MicroRNA Sponge Knockdowns miR-483-5p and Upregulates Serum ALT/AST in Transgenic Mice
Yanfen Zhang1, Dantong Wu2,3, Xuemei Zhang2, Ning Ma2*, and Yanhong Liu1*
1Harbin Medical University, The Second Affiliated Hospital, Department of Clinical Laboratory, Harbin 150086, China; E-mail: 460205012@qq.com2Harbin Medical University, Department of Biochemistry and Molecular Biology, Harbin 150086, China
3Heilongjiang University of Chinese Medicine, The First Affiliated Hospital, Department of Laboratory Diagnostics, Harbin 150086, China
* To whom correspondence should be addressed.
Received August 24, 2017; Revision received September 28, 2017
MicroRNAs are involved in many biological processes. Studying microRNA function requires genetic strategies generating loss-of-function phenotypes, especially in vivo. However, few microRNA loss-of-function models have been reported in mice. Here, we generated several transgenic mouse lines to stably and specifically knockdown miR-483-5p by overexpressing microRNA sponges from CAG promoters. The different levels of expression of microRNA sponges resulted in different levels of mature miR-483-5p, which upregulated serum ALT/AST in these transgenic lines. These results indicate microRNA sponges are effective in mice in vivo, and they can be used in microRNA loss-of-function research.
KEY WORDS: knockdown, loss-of-function, microRNA sponges, transgenic miceDOI: 10.1134/S0006297918010078
Abbreviations: ALT, alanine aminotransferase; AST, aspartate aminotransferase; HE, hematoxylin-eosin (staining); LOF, loss-of-function (research); miRNAs, microRNAs; UTR, untranslated region.
MicroRNAs (miRNAs) are a class of small RNA molecules approximately 22
nucleotides long that regulate gene expression by posttranscriptional
binding to partially or completely complementary sequences in the
3′-untranslated region (UTR) of target mRNAs [1]. Thousands of miRNAs exist in organisms from plants
to humans, yet the biological function of relatively few
miRNA–target interactions has been thoroughly characterized in
vivo.
The traditional approach for studying gene function in vivo is to produce transgenic or knockout animals such as mice. MiRNA transgenic mice were constructed by overexpressing pre-miRNA [2], and knockout mice were constructed in 2003 [3]. However, creating genetic knockouts to determine the function of miRNAs is difficult for the following reasons: (i) individual miRNAs are expressed from multiple genomic loci such as miR-103-1 and miR-103-2, which are located on chromosomes 5 and 20, respectively; (ii) complete miRNA deletion does not conform to pathology, and (iii) severe fetal deaths that have resulted from deletion. It is exigent to develop new strategies to knock down miRNA in loss-of-function (LOF) research. The strategy of miRNA sponges, in which a decoy miRNA target competes with the natural target sites and “absorbs” the mature miRNAs, was developed in 2007 [4]. Until now, this strategy has been used successfully in flies [5] and plants [6, 7], but whether “miRNA sponges” can work well in mice remains almost unknown.
The miRNA miR-483-5p was detected in the livers of mice in 2007; this miRNA, known as an “intronic miRNA”, is embedded in intron 2 of the insulin-like growth factor 2 (Igf2) gene. Several studies have revealed that intronic miRNAs can be coexpressed with their host gene, and indeed, miR-483-5p is coexpressed with the Igf2 gene. Although IGF2 has been identified as a multifunctional factor in many pathways, such as hepatocellular carcinoma, fat metabolism, and development, the function of miR-483-5p remains to be revealed. For further functional studies in vivo, it will be essential to construct a genetically modified mouse model.
