated muscle, J. Muscle Res. Cell Motil., 14,
416-422.
40.Gutierrez-Cruz, G., Van Heerden, A. H., and Wang,
K. (2001) Modular motif, structural folds and affinity profiles of the
PEVK segment of human fetal skeletal muscle titin, J. Biol.
Chem., 276, 7442-7449.
41.Kulke, M., Fujita-Becker, S., Rostkova, E.,
Neagoe, C., Labeit, D., Manstein, D. J., Gautel, M., and Linke, W. A.
(2001) Interaction between PEVK-titin and actin filaments: origin of a
viscous force component in cardiac myofibrils, Circ. Res.,
89, 874-881.
42.Linke, W. A., Kulke, M., Li, H., Fujita Becker,
S., Neagoe, C., Manstein, D. J., Gautel, M., and Fernandez, J. M.
(2002) PEVK domain of titin: an entropic spring with actin-binding
properties, J. Struct. Biol., 137, 194-205.
43.Podlubnaya, Z. A., Shpagina, M. D., Vikhlyantsev,
I. M., Malyshev, S. L., Udaltsov, S. N., Ziegler, C., and Beinbrech, G.
(2003) Comparative electron microscopic study on projectin and titin
binding to F-actin, Insect. Biochem. Mol. Biol., 33,
789-793.
44.Niederlander, N., Raynaud, F., Astier, C., and
Chaussepied, P. (2004) Regulation of the actin–myosin interaction
by titin, Eur. J. Biochem., 271, 4572-4581.
45.Raynaud, F., Astier, C., and Benyamin, Y. (2004)
Evidence for a direct but sequential binding of titin to tropomyosin
and actin filaments, Biochim. Biophys. Acta, 1700,
171-178.
46.Bianco, P., Nagy, A., Kengyel, A., Szatmari, D.,
Martonfalvi, Z., Huber, T., and Kellermayer, M. S. (2007) Interaction
forces between F-actin and titin PEVK domain measured with optical
tweezers, Biophys. J., 93, 2102-2109.
47.Fukushima, H., Chung, C. S., and Granzier, H.
(2010) Titin-isoform dependence of titin–actin interaction and
its regulation by S100A1/Ca2+ in skinned myocardium, J.
Biomed. Biotechnol., 2010, 9.
48.Chung, C. S., Bogomolovas, J., Gasch, A.,
Hidalgo, C. G., Labeit, S., and Granzier, H. L. (2011)
Titin–actin interaction: PEVK-actin-based viscosity in a large
animal, J. Biomed. Biotechnol., 2011, 8.
49.Chung, C. S., Methawasin, M., Nelson, O. L.,
Radke, M. H., Hidalgo, C. G., Gotthardt, M., and Granzier, H. L. (2011)
Titin based viscosity in ventricular physiology: an integrative
investigation of PEVK–actin interactions, J. Mol. Cell
Cardiol., 51, 428-434.
50.Liversage, A. D., Holmes, D., Knight, P. J.,
Tskhovrebova, L., and Trinick, J. (2001) Titin and the sarcomere
symmetry paradox, J. Mol. Biol., 305, 401-409.
51.Labeit, S., and Kolmerer, B. (1995) Titins: giant
proteins in charge of muscle ultrastructure and elasticity,
Science, 270, 293-296.
52.Granzier, H., and Labeit, S. (2004) The giant
protein titin: a major player in myocardial mechanics, signaling, and
disease, Circ. Res., 94, 284-295.
53.Linke, W. (2008) Sense and stretchability: the
role of titin and titin-associated proteins in myocardial
stress-sensing and mechanical dysfunction, Cardiovasc. Res.,
77, 637-648.
54.Voelkel, T., and Linke, W. (2011)
Conformation-regulated mechanosensory control via titin domains in
cardiac muscle, Eur. J. Physiol., 4, 143-154.
55.Gautel, M. (2011) Cytoskeletal protein kinases:
titin and its relations in mechanosensing, Eur. J. Physiol.,
462, 119-134.
