REVIEW: Crowding, Entropic Forces, and Confinement: Crucial Factors for Structures and Functions in the Cell Nucleus
R. Hancock
Biosystems Group, Biotechnology Centre, Silesian University of Technology, 44-100 Gliwice, Poland and Laval University Cancer Research Centre, Québec, Canada G1R2J6; E-mail: ronald.hancock@crhdq.ulaval.ca
Received October 3, 2017; Revision received November 27, 2017
The view of the cell nucleus as a crowded system of colloid particles and that chromosomes are giant self-avoiding polymers is stimulating rapid advances in our understanding of its structure and activities, thanks to concepts and experimental methods from colloid, polymer, soft matter, and nano sciences and to increased computational power for simulating macromolecules and polymers. This review summarizes current understanding of some characteristics of the molecular environment in the nucleus, of how intranuclear compartments are formed, and of how the genome is highly but precisely compacted, and underlines the crucial, subtle, and sometimes unintuitive effects on structures and reactions of entropic forces caused by the high concentration of macromolecules in the nucleus.
KEY WORDS: cell nucleus, macromolecular crowding, confinement, entropic forces, phase separation, chromosomesDOI: 10.1134/S0006297918040041
“The study of the colloidal state is … of special importance since the vast majority, if not all, of the substances of which protoplasm is made up have very large and complicated particles and therefore must give colloidal solutions” [1]. This insight of Alexander Oparin, published close to 100 years ago, was neglected until recently. Now, its implications are leading to rapid progress in our understanding of “protoplasm” and particularly of the cell nucleus, based on the properties of colloids and their interactions with polymers like chromosomes and the experimental and simulation methods that have been developed to study them.
Most of us who work with nuclei or subcellular systems believe, and sometimes state in publications, that our experimental conditions are “physiological” and that our macromolecules are influenced only by van der Waals, electrostatic, hydrogen bonding, and hydrophobic forces [2]. Until recently, most biologists did not realize the significance of the discovery by Asakura and Oosawa in 1958 that “An attractive force appears between particles suspended in solutions of macromolecules … that becomes stronger in solutions of chain macromolecules or of macromolecules of dissymmetrical shape than in solutions of rigid spherical macromolecules … and can have a remarkable influence on the state of suspended particles” [3]. These strong and subtle attractive forces, termed entropic or depletion forces, are negligible in the dilute systems of macromolecules that we usually use, but they become very important at high concentrations when macromolecules are in close proximity. This parameter, termed macromolecular crowding, is a common feature of all parts of biological cells, the cytoplasm [4, 5], membranes [6], and mitochondria [7], and of intercellular spaces in tissues [8] and is an essential but frequently overlooked feature of “physiological” conditions that should be reproduced in experiments. This review describes some characteristics of the molecular environment in the nucleus and underlines how insights and simulations from colloid, polymer, soft matter, and nano sciences are contributing to answering the far-sighted question of Francisco Iborra: “Can visco-elastic phase separation, macromolecular crowding and colloidal physics explain nuclear organization?” [9] and to understanding the structures and properties of the nucleus and the genome.
EFFECTS OF CROWDING ON MACROMOLECULES
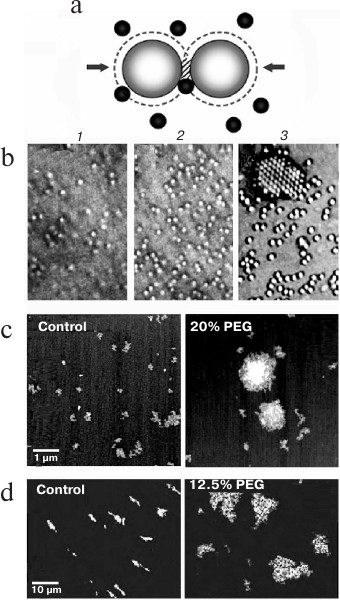
Entropic attractive forces. Entropic forces between macromolecules arise in a concentrated mixture because when the molecules are close enough, others cannot enter the regions between them and the forces that they exert because of random (Brownian) movement are no longer symmetrical (Fig. 1). By the same principle, entropic forces favor the localization of particles on a surface [10] and helical coiling of linear molecules [11].
Fig. 1. Attractive entropic forces. a) In a concentrated mixture, particles or macromolecules are so close that others cannot enter the shaded regions between them and therefore the forces (arrows) caused by random Brownian movement of the black macromolecules are no longer symmetrical. The second law of thermodynamics (states with maximum entropy are favored) is then satisfied because the volumes around the large macromolecules that are not accessible to the black molecules (dashed lines) overlap in the shaded region, increasing the volume available to them. b) Self-association of spherical particles driven by entropic forces as the concentration of smaller particles (not seen) in a mixture increases from (1) to (3) (reproduced from [12] with permission of the Royal Society). c, d) Entropic self-association of nuclear macromolecules: c) a 3 kb mRNA incubated with 1% (control) or 20% PEG (35 kDa) (reproduced from [13] with permission of the American Society for Biochemistry and Molecular Biology); d) polynucleosomes (DNA length ∼ 150 kb) labeled with YOYO-1 incubated without or with 12.5% PEG (8 kDa) and spread on a surface (reproduced from [14] with permission of Springer).
Crowded systems have a second particularity: the effective volume of a macromolecule from which others are excluded exceeds its intrinsic volume, and for a spherical macromolecule this effective volume is 8 times its intrinsic volume. Thus, even when macromolecules appear to occupy only a small part of a total volume, there may remain no space for others of the same size, resulting in a significant increase of their thermodynamic activity.
In studies of effects of crowding in vitro, volume-occupying synthetic polymers such as poly(ethylene glycol) (PEG), dextran, and Ficoll are widely used as “crowding agents”, but it is important to recognize that these do not reproduce perfectly the effects of the complex crowded environments in vivo, as discussed in the last section of this review.
Effects on proteins. Crowding has strong effects on the folding, conformation, stability, interactions, and biological activities of proteins that have been described in several comprehensive reviews [15-18]. The few studies in vivo suggest that these effects depend on the protein and the local environment [19]. Apparently the only nuclear protein that has been studied is histone H1, whose intrinsically disordered C-terminus is converted to a molten globule state in crowded conditions [20].