Here, we report a strategy that resulted in stable and specific knockdown of miR-483-5p by overexpressing miRNA sponges in vivo. We first designed a miR-483-5p sponge that consisted of six copies of an imperfectly complementary fragment to the mature mouse miR-483-5p sequence. The sponges were expressed and knocked down miR-483-5p, but the control miRNA miR-30c and miR-483-3p levels did not change in vitro. Furthermore, liver tumor suppressor of cytokine signaling 3 (Socs3) [8] was upregulated by miR-483-5p sponges at the mRNA and protein levels. To investigate the role of miRNA sponges in vivo, we generated four transgenic mouse lines that overexpressed the sponges. The results suggested that the cytomegalovirus immediate early enhancer-chicken β-actin hybrid (CAG) promoter can drive expression of miRNA sponges and that the sponges can knockdown miR-483-5p in vivo. Pearson correlation analysis indicated that there is a negative correlation between the amount of miR-483-5p and the expression of the transgene. This study provides more evidence that miRNA sponges can function in mice in vivo, and we propose that this knockdown model can be used in miRNA LOF research.
MATERIALS AND METHODS
Animal care. Transgenic and WT mice were housed in a regulated environment (22 ± 2°C, 55 ± 10% humidity, and 12-h light–dark cycle with lights on at 6 a.m.). All animal procedures were approved by the Institutional Animal Care and Use Committee at Harbin Medical University.
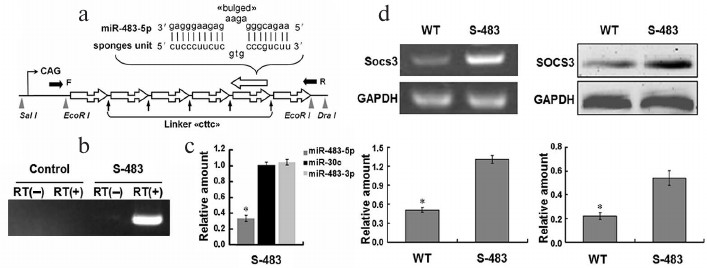
Construction of pCAGGs-S-miR-483-5p recombinant plasmids. The miRNA-interfering constructs expressing miR-483-5p sponges (S-483) were designed in our laboratory, and S-483 was synthesized by Sangon Biological Engineering Technology & Service Company (China). Briefly, S-483 was ligated into two EcoR I sites of a pCAGGs vector, which contains a CAG promoter that can drive expression in most tissues, to produce the pCAGGs-S-miR-483-5p recombinant plasmids (Fig. 1a). S-483 contained six copies of imperfect complementary antisense sequence to mature miR-483-5p with a linker between the two antisense sequences. We used this method to knock down miR-483-5p in vitro and in vivo.
Fig. 1. Design of miR-483-5p sponges and pCAGGs-S-483-5p knockdown of mature miR-483-5p in vitro. a) MiR-483-5p sponges were constructed by subcloning the miR-483-5p binding-site regions into a vector containing the CAG promoter via two EcoR I sites. Imperfect pairing between the miRNA and sponge forms a bulge and “cttc” linker between the two binding-site regions. The sponges can soak up mature miR-483-5p and stably interact with the available miR-483-5p. A microinjection fragment was obtained by digestion of pCAGGs-S-miR-483-5p with Sal I and Dra I. The black arrow (F/R) shows screening primers. b) Reverse transcription PCR analyses of miRNA sponge expression in Hepa1-6 cells treated with pCAGGs or pCAGGs-S-miR-483-5p plasmid. The plasmids were expressed in vitro. c) Real-time PCR analyses of amount of mature miR-483-5p/3p and miR-30c in cells transfected with the plasmids were used as the standard (1.00) (n = 3; * p < 0.05). d) Overexpression of miR-483-5p sponges upregulated Socs3 expression at both mRNA (left) and protein (right) levels in Hepa1-6. The mRNA level of Socs3 was quantified using semiquantitative RT-PCR, and the protein level of SOCS3 was determined by Western blotting (n = 3; * p < 0.01).
Cell line. Murine cell line Hepa1-6 was cultured with Dulbecco’s modified Eagle’s medium (DMEM) supplemented 10% heat-inactivated fetal bovine serum (Sigma, China) and 2 mM L-glutamine (Invitrogen, China). The cells were grown at 37°C in a 5% CO2 atmosphere.