56.Peckham, M., Young, P., and Gautel, M. (1997)
Constitutive and variable regions of Z-disk titin/connectin in
myofibril formation: a dominant-negative screen, Cell Struct.
Funct., 22, 95-101.
57.Gregorio, C. C., Trombitas, K., Centner, T.,
Kolmerer, B., Stier, G., Kunke, K., Suzuki, K., Obermayr, F., Herrmann,
B., Granzier, H., Sorimachi, H., and Labeit, S. (1998) The
NH2 terminus of titin spans the Z-disc: its interaction with
a novel 19-kDa ligand (T-cap) is required for sarcomeric integrity,
J. Cell Biol., 143, 1013-1027.
58.Ayoob, J. C., Turnacioglu, K. K., Mittal, B.,
Sanger, J. M., and Sanger, J. W. (2000) Targeting of cardiac muscle
titin fragments to the Z-bands and dense bodies of living muscle and
non-muscle cells, Cell Motil. Cytoskeleton, 45,
67-82.
59.Van der Ven, P. F., Bartsch, J. W., Gautel, M.,
Jockusch, H., and Furst, D. O. (2000) A functional knock-out of titin
results in defective myofibril assembly, J. Cell Sci.,
113, 1405-1414.
60.Person, V., Kostin, S., Suzuki, K., Labeit, S.,
and Schaper, J. (2000) Antisense oligonucleotide experiments elucidate
the essential role of titin in sarcomerogenesis in adult rat
cardiomyocytes in long-term culture, J. Cell Sci., 113,
3851-3859.
61.Horowits, R., Kempner, E. S., Bisher, M. E., and
Podolsky, R. J. (1986) A physiological role for titin and nebulin in
skeletal muscle, Nature, 323, 160-164.
62.Higuchi, H. (1992) Changes in contractile
properties with selective digestion of connectin (titin) in skinned
fibers of frog skeletal muscle, J. Biochem., 111,
291-295.
63.Lee, E. J., Peng, J., Radke, M., Gotthardt, M.,
and Granzier, H. L. (2010) Calcium sensitivity and the
Frank–Starling mechanism of the heart are increased in titin N2B
region-deficient mice, J. Mol. Cell Cardiol., 49,
449-458.
64.Kimura, S., Maruyama, K., and Huang, Y. P. (1984)
Interactions of muscle beta-connectin with myosin, actin, and
actomyosin at low ionic strengths, Biochem. J., 96,
499-506.
65.Vikhliantsev, I. M., and Podlubnaia, Z. A. (2003)
Phosphorylation of sarcomeric cytoskeletal proteins – an adaptive
factor for inhibiting the contractile activity of muscle during
hibernation, Biofizika, 48, 499-504.
66.Granzier, H. L., and Labeit, S. (2006) The giant
muscle protein titin is an adjustable molecular spring, Exerc. Sport
Sci. Rev., 34, 50-53.
67.Linke, W. (2008) Sense and stretchability: the
role of titin and titin-associated proteins in myocardial
stress-sensing and mechanical dysfunction, Cardiovasc. Res.,
77, 637-648.
68.Linke, W. A., and Kruger, M. (2010) The giant
protein titin as an integrator of myocyte signaling pathways,
Physiology (Bethesda), 25, 186-198.
69.LeWinter, M. M., and Granzier, H. (2010) Cardiac
titin: a multifunctional giant, Circulation, 121,
2137-2145.
70.Ottenheijm, C. A., and Granzier, H. (2010) Role
of titin in skeletal muscle function and disease, Adv. Exp. Med.
Biol., 682, 105-122.
71.Gautel, M. (2011) The sarcomeric cytoskeleton:
who picks up the strain? Curr. Opin. Cell Biol., 23,
39-46.
72.Gautel, M. (2011) Cytoskeletal protein kinases:
titin and its relations in mechanosensing, Pflugers Arch.,
462, 119-134.
73.Tskhovrebova, L., and Trinick, J. (2005) Muscle
disease: a giant feels the strain, Nat. Med., 11,
478-479.