Effects on DNA and RNA conformation. Numerous changes of DNA conformation in crowded conditions have been reported, implying that conformations in living cells may be different from those in in vitro, but their implications are not yet clear because complex interactions occur between ionic effects and crowding [21]. A model DNA duplex (5′-TAGGTTATAA-3′/5′-TTATAACCTA-3′) shows a 7°C decrease of melting temperature when crowded in 20% PEG (200 Da) [22]. Triplex formation by poly(dA)-poly(dT) in vitro is promoted strongly in crowded conditions [23], and an intramolecular i-motif structure of triplet-repeat DNA oligomers is induced by crowding [24, 25]. The formation of G-quadruplexes in double-stranded DNA is favored by crowding [26], but its influence on the formation of telomeric G-quadruplexes in vivo is not yet clear. Folding of RNA is also promoted by crowding, with dramatic stabilization of folded states [27, 28].
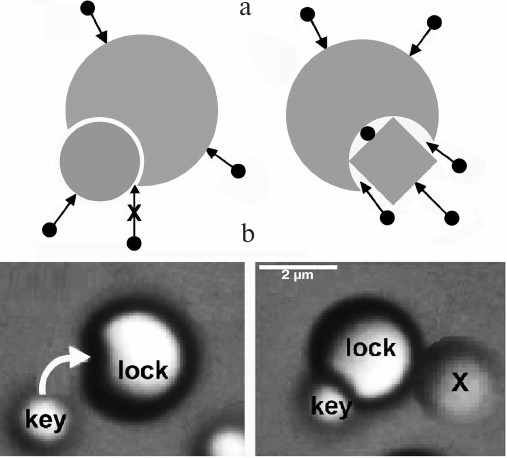
Entropic interactions can be highly specific. Entropic forces are highly sensitive to the local shape of interacting surfaces, and therefore show selectivity of a “lock and key” type (Fig. 2) [29]. Their ability to produce torque could be significant when a macromolecule has to adopt a particular orientation for its function [29]. Because of their ability to organize particles and molecules precisely, entropic interactions are studied widely for the design of nanostructures [30, 31].
Fig. 2. Entropic interactions are highly sensitive to shape. a) Left: a “key” molecule that fits precisely into a cavity on another molecule feels a strong entropic attraction, because other molecules cannot enter the interface. If the fit is not precise (right), other molecules can enter the interface and weaken the entropic attraction. b) Snapshots that show this selectivity during self-assembly of nanoparticles (reproduced from [31] with permission of Nature Publishing Group).
CROWDING AND THE STRUCTURE OF THE NUCLEUS
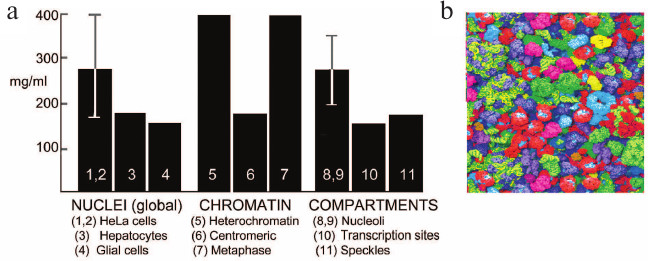
Concentration of macromolecules in the nucleus. Different methods confirm that nuclei contain macromolecules at a global concentration of at least 100-200 mg dry mass/ml (Fig. 3). This high concentration is maintained by confining the contents of the nucleus within the mechanically strong but somewhat elastic nuclear envelope [32] (Fig. 4b).
Fig. 3. a) Concentrations of macromolecules in the nucleus and its internal compartments; references are from [33]. b) Snapshot from a simulation of a similar macromolecular environment, the cytoplasm of E. coli, where the global concentration is somewhat higher than that in the nucleus but similar to that in the nucleolus (reproduced from [34], image kindly provided by Adrian Elcock).
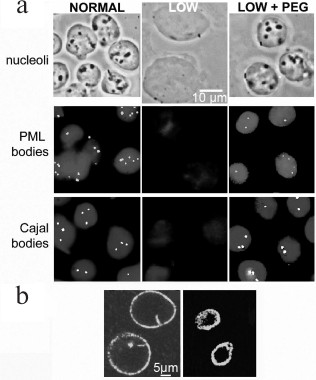
Fig. 4. Formation of compartments in the nucleus by crowding: a) the upper row shows phase contrast images of isolated nuclei, and the center and lower rows show PML or Cajal bodies visualized using fluorescent antibodies to their major protein. Left column, nuclei in normal buffer; center column, disassembly of compartments when nuclei expand after incubation in low ionic strength buffer; right column, reassembly of compartments after further incubation with addition of 12% PEG (8 kDa); b) the contents of the nucleus are confined and compressed by the nuclear lamina, visualized here by immunolabeling of lamin A. The lamina of isolated nuclei (left) contracts after DNA is digested with restriction enzymes and chromatin is removed by electroelution (right) [R. Hancock, unpublished data].
Compartments in the nucleus. At least 20 types of compartments have been visualized by optical microscopy within nuclei under different conditions, of which the most ubiquitous are nucleoli, Cajal and PML bodies, splicing speckles, gems, and sites (“factories”) where RNA transcription or DNA replication occur [35]. These compartments have characteristic properties: absence of a surrounding membrane, mobility within the nucleus, and dynamic exchange of their components with the surrounding nuclear space.
A crucial clue to the formation of these compartments was provided by Oksana Dudnik, Olga Zatsepina, and Yuri Chentsov who discovered by optical microscopy that nucleoli disassemble almost completely when living cells are incubated in a hypotonic medium, and they reform when the normal medium is restored [36]. Subsequently, it was found that Cajal and PML bodies [37], DNA replication foci [38], and interactions of chromatin that regulate gene expression [39] (see further Fig. 7) also disassemble in hypotonic conditions. Isolated nuclei increase considerably in volume in hypotonic media, suggesting that these effects are caused by a decrease of the concentration of the macromolecular components of these compartments. This model is supported by the observation that they reassemble when the nuclear volume is restored by adding a macromolecular crowding agent [37, 39] (Fig. 4; see further Fig. 7).