Transfections with pCAGGs-S-miR-483-5p recombinant plasmids. Knockdown expressions of miR-483-5p were performed by transfection with pCAGGs and pCAGGs-S-miR-483-5p recombinant plasmids. A total of 5·104 cells were plated in 35-mm culture dishes for 24 h and then transfected with 600 ng of the plasmids with Lipofectamine 2000 (Invitrogen). After 24 h, the cells were subjected to RNA extraction.
RNA extractions. Total RNA, including miRNA fraction, was isolated from cell lines and mouse livers using an miRNeasy Mini Kit (Qiagen, China) according to the manufacturer’s protocol. After extraction, the quality of each sample was checked using NanoDrop spectrophotometry.
Real-time reverse transcription (RT)-PCR assays for mature miRNAs. To quantify expression of mature miRNAs, reverse transcription was performed using 1.5 µg of poly(A)-tailed small RNA from cells and tissues with 200 U of Super-Script III (Invitrogen) according to the method described above. Levels of miR-483-5p were assessed by quantitative real-time PCR using a SYBR-green PCR Master Mix in a 7500 Fast Real-Time PCR System with special primers (Table S1, see Supplement to this paper on the site of the journal http://protein.bio.msu.ru/biokhimiya and Springer site Link.springer.com). To normalize expression level of the miRNAs, U6 was used as endogenous control. In reverse transcription-PCR experiments performed to assess expression of the S-483 transgene in vitro and in vivo, β-actin was used as endogenous control.
Analyses of transgenic mice. The vector-free, Sal I- and Dra I-digested CAG-S-483 inserts were microinjected into C57Bl/6 oocytes. Genomic insertion of the transgene was analyzed by PCR amplification of tail-extracted DNA using a set of oligonucleotides (Fig. 1a and Supplement (Table S1 and Scheme)) located within the CAG promoter sequence and S-483. Four transgenic mice per line and wild-type (WT) C57Bl/6 mice (8 weeks old) were sacrificed, and liver samples were quickly removed for RNA extraction and analysis of transgene and miR-483-5p expression. All mice are heterozygous.
Western blot. Total protein from Hepa1-6 cells was extracted and analyzed by Western blotting. Total protein extracts were separated by 12% SDS-PAGE and transferred to PVDF membranes. Level of SOCS3 expression was evaluated using a rabbit polyclonal anti-SOCS3 antibody (Santa, China), which is immunogen affinity purified. Bands were quantified with Image-Pro Plus software. GAPDH served as loading control.
Liver hematoxylin-eosin stain and serum ALT/AST analysis. All mice were sacrificed at three months old and their livers removed and separated into individual lobes. The large lobes were fixed in 4% paraformaldehyde overnight and paraffin embedded. Sections (5 µm) were hematoxylin-eosin (HE) stained and examined under microscope by two independent pathologists.
Blood was collected from angular artery and centrifugated at 3000 rpm for 5 min. Serum ALT/AST was analyzed using a blood biochemistry analyzer.
Statistical analysis. The data are expressed as mean ± SEM from at least three independent experiments in vitro and four independent experiments in vivo. The difference between the two groups in the real-time PCR analysis was evaluated by two-tailed Student’s t-test. The difference was considered significant for p values < 0.05. The correlation of miR-483-5p with S-483 transgene expression in livers of WT and transgenic mice was examined by Pearson correlation. Differences were considered significant for p values <0.01.