74.Lange, S., Xiang, F., Yakovenko, A., Vihola, A.,
Hackman, P., Rostkova, E., Kristensen, J., Brandmeier, B., Franzen, G.,
Hedberg, B., Gunnarsson, L. G., Hughes, S. M., Marchand, S., Sejersen,
T., Richard, I., Edstrom, L., Ehler, E., Udd, B., and Gautel, M. (2005)
The kinase domain of titin controls muscle gene expression and protein
turnover, Science, 308, 1599-1603.
75.Puchner, E. M., Alexandrovich, A., Kho, A. L.,
Hensen, U., Schafer, L. V., Brandmeier, B., Grater, F., Grubmuller, H.,
Gaub, H. E., and Gautel, M. (2008) Mechanoenzymatics of titin kinase,
Proc. Natl. Acad. Sci. USA, 105, 13385-13390.
76.Buyandelger, B., Ng, K. E., Miocic, S., Gunkel,
S., Piotrowska, I., Ku, C. H., and Knoll, R. (2011) Genetics of
mechanosensation in the heart, J. Cardiovasc. Transl. Res.,
4, 238-244.
77.Stahl, S. W., Puchner, E. M., Alexandrovich, A.,
Gautel, M., and Gaub, H. E. (2011) A conditional gating mechanism
assures the integrity of the molecular force-sensor titin kinase,
Biophys. J., 101, 1978-1986.
78.Erickson, H. P. (1997) Stretching single protein
molecules: titin is a weird spring, Science, 276,
1090-1092.
79.Linke, W. A., Popov, V. I., and Pollack, G. H.
(1994) Passive and active tension in single cardiac myofibrils,
Biophys. J., 67, 782-792.
80.Granzier, H. L., and Irving, T. C. (1995) Passive
tension in cardiac muscle: contribution of collagen, titin,
microtubules, and intermediate filaments, Biophys. J.,
68, 1027-1044.
81.Linke, W. A., Bartoo, M. L., Ivemeyer, M., and
Pollack, G. H. (1996) Limits of titin extension in single cardiac
myofibrils, J. Muscle Res. Cell Motil., 17, 425-438.
82.Helmes, M., Trombitas, K., and Granzier, H.
(1996) Titin develops restoring force in rat cardiac myocytes, Circ.
Res., 79, 619-626.
83.Kellermayer, M. S., Smith, S. B., Granzier, H.
L., and Bustamante, C. (1997) Folding–unfolding transitions in
single titin molecules characterized with laser tweezers,
Science, 276, 1112-1116.
84.Lee, E. H., Gao, M., Pinotsis, N., Wilmanns, M.,
and Schulten, K. (2006) Mechanical strength of the titin
Z1Z2/telethonin complex, Structure, 14, 497-509.
85.Grutzner, A., Garcia-Manyes, S., Kotter, S.,
Badilla, C. L., Fernandez, J. M., and Linke, W. A. (2009) Modulation of
titin-based stiffness by disulfide bonding in the cardiac titin N2-B
unique sequence, Biophys. J., 97, 825-834.
86.Li, H., Linke, W. A., Oberhauser, A. F.,
Carrion-Vazquez, M., Kerkvliet, J. G., Lu, H., Marszalek, P. E., and
Fernandez, J. M. (2002) Reverse engineering of the giant muscle protein
titin, Nature, 418, 998-1002.
87.Rief, M., Gautel, M., Schemmel, A., and Gaub, H.
E. (1998) The mechanical stability of immunoglobulin and fibronectin
III domains in the muscle protein titin measured by atomic force
microscopy, Biophys. J., 75, 3008-3014.
88.Watanabe, K., Muhle-Goll, C., Kellermayer, M. S.,
Labeit, S., and Granzier, H. (2002). Different molecular mechanics
displayed by titin’s constitutively and differentially expressed
tandem Ig segments, J. Struct. Biol., 137, 248-258.
89.Wang, S. M., Jeng, C. J., and Sun, M. C. (1992)
Studies on the interaction between titin and myosin, Histol.