Simulations of systems of concentrated macromolecules support the idea that nuclear compartments are assembled from their component macromolecules by crowding [40, 41] (Fig. 5a). Entropic forces favor the localization of particles on a surface [10], and simulations explain why nuclear compartments are frequently associated with the surface of chromosomes [41] (Fig. 5b). Nuclear compartments therefore appear to be the results of purely physicochemical interactions in the crowded nucleoplasm, suggesting that the continuing search for specific functions of compartments such as PML and Cajal bodies may not be realistic.
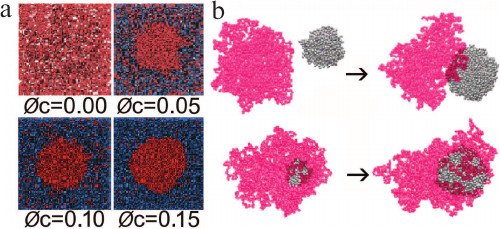
Fig. 5. Simulations by molecular dynamics to understand the formation of compartments in the nucleus and their tendency to associate with chromosomes. a) Clusters of spherical particles (red) are predicted to form when the volume fraction (φc) of crowding particles (blue) increases (reproduced from [40] with permission from Elsevier). b) In the presence of a polymer chain (red), clusters (gray) are predicted to form and then associate with the polymer when the particle density is high and particle–polymer interactions are moderately weak (above), while for strong interactions and lower particle densities the clusters grow in contact with the polymer (below) (reproduced from [41] with permission of the Royal Society of Chemistry).
Nuclear compartments as liquid drops produced by phase separation. When attraction due to entropic and other forces is strong enough in a mixture of particles or particles and polymers, the mixture may separate and form discrete phases, and crowding stimulates phase separation by increasing the effective concentrations in a mixture [42, 43]; this is seen in the simulations in Fig. 5 when clusters of particles are formed. The idea that compartments in the nucleus are formed by phase separation (of which coacervation is a particular type) was proposed in quite early studies; Albert Frey-Wyssling wrote in 1938 that “the nucleus … may behave like a liquid drop … nucleolus formation must be considered as an accumulation of the karyolymph proteins … until a coacervate droplet rich in proteins is formed” [44], and in 1946 Lars Ehrenberg concluded that “the nucleolus is a separated phase out of a saturated solution… Its globular shape and the absence of discontinuities as well as the mode of deformation of the persisting nucleolus by the spindle suggest that it is a drop” [45]. Phase separation requires a critical concentration of some component(s) [42, 46, 47] and is therefore closely dependent on local level of crowding. The concept that “the nucleus … may behave like a liquid drop” and that “the nucleolus is a separated phase” with “coacervate nature” has now become a major theme in cell biology [46, 47].
CROWDING, CONFINEMENT, AND THE CONFORMATION OF THE GENOME
The dense crowding of ∼65 cm of genomic chromatin into a ∼6 μm diameter nucleus implies that there must be abundant contacts between different regions of the genome. Earlier, at least some contacts were believed to be stable and to result in the formation of the loops seen by optical microscopy, for example [48].
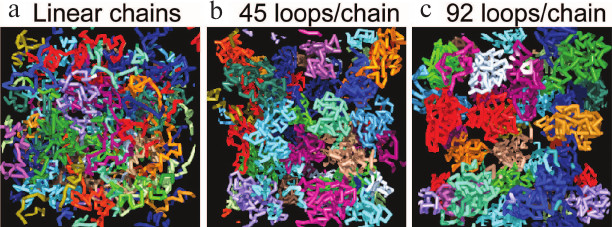
Simulations show that entropic forces favor the formation of loops in a self-avoiding polymer chain like a chromatin fiber [49-53]. In some models, inter-chain contacts are proposed to be mediated through protein “binders” [54], but most simulations find that loops form spontaneously when a self-avoiding chain folds in a confined space, without the need to postulate “binders” [55-58]. Comprehensive simulations have led to the Dynamic Loop model [51, 52] and show that the formation of loops results solely from diffusional motion of a polymer, without other long-range interactions. Introducing loops in polymer chains strongly increases the probability of contacts between distant regions compared to linear chains, and it also results in a multi-fold higher repulsion between them due to entropic forces compared to linear chains, reproducing the discrete form of chromosome territories in vivo (Fig. 6).
Fig. 6. Simulations showing that in a confined volume, chromosomes with larger portion of loops are separated more completely, like chromosome territories in vivo. a) Linear polymer chains have abundant contacts with others; the fraction of inter-chain contacts is 15.4%. b, c) Increasing the portion of polymer organized in loops (the number of loops) results in more and more confined structures with fewer inter-chain interactions. For chains with an average of 45 functional loops (b) the fraction of contacts is reduced to 1.8% (reproduced from [51]).
The dynamic nature of loops was observed quite early [56], and only dynamic loops reproduce the chromatin interaction frequencies observed by chromosome conformation capture [57]. Entropic forces and confinement are also predicted to cause the formation of topologically associating domains (TADS), regions of the genome where chromosomal interactions are particularly frequent [58].
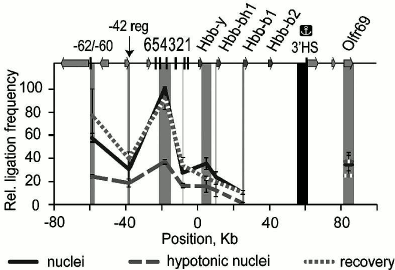
The formation by crowding of functional loops in chromatin in vivo to bring a gene into contact with its remote regulatory elements has been shown clearly by chromosome conformation capture (3C) experiments on the β-globin gene domain of the mouse genome [39]. In nuclei in isotonic buffer the frequency of contacts between upstream regulatory elements and the gene domain is high, but these contacts are disrupted when nuclei expand in hypotonic conditions. The contacts are re-established when the crowder PEG is added to the expanded nuclei (Fig. 7).
Fig. 7. Crowding in nuclei drives the interaction between the β-globin gene domain of the genome and its remote regulatory elements. Chromosome conformation capture was performed on nuclei in isotonic conditions (“nuclei”), expanded in hypotonic buffer (“hypotonic nuclei”), or in hypotonic buffer with addition of 12.5% PEG (8 kDa) (“recovery”). The vertical scale shows the relative frequency of ligation of the fragment harboring the 3′-insulator (colocalized with the 3′ DNase I-hypersensitive site (3′HS)) with other fragments of the domain, reflecting the frequency of contact between these regions. The map (top) shows the β-globin genes, olfactory receptor genes, and HS; 0 kb corresponds to the start of the Hbb-y gene. The dark gray rectangle shows the anchor fragment and the light gray rectangles indicate test fragments (adapted from [39]).