RESULTS AND DISCUSSION
MiRNA sponge knockdowns miR-483-5p in vitro. To knockdown miR-483-5p in vivo, we designed a miRNA sponge comprised of partially complementary sequences to mature mouse miR-483-5p. To assess its efficacy, the pCAGGs construct carrying the miR-483-5p sponges and pCAGGs plasmids was transfected into Hepa1-6 cells, a cell line that overexpresses miR-483-5p. Cotransfected eGFP plasmid was used as transfection control. The pCAGGs-S-miR-483-5p recombinant plasmids were expressed in Hepa1-6 cells. Efficient knockdown of miR-483-5p was observed in Hepa1-6 cells transfected with pCAGGs-S-miR-483-5p recombinant plasmids (Fig. 1, b and c). However, levels of the controls miR-30c and miR-483-3p, which comes from the same pre-miRNA, did not change. Overall, these data suggest that pCAGGs plasmids carrying the CAG promoter are efficient miRNA knockdown vectors and that S-483 does not have off-target effects. Therefore, the vector was then tested in transgenic mice.
To identify effectiveness of miR-483-5p sponges, level of suppressor of cytokine signaling 3 (Socs3) [8] was detected. Total RNA and protein were isolated from Hepa1-6 cells after transfecting sponges. The mRNA level of Socs3 was quantified using semiquantitative RT-PCR, and the SOCS3 protein level was detected by Western blot analysis (Fig. 1d). The results showed that both mRNA and protein levels of Socs3 were elevated in the utilization of the miRNA sponges. We will analyze this phenotype further in applications such as liver development and liver disease.
Here, we constructed miR-483-5p sponges by inserting tandemly arrayed miRNA binding sites into pCAGGs vector driven by CAG promoter. Binding sites were imperfectly complementary in the seed region, with a bulge at positions 9-12 to prevent RNA interference-type cleavage and degradation of sponge RNA. The miRNA sponge strategy was developed in 2007 [4] and many investigators [9-12] are confident with miRNA sponges, especially in genetically modified mice [13]. However, reports [14] are limited on this subject until now.
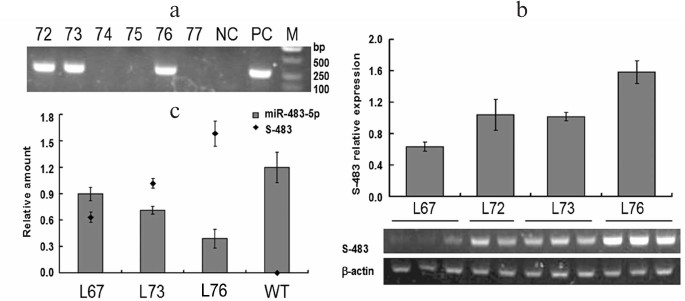
Generation of transgenic mice. The insert fragment was microinjected into C57Bl/6 oocytes. Four transgenic founders (No. 67, 72, 73, and 76) were detected by PCR analysis of their tail genomic DNA (Fig. 2a, Nos. 72, 73, and 76). These founder mice transmitted the transgene to their progeny, and the derived lines (L67, L72, L73, and L76) were further analyzed. In all lines, the transgene transmission rate was 50% (Table S2, see Supplement), suggesting the existence of a single integration site in each line.
Fig. 2. Screening of miR-483-5p knockdown transgenic mice and detection of expression of miR-483-5p sponges in transgenic mice. a) PCR analyses of tail-extracted genomic DNA. Four F0 mice (Nos. 72, 73, 76, and 67 (data not shown)) integrated the transgene. b) Detection of miRNA sponges produced by CAG-S-483 construct in livers of four transgenic mouse lines. RT-PCR products obtained by amplification with the primers (Table S1, see Supplement) were separated on 0.8% agarose gel via electrophoresis. Amplification of β-actin transcripts was used as internal control. c) To test the efficacy of inhibiting endogenous miR-483-5p by miRNA sponges, four transgenic mice per line and WT C57Bl/6 mice (8 weeks old) were sacrificed, liver samples quickly removed, and real-time PCR performed to assess amounts of mature miR-483-5p. Expression levels are presented as ratios with respect to U6. Extent of miR-483-5p downregulation in mice appears to be correlated with sponge expression level. The data were analyzed by Pearson correlation in different transgenic mouse lines and WT mice (Pearson correlation = –0.99, p < 0.01).