Histopathol., 7, 333-337.
90.Schleeger, M., Van den Akker, C. C.,
Deckert-Gaudig, T., Deckert, V., Velikov, K. P., Koenderink, G., and
Bonn, M. (2013) Amyloids: from molecular structure to mechanical
properties, Polymer, 54, 2473-2488.
91.Volpatti, L. R., and Knowles, T. P. J. (2014)
Polymer physics inspired approaches for the study of the mechanical
properties of amyloid fibrils, J. Polymer Sci., 52,
281-292.
92.Vikhlyantsev, I. M., and Podlubnaya, Z. A. (2012)
New titin (connectin) isoforms and their functional role in striated
muscles of mammals: facts and suppositions, Biochemistry
(Moscow), 77, 1515-1535.
93.Opitz, C. A., and Linke, W. A. (2005) Plasticity
of cardiac titin/connectin in heart development, J. Muscle Res. Cell
Motil., 26, 333-342.
94.Neagoe, C., Kulke, M., Del Monte F., Gwathmey, J.
K., De Tombe, P. P., Hajjar, R., and Linke, W. A. (2002) Titin isoform
switch in ischemic human heart disease, Circulation, 106,
1333-1341.
95.Neagoe, C., Opitz, C. A., Makarenko, I., and
Linke, W. A. (2003) Gigantic variety: expression patterns of titin
isoforms in striated muscles and consequences for myofibrillar passive
stiffness, J. Muscle Res. Cell Motil., 24, 175-189.
96.Opitz, C. A., Leake, M. C., Makarenko, I., Benes,
V., and Linke, W. A. (2004) Developmentally regulated switching of
titin size alters myofibrillar stiffness in the perinatal heart,
Circ. Res., 94, 967-975.
97.Lahmers, S., Wu, Y., Call, D. R., Labeit, S., and
Granzier, H. (2004) Developmental control of titin isoform expression
and passive stiffness in fetal and neonatal myocardium, Circ.
Res., 94, 505-513.
98.Warren, C. M., Krzesinski, P. R., Campbell, K.
S., Moss, R. L., and Greaser, M. L. (2004) Titin isoform changes in rat
myocardium during development, Mech. Dev., 121,
1301-1312.
99.Opitz, C. A., Kulke, M., Leake, M. C., Neagoe,
C., Hinssen, H., Hajjar, R. J., and Linke, W. A. (2003) Damped elastic
recoil of the titin spring in myofibrils of human myocardium, Proc.
Natl. Acad. Sci. USA, 100, 12688-12693.
100.Freiburg, A., Trombitas, K., Hell, W., Cazorla,
O., Fougerousse, F., Centner, T., Kolmerer, B., Witt, C., Beckmann, J.
S., Gregorio, C. C., Granzier, H., and Labeit, S. (2000) Series of
exon-skipping events in the elastic spring region of titin as the
structural basis for myofibrillar elastic diversity, Circ. Res.,
86, 1114-11121.
101.Linke, W. A., and Hamdani, N. (2014) Gigantic
business: titin properties and function through thick and thin,
Circ. Res., 114, 1052-1068.
102.Kotter, S., Gout, L., Von Frieling-Salewsky,
M., Muller, A. E., Helling, S., Marcus, K., Dos Remedios, C., Linke, W.
A., and Kruger, M. (2013) Differential changes in titin domain
phosphorylation increase myofilament stiffness in failing human hearts,
Cardiovasc. Res., 99, 648-656.
103.Kruger, M., and Linke, W. A. (2006) Protein
kinase-A phosphorylates titin in human heart muscle and reduces
myofibrillar passive tension, J. Muscle Res. Cell Motil.,
27, 435-444.
104.Beckendorf, L., and Linke, W. A. (2015)
Emerging importance of oxidative stress in regulating striated muscle
elasticity, J. Muscle Res. Cell Motil., 36, 25-36.
105.Wright, C. F., Teichmann, S. A., Clarke, J.,
and Dobson, C. M. (2005) The importance of sequence diversity in the
aggregation and evolution of proteins, Nature, 438,
878-881.