EFFECTS OF CROWDING ON FUNCTIONS IN THE NUCLEUS
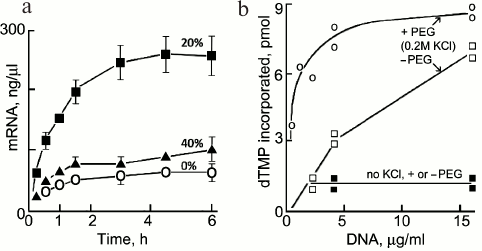
Transcription. Transcription of mRNAs in in vitro systems is stimulated by crowding agents. For example, the production of mRNA for Renilla luciferase in wheat germ extract is enhanced ∼4-fold in the presence of 20% Ficoll (70 kDa) (Fig. 8a); the stimulation by 40% Ficoll is smaller, probably due to increased viscosity and slower dissociation of some enzyme–substrate intermediates [59]. Crowding increases the rate of mRNA production significantly in picolitre water-in-oil droplets containing a coacervated lysate of E. coli [60]. A good example of sometimes subtle effects of crowding is seen in a model of the sporadic pulses or “bursts” of transcription that occur in vivo [61]; if polymerase molecules working along a gene meet, entropic attraction between them causes them to form a stable peloton or cluster that moves together along the gene and results in a burst of transcription [62].
Fig. 8. Stimulation by crowding of reactions involved in transcription and replication: a) effect of Ficoll (70 kDa) on synthesis of mRNA for Renilla luciferase (Rluc) in an in vitro transcription system (reproduced from [59]); b) effect of 8 kDa PEG (12%) on the affinity of DNA polymerase I for DNA (reproduced from [64]).
DNA replication. Crowding stimulates several individual steps related to DNA replication [63] including the activity of DNA helicase, the formation of the T4 DNA polymerase and other intermolecular complexes, and the affinity of DNA polymerase I for DNA (Fig. 8b) [64].
In growing cells, the sites of DNA replication are seen as foci in nuclei, which were earlier interpreted as replication “factories” through which the DNA of multiple replicons moves and is replicated. The observation that the number of foci increases markedly when nuclei expand upon incubation of cells in hypotonic medium [38], however, challenged this model and showed that foci are in fact assemblies of smaller structures that are probably associated reversibly by crowding effects like other intranuclear compartments. Now, super-resolution microscopy in three dimensions resolves several thousand replication foci per nucleus, each corresponding to a single replicon [65].
Diffusion, signaling, and finding targets. In the nucleus, macromolecules diffuse several-fold more slowly than in aqueous solution and even more slowly within the nucleolus. Their subdiffusion is anomalous, showing that their movement is hindered by obstacles [66-68]. Surprisingly, simulations show that this increases the probability of finding a nearby target and consequently facilitates the propagation of signals [69]. Crowding has a potentially important influence on signaling pathways, which frequently include a cascade of steps where intermediates are phosphorylated at two sites; since protein kinases diffuse slowly in a crowded environment, after a first phosphorylation they are more likely to rebind to intermediates and to phosphorylate them again, resulting in more processive responses, which have been confirmed by experiments in vitro [70].
Factors tht bind to and regulate activities of DNA find their targets rapidly, although competing nonspecific binding sites are in very large excess, and they are believed to search by nonspecific binding and then sliding and hops and jumps [71]. The compact and looped conformations of DNA in vivo imposed by crowding and confinement facilitate jumping between DNA segments in close proximity, and looping helps them to bypass other factors bound to DNA while searching [72].
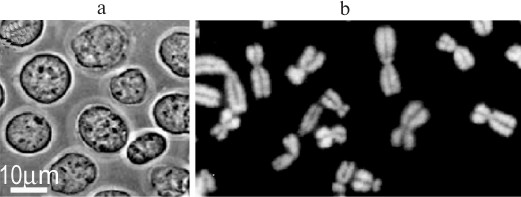
Crowding and the structure of nuclei and mitotic chromosomes in vivo. Nuclei and chromosomes in vivo are expected to feel strong crowding effects from the surrounding cytoplasm, which contains diffusible macromolecules at ~130 mg/ml [73, 74]. They are usually isolated in media without crowding agents to reproduce these conditions, but which contain Mg2+ and/or other cations at millimolar concentrations. However, nuclei and chromosomes are perfectly stable in solutions that contain only a crowding macromolecule to mimic the cytoplasmic environment, without cations and/or polyamines (Fig. 9), raising the question if the conditions generally used for their isolation are really “physiological” and reproduce the situation in the cell. This paradox is illustrated by the fact that chromatin incubated with cations in vitro has a different conformation from that in nuclei or chromosomes in vivo [75]. Together, these arguments support the hypothesis that the integrity of nuclei and chromosomes in vivo is strongly influenced by crowding from cytoplasmic macromolecules. Several other cases are known where crowding can replace or vastly reduce a requirement for Mg2+ to stabilize macromolecular assemblies, for example ribosomes [76] and bacterial chromosomes [77], or to stimulate their reactions, for example ribozymes [27] and recA protein [78]. The quantities of ions in the nucleus and cytoplasm have been measured, but the concentrations of osmotically active ions cannot be calculated because the volume, in which they are dissolved, and the fractions bound to macromolecules are not known, and it has been argued that most cations in the cell are bound to macromolecules [79].
Fig. 9. Nuclei and metaphase chromosomes isolated in a medium that mimics crowding by the cytoplasm in vivo but contains only 100 μM K-Hepes, pH 7.4, and 0.5% Triton X-100: a) nuclei of K562 cells in 70 kDa dextran (35%); b) chromosomes of Chinese hamster fibroblasts in 8 kDa PEG (12%), DNA stained with DAPI [R. Hancock, unpublished data].