Genetically modified mice are an important tool in miRNA function research. However, lack of an LOF model restricts function research in vivo. Although the role of excess miR-1 in heart development was identified using knockout mice [3], miRNA knockout strategy is not a satisfactory method yet. Artificial miRNA inhibitors, another miRNA knockdown strategy based on base complementarity, are widely used in LOF research [15], but they are only functional for up to a week. And they cannot be inherited, although some sponges can integrate into the genome [16-19]. A novel method is required for LOF research in vivo.
To accelerate study of miRNA function and mechanism in vivo, we developed a method to disrupt individual miRNAs using miRNA sponges in transgenic mice. The results indicate that the miRNA sponges are inheritable in all transgenic lines, which is a dominant feature that distinguishes this strategy from knockdown strategies used in the past.
MiRNA sponges induce miR-483-5p downregulation in several lines. Reverse transcription (RT)-PCR analysis of liver cDNA samples was used to detect the expression of the transgene in different lines (Fig. 2b); the expression of β-actin served as control. RT-PCR without reverse transcriptase revealed that there was no genomic DNA pollution in the RNA samples (data not shown). High expression of the transgene was observed in line 76; however, expression was low in line 67 and was mediocre in lines 72 and 73. These results suggest that the CAG promoter can function efficiently in this model.
Amount of miR-483-5p is negatively correlated with sponge expression level. To further investigate the role of miRNA sponges, we analyzed expression of the miRNA sponges, and expression of mature miRNA was analyzed by real-time RT-PCR (Fig. 2c). Different levels of mature miR-483-5p knockdown that were observed in the transgenic lines could reflect different expression levels of the miR-483-5p sponges. Pearson correlation analysis revealed that the relative amounts of miR-483-5p were directly and negatively related to levels of sponge expression (Pearson’s correlation coefficient = -0.99).
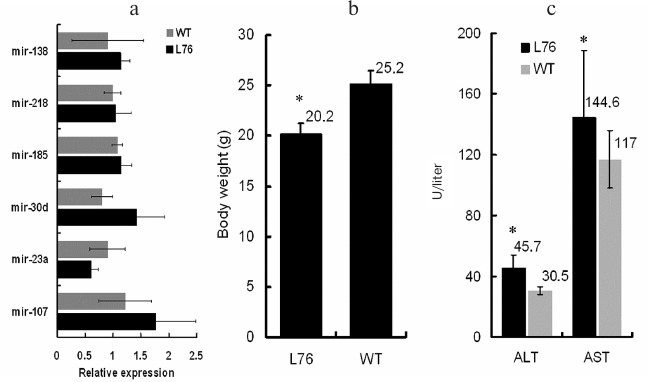
To test for off-target effect of the miR-483-5p sponges, some miRNAs were quantified by real-time PCR. Amounts of miR-138, miR-218, miR-185, miR-30d, miR-23a, and miR-107 did not differ between WT mice and miR-483-5p knockdown mice (Fig. 3a).
Fig. 3. Off-target effect and phenotype in transgenic mice. a) Amounts of miR-138, miR-218, miR-185, miR-30d, miR-23a, and miR-107 were detected by real-time PCR. There was no difference between transgenic mice and WT mice. b) Body weight of two-month-old transgenic mice and WT mice were recorded, and the body weight of miR-483-5p knockdown mice was lower than that of WT mice (n = 6). c) Serum ALT/AST were upregulated in the transgenic mice (n = 4).
Previous studies have indicated that there might be Pol III elements other than U6 that would produce sponge RNAs at high level [4]. Our data showed that the CAG promoter was highly effective and efficient in mice in vivo. As an intronic miRNA, miR-483-5p was coexpressed with its host gene Igf2, and we hypothesize that it would play its role as the partner of IGF2. The abnormal expression patterns of miR-483-5p in osteoarthritis [20] and tumors [21] are consistent with expression patterns of Igf2. MiR-483-5p may promote cell proliferation and organ development via the Hedgehog signaling pathway or the JAK/STAT pathway, which is key in IGF2 function. Further function research in vivo is needed for gain-of-function [22] and LOF levels.