106.Sonnen, A. F.-P., Yu, C., Evans, E. J., Stuart,
D. I., Davis, S. J., and Gilbert, R. J. C. (2010) Domain metastability:
a molecular basis for immunoglobulin deposition? J. Mol. Biol.,
399, 207-213.
107.Oberhauser, A. F., Marszalek, P. E.,
Carrion-Vazquez, M., and Fernandez, J. M. (1999) Single protein
misfolding events captured by atomic force microscopy, Nat. Struct.
Mol. Biol., 6, 1025-1028.
108.Minajeva, A., Kulke, M., Fernandez, J. M., and
Linke, W. A. (2001) Unfolding of titin domains explains the
viscoelastic behavior of skeletal myofibrils, Biophys. J.,
80, 1442-1451.
109.Rief, M., Gautel, M., Oesterhelt, F.,
Fernandez, J. M., and Gaub, H. E. (1997) Reversible unfolding of
individual titin immunoglobulin domains by AFM, Science,
276, 1109-1112.
110.Kotter, S., Unger, A., Hamdani, N., Lang, P.,
Vorgerd, M., Nagel-Steger, L., and Linke, W. A. (2014) Human myocytes
are protected from titin aggregation-induced stiffening by small heat
shock proteins, J. Cell Biol., 204, 187-202.
111.Monsellier, E., and Chiti, F. (2007) Prevention
of amyloid-like aggregation as a driving force of protein evolution,
EMBO Rep., 8, 737-742.
112.Galvez, A. S., Diwan, A., Odley, A. M., Hahn,
H. S., Osinska, H., Melendez, J. G., Robbins, J., Lynch, R. A.,
Marreez, Y., and Dorn, G. W., 2nd (2007) Cardiomyocyte degeneration
with calpain deficiency reveals a critical role in protein homeostasis,
Circ. Res., 100, 1071-1078.
113.Willis, M. S., Schisler, J. C., Portbury, A.
L., and Patterson, C. (2009) Build it up-tear it down: protein quality
control in the cardiac sarcomere, Cardiovasc. Res., 81,
439-448.
114.Selivanova, O. M., and Galzitskaya, O. V.
(2012) Structural polymorphism and possible pathways of amyloid fibril
formation on the example of insulin protein, Biochemistry
(Moscow), 77, 1237-1248.
115.Gross, M. (2000) Proteins that convert from
alpha helix to beta sheet: implications for folding and disease,
Curr. Protein Pept. Sci., 1, 339-347.
116.Engel, M. F., Khemtemourian, L., Kleijer, C.
C., Meeldijk, H. J. D., Jacobs, J., Verkleij, A. J., De Kruijff, B.,
Killian, J. A., and Hoppener, J. W. M. (2008) Membrane damage by human
islet amyloid polypeptide through fibril growth at the membrane,
Proc. Natl. Acad. Sci. USA, 105, 6033.
117.Fitzpatrick, A. W. P., Park, S. T., and Zewail,
A. H. (2013) Exceptional rigidity and biomechanics of amyloid revealed
by 4D electron microscopy, Proc. Natl. Acad. Sci. USA,
110, 10976.
118.Pepys, M. B. (2001) Pathogenesis, diagnosis and
treatment of systemic amyloidosis, Philos. Trans. R. Soc. Lond. B
Biol. Sci., 356, 203-210.
119.Iizuka, E. (1965) Degree of crystallinity and
modulus relationships of silk thread from cocoons of Bombyx mori
L. and other moths, Biorheology, 3, 1-8.
120.Gosline, J. M., Guerette, P. A., Ortlepp, C.
S., and Savage, K. N. (1999) The mechanical design of spider silks:
from fibroin sequence to mechanical function, J. Exp. Biol.,
202, 3295-3303.
121.Kruger, M., and Linke, W. A. (2011) The giant
protein titin: a regulatory node that integrates myocyte signaling
pathways, J. Biol. Chem., 286, 9905-9912.