EVOLUTION OF THE CROWDED NUCLEUS
All known types of DNA genomes appear to require a crowded environment for their structure and activities, and crowded and confined conditions appear to have been fundamental for the evolution of life [80]. Bacterial cells are even more crowded than eukaryotic nuclei, and entropic forces drive the separation of their daughter genomes [81]. The intracellular milieu of dinoflagellates is so crowded that the genome has a liquid crystal conformation [82]. The strong and subtle effects of crowding on structures and activities in the nucleus that are discussed here suggest some plausible reasons why a crowded milieu was essential for evolution: the conformations and interactions of macromolecules and chromosomes are exquisitely sensitive to entropic forces; in crowded conditions, the thermodynamic activity of macromolecules is enhanced so that interactions proceed efficiently with a smaller number of molecules; and molecular interactions under entropic forces may be more readily reversible than those caused by ionic forces, for example DNA condensed by PEG remains more flexible and less compact than when it is condensed by electrostatic interactions [83]. Crowded conditions could provide a form of “metabolic buffering” to stabilize the complex network of molecular interactions in the nucleus [66].
QUESTIONS FOR THE FUTURE
Measurement of crowding in vivo. Local levels of crowding in the nucleus are expected to vary depending on the local macromolecular environment. Sensors that respond to crowding are being developed and should give useful information if they can be targeted to specific regions [84-86]. In-cell NMR [19, 87, 88] has not yet been applied to examine proteins in the nucleus.
Crowders to mimic the conditions in vivo. Common, so-called inert crowders like PEG are not hard spherical particles like those used for theoretical simulations. They have subtle effects on the structure, stability, and functional activity of macromolecules that cannot yet be predicted and can show weak attractive interactions that destabilize protein complexes, in contrast to the stabilization predicted by entropic effects [17, 89]. For example, when telomeric repeat DNA is crowded by PEG, its conformation is not the same as that formed with Ficoll or in an extract of Xenopus laevis eggs, suggesting that in this case PEG does not correctly reproduce the effects of the crowded milieu in vivo [90]. Some studies go so far as to suggest that commonly used crowders do not yield physiologically relevant information [89, 91].
Crowding and the movement of macromolecules. A novel aspect of crowding has been revealed by computational methods [92]; local heterogeneity of the level of crowding is predicted to cause directional movement of other molecules. This effect has potentially important implications for understanding movements of molecules in vivo, for example movements of chromatin [93] and of newly-synthesized mRNAs away from chromatin [94].
“Lock and key” interactions. When considering the mechanisms by which covalent modifications of proteins or DNA influence their interactions with other macromolecules, the fact that local shapes of molecular surfaces strongly influence entropic interactions (Fig. 2) is often overlooked. For example, the effects of phosphorylation of a protein are generally interpreted in terms of modified hydrogen bonds or ionic interactions through the phosphate group [95], but they could be the result of a change of surface shape that is recognized and bound entropically by a “key” motif on binding partners. Similarly, methylation of DNA sequences causes a change of their conformation [96], which could be recognized by entropic “keys”.
Crowding homeostasis. Structures and activities in the nucleus are clearly highly dependent on and sensitive to the level of crowding, implying that “crowding homeostasis” must be essential [97, 98]. Signaling pathways that control the level of crowding by precisely equilibrating the exit of messenger RNAs with the entry and exit of proteins through nuclear pores must exist but have not yet been explored.
REFERENCES
1.Oparin, A. I. (1924) Origin of Life [in
Russian], Moskovskii Rabochii, Moscow; Oparin, A. I. (2003) Origin
of Life (translated by Morgulis, S.), Dover Publications, Mineola,
New York.
2.Leckband, D., and Israelachvili, J. (2001)
Intermolecular forces in biology, Quart. Rev. Biophys.,
34, 105-267.
3.Asakura, S., and Oosawa, F. (1958) Interaction
between particles suspended in solutions of macromolecules, J.
Polym. Sci., 33, 126-183.
4.Walter, H., and Brooks, D. E. (1995) Phase
separation in cytoplasm, due to macromolecular crowding, is the basis
for microcompartmentation, FEBS Lett., 361, 135-139.
5.Johansson, H. O., Brooks, D. E., and Haynes, C. A.
(2000) Macromolecular crowding and its consequences, Int. Rev.
Cytol., 192, 155-170.
6.Zhou, H.-X. (2009) Crowding effects of membrane
proteins, J. Phys. Chem. B, 113, 7995-8005.
7.Srere, P. A. (1980) The infrastructure of the
mitochondrial matrix, Trends Biochem. Sci., 5,
120-121.
8.Kumar, P., Satyam, A., Fan, X., Collin, E., Rochev,
Y., Rodriguez, B. J., Gorelov, A., Dillon, S., Joshi, L., Raghunath,
M., Pandit, A., and Zeugolisa, D. I. (2015) Macromolecularly crowded
in vitro microenvironments accelerate the production of
extracellular matrix-rich supramolecular assemblies, Sci. Rep.,
5, 8729.
9.Iborra, F. J. (2007) Can visco-elastic phase
separation, macromolecular crowding and colloidal physics explain
nuclear organisation? Theor. Biol. Med. Model., 4,
15.
10.Dinsmore, A. D., Wong, D. T., Nelson, P., and
Yodh, A. G. (1998) Hard spheres in vesicles: curvature-induced forces
and particle-induced curvature, Phys. Rev. Lett., 80,
409-412.
11.Snir, Y., and Kamien, R. D. (2005) Entropically
driven helix formation, Science, 307, 1067.
12.Yodh, A. G., Lin, K., Crocker, J. C., Dinsmore,
A. D., Verma, R., and Kaplan, P. D. (2001) Entropically driven
self-assembly and interaction in suspension, Phil. Trans. R. Soc.
Lond. A, 359, 921-937.
13.Bounedjah, O., Hamon, L., Savarin, P., Desforges,
B., Curmi, P. A., and Pastre, D. (2012) Macromolecular crowding
regulates assembly of mRNA stress granules after osmotic stress: new
role for compatible osmolytes, J. Biol. Chem., 287,
2446-2458.
14.Hancock, R. (2008) Self-association of
polynucleosome chains by macromolecular crowding, Europ. Biophys.
J., 37, 1059-1064.
15.Chebotareva, N. A., Kurganov, B. I., and
Livanova, N. B. (2004) Biochemical effects of molecular crowding,
Biochemistry (Moscow), 69, 1239-1251.