MiR-483-5p upregulates serum ALT/AST in transgenic mice. Because miR-483 was found in murine fetus liver, we hypothesized it promoted organism development. We measured body weight of the mice, and surprisingly that of the transgenic mice was lower than that of WT mice at two-month-old (Fig. 3b). We then detected serum alanine aminotransferase and aspartate aminotransferase (ALT/AST), two markers of liver damage (Fig. 3c). They were upregulated in transgenic mice, which suggests miR-483-5p might promote damage of hepatocytes.
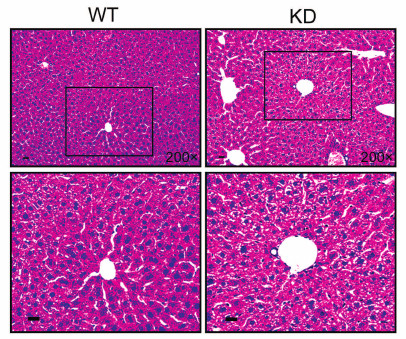
To investigate the role of miR-483 in liver, we detected it in liver of three-month-old mice by HE staining. However, there was no apparent difference between transgenic mice and WT mice (Fig. 4). This might result from: (i) the mice were young, and although the amount of miR-483-5p changed in transgenic mice, the effect did not accumulate enough to emerge in phenotype; (ii) liver is an organ that has the ability of compensation, and it should give some intervention to these transgenic mice, for example, hepatoma-induced model or hepatic-fibrosis induced model.
Fig. 4. HE staining of transgenic mice (miR-483-5p knockdown, KD) and WT mice. To investigate the role of miR-483-5p in liver, we detected in livers of three-month-old mice by HE staining. However, there was no apparent difference between transgenic mice and WT mice. Scale bar, 20 µm.
Acknowledgments
This work was supported by the Foundation of Education Department of Heilongjiang Province (12541363), the Foundation of Health Department of Heilongjiang Province (2012-525), Yu Weihan Academician Fund for Distinguished Young Scholars of Harbin Medical University, the Postdoctoral Fund of Heilongjiang Province (LBh-q15108), the National Natural Science Foundation of China (81541031).
REFERENCES
1.Bartel, D. P. (2004) MicroRNAs: genomics,
biogenesis, mechanism, and function, Cell, 116,
281-297.
2.Costinean, S., Zanesi, N., Pekarsky, Y., Tili, E.,
Volinia, S., Heerema, N., and Croce, C. M. (2006) Pre-B cell
proliferation and lymphoblastic leukemia/high-grade lymphoma in
E(mu)-miR155 transgenic mice, Proc. Natl. Acad. Sci. USA,
103, 7024-7029.
3.Zhao, Y., Samal, E., and Srivastava, D. (2005)
Serum response factor regulates a muscle-specific microRNA that targets
Hand2 during cardiogenesis, Nature, 436, 214-220.
4.Ebert, M. S., Neilson, J. R., and Sharp, P. A.
(2007) MicroRNA sponges: competitive inhibitors of small RNAs in
mammalian cells, Nat. Methods, 4, 721-726.
5.Loya, C. M., Lu, C. S., Van Vactor, D., and Fulga,
T. A. (2009) Transgenic microRNA inhibition with spatiotemporal
specificity in intact organisms, Nat. Methods, 6,
897-903.
6.Franco-Zorrilla, J. M., Valli, A., Todesco, M.,
Mateos, I., Puga, M. I., Rubio-Somoza, I., Leyva, A., Weigel, D.,
Garcia, J. A., and Paz-Ares, J. (2007) Target mimicry provides a new
mechanism for regulation of microRNA activity, Nat. Genet.,
39, 1033-1037.
7.Chitwood, D. H., and Timmermans, M. C. (2007)
Target mimics modulate miRNAs, Nat. Genet., 39,
935-936.