16.Turoverov, K. K., Kuznetsova, I. M., and Uversky,
V. N. (2014) What macromolecular crowding can do to a protein, Int.
J. Mol. Sci., 15, 23090-23140.
17.Kuznetsova, I. M., Zaslavsky, B. Y., Breydo, L.,
Turoverov, K. K., and Uversky, V. N. (2015) Beyond the excluded volume
effects: mechanistic complexity of the crowded milieu,
Molecules, 20, 1377-1409.
18.Rivas, G., and Minton, A. P. (2016)
Macromolecular crowding in vitro, in vivo, and in
between, Trends Biochem. Sci., 41, 970-981.
19.Danielsson, J., Mu, X., Lang, L., Wang, H.,
Binolfi, A., Theillet, F. X., Bekei, B., Logan, D. T., Selenko, P.,
Wennerstrom, H., and Oliveberg, M. (2015) Thermodynamics of protein
destabilization in live cells, Proc. Natl. Acad. Sci. USA,
112, 12402-12407.
20.Roque, A., Ponte, I., and Suau, P. (2016)
Interplay between histone H1 structure and function, Biochim.
Biophys. Acta, 1859, 444-454.
21.Zhou, J., Tateishi-Karimata, H., Mergny, J. L.,
Cheng, M., Feng, Z., Miyoshi, D., Sugimoto, N., and Li, C. (2016)
Reevaluation of the stability of G-quadruplex structures under crowding
conditions, Biochimie, 121, 204-208.
22.Gu, X. B., Nakano, S., and Sugimoto, N. (2006)
The effect of the structure of cosolutes on the DNA duplex formation,
Nucleic Acids Symp. Ser. (Oxf.), 50, 205-206.
23.Spink, C. H., and Chaires, J. B. (1995) Selective
stabilization of triplex DNA by poly(ethylene glycols), J. Am. Chem.
Soc., 117, 12887-12888.
24.Jiang, H. X., Cui, Y., Zhao, T., Fu, H. W.,
Koirala, D., Punnoose, J. A., Kong, D. M., and Mao, H. (2015) Divalent
cations and molecular crowding buffers stabilize G-triplex at
physiologically relevant temperatures, Sci. Rep., 5,
9255.
25.Cui, J., Waltman, P., Le, V. H., and Lewis, E. A.
(2013) The effect of molecular crowding on the stability of human c-MYC
promoter sequence i-motif at neutral pH, Molecules, 18,
12751-12767.
26.Kumar, N., and Maiti, S. (2008) Role of molecular
crowding in perturbing quadruplex/Watson–Crick duplex
equilibrium, Nucleic Acids Symp. Ser. (Oxf.), 52,
157-158.
27.Denesyuk, N. A., and Thirumalai, D. (2013)
Entropic stabilization of the folded states of RNA due to
macromolecular crowding, Biophys. Rev., 5, 225-232.
28.Strulson, C. A., Boyer, J. A., Whitman, E. E.,
and Bevilacqua, P. C. (2014) Molecular crowders and cosolutes promote
folding cooperativity of RNA under physiological ionic conditions,
RNA, 20, 331-347.
29.Kinoshita, M., and Oguni, T. (2002) Depletion
effects on the lock and key steric interactions between macromolecules,
Chem. Phys. Lett., 351, 79-84.
30.Bishop, K. J. M., Wilmer, C. E., Soh, S., and
Grzybowski, B. A. (2009) Nanoscale forces and their uses in
self-assembly, Small, 5, 1600-1630.
31.Sacanna, S., Irvine, W. T., Chaikin, P. M., and
Pine, D. J. (2010) Lock and key colloids, Nature, 464,
575-578.
32.Rowat, A. C., Foster, L. J., Nielsen, M. M.,
Weiss, M., and Ipsen, J. H. (2005) Characterization of the
elastic properties of the nuclear envelope, J. R. Soc.
Interface, 2, 63-69.
33.Hancock, R. (2011) in Genome Organization and
Function in the Cell Nucleus (Rippe, K., ed.) WILEY-VCH Verlag
Weinheim, pp. 171-182.
34.McGuffee, S. R., and Elcock, A. H. (2010)
Diffusion, crowding and protein stability in a dynamic molecular model
of the bacterial cytoplasm, PLoS Comput. Biol., e1000694.
35.Hancock, R. (2014) The crowded nucleus, in New
Models of the Cell Nucleus: Crowding, Entropic Forces,
Phase Separation, and Fractals (Hancock, R., and Jeon,
K., eds.) Int. Rev. Cell Mol. Biol., 307, 15-26.
36.Dudnik, O. A., Zatsepina, O. V., and Chentsov, Y.
S. (1993) The effect of low ionic strength solutions on the structure
and function of the nucleoli in living ESK cells, Tsitologiia,
35, 10-16.
37.Hancock, R. (2004) A role for macromolecular
crowding effects in the assembly and function of compartments in the
nucleus, J. Struct. Biol., 146, 281-290.
38.Chagin, V. O., Rozanov, Iu. M., Solov’eva,
L. V., and Tomilin, N. V. (2004) High resolution analysis of
replication foci by conventional fluorescent microscopy. I. A study of
complexity and DNA content of the foci, Tsitologiia, 46,
229-243.
39.Golov, A. K., Gavrilov, A. A., and Razin, S. V.
(2015) The role of crowding forces in juxtaposing β-globin gene
domain remote regulatory elements in mouse erythroid cells, PLoS
One, 10, e0139855.
40.Cho, E. J., and Kim, J. S. (2012) Crowding
effects on the formation and maintenance of nuclear bodies: insights
from molecular-dynamics simulations of simple spherical model
particles, Biophys. J., 103, 424-433.
41.Oh, I., Choi, S., Jung, Y., and Kim, J. S. (2015)
Phase separation of a Lennard–Jones fluid interacting with a
long, condensed polymer chain: implications for the nuclear body
formation near chromosomes, Soft Matter, 11,
6450-6459.
42.Aumiller, W. M., Davis, B. W., and Keating, C. D.
(2014) Phase separation as a possible means of nuclear
compartmentalization, in New Models of the Cell Nucleus:
Crowding, Entropic Forces, Phase Separation, and
Fractals (Hancock, R., and Jeon, K., eds.) Int. Rev. Cell Mol.