8.Riehle, K. J., Campbell, J. S., McMahan, R. S.,
Johnson, M. M., Beyer, R. P., Bammler, T. K., and Fausto, N. (2008)
Regulation of liver regeneration and hepatocarcinogenesis by suppressor
of cytokine signaling 3, J. Exp. Med., 205, 91-103.
9.Horwich, M. D., and Zamore, P. D. (2008) Design and
delivery of antisense oligonucleotides to block microRNA function in
cultured Drosophila and human cells, Nat. Protoc.,
3, 1537-1549.
10.Medina, P. P., and Slack, F. J. (2009) Inhibiting
microRNA function in vivo, Nat. Methods, 6,
37-38.
11.Cohen, S. M. (2009) Use of microRNA sponges to
explore tissue-specific microRNA functions in vivo, Nat.
Methods, 6, 873-874.
12.Zhiguo, W. (2009) MicroRNA Interference
Technologies, Springer, Heidelberg, Germany.
13.Ebert, M. S., and Sharp, P. A. (2010) MicroRNA
sponges: progress and possibilities, RNA, 16,
2043-2050.
14.Zhu, Q., Sun, W., Okano, K., Chen, Y., Zhang, N.,
Maeda, T., and Palczewski, K. (2011) Sponge transgenic mouse model
reveals important roles for the microRNA-183 (miR-183)/96/182 cluster
in postmitotic photoreceptors of the retina, J. Biol. Chem.,
286, 31749-31760.
15.Lu, Y., Xiao, J., Lin, H., Bai, Y., Luo, X.,
Wang, Z., and Yang, B. (2009) A single anti-microRNA antisense
oligodeoxyribonucleotide (AMO) targeting multiple microRNAs offers an
improved approach for microRNA interference, Nucleic Acids Res.,
37, e24.
16.Davis, S., Lollo, B., Freier, S., and Esau, C.
(2006) Improved targeting of miRNA with antisense oligonucleotides,
Nucleic Acids Res., 34, 2294-2304.
17.Scherr, M., Venturini, L., Battmer, K.,
Schaller-Schoenitz, M., Schaefer, D., Dallmann, I., Ganser, A., and
Eder, M. (2007) Lentivirus-mediated antagomir expression for specific
inhibition of miRNA function, Nucleic Acids Res., 35,
e149.
18.Krutzfeldt, J., Rajewsky, N., Braich, R., Rajeev,
K. G., Tuschl, T., Manoharan, M., and Stoffel, M. (2005) Silencing of
microRNAs in vivo with “antagomirs”, Nature,
438, 685-689.
19.Gentner, B., Schira, G., Giustacchini, A.,
Amendola, M., Brown, B. D., Ponzoni, M., and Naldini, L. (2009)
Knockdown of microRNA in vivo by lentiviral vectors, Nat.
Methods, 6, 63-66.
20.Iliopoulos, D., Malizos, K. N., Oikonomou, P.,
and Tsezou, A. (2008) Integrative microRNA and proteomic approaches
identify novel osteoarthritis genes and their collaborative metabolic
and inflammatory networks, PLoS One, 3, e3740.
21.Soon, P. S., Tacon, L. J., Gill, A. J., Bambach,
C. P., Sywak, M. S., Campbell, P. R., Yeh, M. W., Wong, S. G.,
Clifton-Bligh, R. J., Robinson, B. G., and Sidhu, S. B. (2009) miR-195
and miR-483-5p identified as predictors of poor prognosis in
adrenocortical cancer, Clin. Cancer Res., 15,
7684-7692.
22.Ma, N., Wang, X.-D., Qiao, Y., Li, F.-Y., Yang,
B.-F., Lv, Y.-J., Shan, H.-L., Zheng, X.-F., and Gao, X. (2010)
Construction of pCAGGs-miR-483 plasmid and establishment of a miR-483
transgenic mouse model, Chinese J. Biochem. Mol. Biol.,
26, 478-483.
Supplemental Tables S1-S2 and Scheme (PDF)