Biol., 307, 109-149.
43.Hyman, A. A., Weber, C. A., and Julicher, F.
(2014) Liquid–liquid phase separation in biology, Annu. Rev.
Cell Dev. Biol., 30, 39-58.
44.Frey-Wyssling, A. (1938) Submikroskopische
Morphologie des Protoplasmas und seiner Derivate, Gebruder
Borntraeger, Berlin.
45.Ehrenberg, L. (1946) Influence of temperature on
the nucleolus and its coacervate nature, Hereditas, 32,
407-418.
46.Jacobs, W. M., and Frenkel, D. (2017) Phase
transitions in biological systems with many components, Biophys.
J., 112, 683-691.
47.Shin, Y., and Brangwynne, C. P. (2017) Liquid
phase condensation in cell physiology and disease, Science,
357, eaaf4382.
48.Iarovaia, O. V., Bystritskiy, A., Ravcheev, D.,
Hancock, R., and Razin, S. V. (2004) Visualization of individual DNA
loops and a map of loop domains in the human dystrophin gene,
Nucleic Acids Res., 32, 2079-2086.
49.Zhang, G., Winnik, F. M., and Wu, C. (2003)
Structure of a collapsed polymer chain with stickers: a single- or
multiflower? Phys. Rev. Lett., 90, 035506.
50.Marenduzzo, D., and Orlandini, E. (2009)
Topological and entropic repulsion in biopolymers, J. Stat.
Mech., L09002.
51.Bohm, M., and Heermann, D. W. (2010)
Diffusion-driven looping provides a consistent framework for chromatin
organization, PLoS One, 5, e12218.
52.Bohm, M., and Heermann, D. W. (2011) Repulsive
forces between looping chromosomes induce entropy-driven segregation,
PLoS One, 6, e14428.
53.St-Jean, P., Vaillant, C., Audit, B., and
Arneodo, A. (2008) Spontaneous emergence of sequence-dependent
rosettelike folding of chromatin fiber, Phys. Rev. E, 77,
061923.
54.Johnson, J., Brackley, C. A., Cook, P. R., and
Marenduzzo, D. (2015) A simple model for DNA bridging proteins and
bacterial or human genomes: bridging-induced attraction and genome
compaction, J. Phys. Condens. Matter, 27, 064119.
55.Gursoy, G., Xu, Y., Kenter, A. L., and Liang, J.
(2014) Spatial confinement is a major determinant of the folding
landscape of human chromosomes, Nucleic Acids Res., 42,
8223-8230.
56.Gerdes, M. G., Carter, K. C., Moen, P. T., and
Lawrence, J. B. (1994) Dynamic changes in the higher-level chromatin
organization of specific sequences revealed by in situ
hybridization to nuclear halos, J. Cell Biol., 126,
289-304.
57.Gibcus, J. H., and Dekker, J. (2013) Connecting
the genome: dynamics and stochasticity in a new hierarchy for
chromosome conformation, Mol. Cell, 49, 773-782.
58.Vasquez, P. A., Hult, C., Adalsteinsson, D.,
Lawrimore, J., Forest, M. G., and Bloom, K. (2016) Entropy gives rise
to topologically associating domains, Nucleic Acids Res.,
44, 5540-5549.
59.Ge, X., Luo, D., and Xu, J. (2011) Cell-free
protein expression under macromolecular crowding conditions, PLoS
One, 6, e28707.
60.Sokolova, E., Spruijt, E., Hansen, M. M., Dubuc,
E., Groen, J., Chokkalingam, V., Piruska, A., Heus, H. A., and Huck, W.
T. (2013) Enhanced transcription rates in membrane-free protocells
formed by coacervation of cell lysate, Proc. Natl. Acad. Sci.
USA, 110, 11692-11697.
61.Blake, W. J., Kaern, M., Cantor, C. R., and
Collins, J. J. (2003) Noise in eukaryotic gene expression,
Nature, 422, 633-637.
62.Van den Berg, A. A., and Depken, M. (2017)
Crowding-induced transcriptional bursts dictate polymerase and
nucleosome density profiles along genes, Nucleic Acids Res.,
45, 7623-7632.
63.Akabayov, B., Akabayov, S. R., Lee, S. J.,
Wagner, G., and Richardson, C. C. (2013) Impact of macromolecular
crowding on DNA replication, Nat. Commun., 4, 1615.
64.Zimmerman, S. B., and Harrison, B. (1987)
Macromolecular crowding increases binding of DNA polymerase to DNA: an
adaptive effect, Proc. Natl. Acad. Sci. USA, 84,
1871-1875.
65.Baddeley, D., Chagin, V. O., Schermelleh, L.,
Martin, S., Pombo, A., Carlton, P. M., Gahl, A., Domaing, P., Birk, U.,
Leonhardt, H., Cremer, C., and Cardoso, M. C. (2010) Measurement of
replication structures at the nanometer scale using super-resolution
light microscopy, Nucleic Acids Res., 38, e8.
66.Politz, J. C., Tuft, R. A., and Pederson, T.
(2003) Diffusion-based transport of nascent ribosomes in the nucleus,
Mol. Biol. Cell, 14, 4805-4812.
67.Guigas, G., Kalla, C., and Weiss, M. (2007)
Probing the nanoscale viscoelasticity of intracellular fluids in living
cells, Biophys. J., 93, 316-323.
68.Bancaud, A., Huet, S., Daigle, N., Mozziconacci,
J., Beaudouin, J., and Ellenberg, J. (2009) Molecular crowding affects
diffusion and binding of nuclear proteins in heterochromatin and
reveals the fractal organization of chromatin, EMBO J.,
28, 3785-3798.
69.Guigas, G., and Weiss, M. (2008) Sampling the
cell with anomalous diffusion – the discovery of slowness,
Biophys. J., 94, 90-94.
70.Aoki, K., Takahashi, K., Kaizu, K., and Matsuda,
M. (2013) A quantitative model of ERK MAP kinase phosphorylation in
crowded media, Sci. Rep., 3, 1541.
71.Mirny, L., Slutsky, M., Wunderlich, Z., Tafvizi,
A., Leith, J., and Kosmrlj, A. (2009) How a protein searches for its
site on DNA: the mechanism of facilitated diffusion, J. Phys. A:
Math. Theor., 42, 1751-8121.
72.Li, G., Berg, O. G., and Elf, J. (2009) Effects
of macromolecular crowding and DNA looping on gene regulation kinetics,
Nat. Phys., 5, 294-297.
73.Hancock, R., and Hadj-Sahraoui, Y. (2009)
Isolation of cell nuclei using inert macromolecules to mimic the
crowded cytoplasm, PLoS One, 4, e7560.
74.Hancock, R. (2012) Structure of metaphase
chromosomes: a role for effects of macromolecular crowding, PLoS
One, 7, e36045.
75.Maeshima, K., Imai, R., Tamura, S., and Nozaki,
T. (2014) Chromatin as dynamic 10-nm fibers, Chromosoma,
123, 225-237.
76.Zimmerman, S. B., and Trach, S. O. (1988) Effects
of macromolecular crowding on the association of E. coli
ribosomal particles, Nucleic Acids Res., 16,
6309-6326.
77.Cunha, S., Woldringh, C. L., and Odijk, T. (2001)
Polymer-mediated compaction and internal dynamics of isolated
Escherichia coli nucleoids, J. Struct. Biol., 136,
53-66.
78.Lavery, P. E., and Kowalczykowski, S. C. (1992)
Enhancement of recA protein-promoted DNA strand exchange activity by
volume-occupying agents, J. Biol. Chem., 267,
9307-9314.
79.Spitzer, J. J., and Poolman, B. (2005)
Electrochemical structure of the crowded cytoplasm, Trends Biochem.
Sci., 30, 536-541.
80.Spitzer, J. (2017) Emergence of life on Earth: a
physicochemical jigsaw puzzle, J. Mol. Evol., 84,
1-7.
81.Jun, S., and Mulder, B. (2006) Entropy-driven
spatial organization of highly confined polymers: lessons for the
bacterial chromosome, Proc. Natl. Acad. Sci. USA, 103,
12388-12393.
82.Bouligand, Y., Soyer, M. O., and Puiseaux-Dao, S.
(1968) La structure fibrillaire et l’orientation des chromosomes
chez les Dinoflagellés, Chromosoma, 24,
251-287.
83.Kombrabail, M. H., and Krishnamoorthy, G. (2005)
Fluorescence dynamics of DNA condensed by the molecular crowding agent
poly(ethylene glycol), J. Fluoresc., 15, 741-747.
84.Soleimaninejad, H., Chen, M. Z., Lou X., Smith,
T. A., and Hong, Y. (2017) Measuring macromolecular crowding in cells
through fluorescence anisotropy imaging with an AIE fluorogen, Chem.
Commun., 53, 2874-2877.
85.Machiyama, H., Morikawa, T. J., Okamoto, K.,
Watanabe, T. M., and Fujita, H. (2017) The use of a genetically encoded
molecular crowding sensor in various biological phenomena, Biophys.
Physicobiol., 14, 119-125.
86.Currie, M., Leopold, H., Schwarz, J., Boersma, A.
J., Sheets, E. D., and Heikal, A. A. (2017) Fluorescence dynamics of a
FRET probe designed for crowding studies, J. Phys. Chem. B,
121, 5688-5698.
87.Smith, A. E., Zhou, L. Z., Gorensek, A. H.,
Senske, M., and Pielak, G. J. (2016) In-cell thermodynamics and a new
role for protein surfaces, Proc. Natl. Acad. Sci. USA,
113, 1725-1730.
88.Hansel, R., Luh, L. M., Corbeski, I., Trantirek,
L., and Dotsch, V. (2014) In-cell NMR and EPR spectroscopy of
biomacromolecules, Angew. Chem. Int. Ed. Engl., 53,
10300-10314.
89.Shahid, S., Hassan, M. I., Islam, A., and Ahmad,
F. (2017) Size-dependent studies of macromolecular crowding on the
thermodynamic stability, structure and functional activity of proteins:
in vitro and in silico approaches, Biochim. Biophys.
Acta, 1861, 178-197.
90.Hansel, R., Lohr, F., Foldynova-Trantirkova, S.,
Bamberg, E., Trantírek, L., and Dotsch, V. (2011) The parallel
G-quadruplex structure of vertebrate telomeric repeat sequences is not
the preferred folding topology under physiological conditions,
Nucleic Acids Res., 39, 5768-5775.
91.Tyrrell, J., Weeks, K. M., and Pielak, G. J.
(2015) Challenge of mimicking the influences of the cellular
environment on RNA structure by PEG-induced macromolecular crowding,
Biochemistry, 54, 6447-6453.
92.Smith, S., Cianci, C., and Grima, R. (2017)
Macromolecular crowding directs the motion of small molecules inside
cells, J. R. Soc. Interface, 14, 20170047.
93.Nozaki, T., Imai, R., Tanbo, M., Nagashima, R.,
Tamura, S., Tani, T., Joti, Y., Tomita, M., Hibino, K., Kanemaki, M.
T., Wendt, K. S., Okada, Y., Nagai, T., and Maeshima, K. (2017) Dynamic
organization of chromatin domains revealed by super-resolution
live-cell imaging, Mol. Cell, 67, 282-293.
94.Vargas, D. Y., Raj, A., Marras, S. A., Kramer, F.
R., and Tyagi, S. (2005) Mechanism of mRNA transport in the nucleus,
Proc. Natl. Acad. Sci. USA, 102, 17008-17013.
95.Hunter, T. (2012) Why nature chose phosphate to
modify proteins, Philos. Trans. R. Soc. Lond. B. Biol. Sci.,
367, 2513-2516.
96.Shimooka, Y., Nishikawa, J., and Ohyama, T.
(2013) Most methylation-susceptible DNA sequences in human embryonic
stem cells undergo a change in conformation or flexibility upon
methylation, Biochemistry, 52, 1344-1353.
97.Guigas, G., Kalla, C., and Weiss, M. (2007) The
degree of macromolecular crowding in the cytoplasm and nucleoplasm of
mammalian cells is conserved, FEBS Lett., 581,
5094-5098.
98.Van den Berg, J., Boersma, A. J., and Poolman, B.
(2017) Microorganisms maintain crowding homeostasis, Nat. Rev.
Microbiol., 15, 309-318.