REVIEW: Banding Pattern of Polytene Chromosomes as a Representation of Universal Principles of Chromatin Organization into Topological Domains
T. D. Kolesnikova1,2
1Institute of Molecular and Cellular Biology, Siberian Branch of the Russian Academy of Sciences, 630090 Novosibirsk, Russia; E-mail: kolesnikova@mcb.nsc.ru2Novosibirsk State University, 630090 Novosibirsk, Russia
Received November 14, 2017; Revision received December 3, 2017
Drosophila polytene chromosomes are widely used as a model of eukaryotic interphase chromosomes. The most noticeable feature of polytene chromosome is transverse banding associated with alternation of dense stripes (dark or black bands) and light diffuse areas that encompass alternating less compact gray bands and interbands visible with an electron microscope. In recent years, several approaches have been developed to predict location of morphological structures of polytene chromosomes based on the distribution of proteins on the molecular map of Drosophila genome. Comparison of these structures with the results of analysis of the three-dimensional chromatin organization by the Hi-C method indicates that the morphology of polytene chromosomes represents direct visualization of the interphase nucleus spatial organization into topological domains. Compact black bands correspond to the extended topological domains of inactive chromatin, while interbands are the barriers between the adjacent domains. Here, we discuss the prospects of using polytene chromosomes to study mechanisms of spatial organization of interphase chromosomes, as well as their dynamics and evolution.
KEY WORDS: polytene chromosomes, topologically associating domain, TADs, spatial organization of interphase chromosome, 3D chromatin organization, interband, Hi-CDOI: 10.1134/S0006297918040053
Abbreviations: IH, intercalary heterochromatin; PCh, polytene chromosomes; TAD, topologically associating domain.
BANDING PATTERN OF POLYTENE CHROMOSOMES
Polytene chromosomes (PChs) of Drosophila salivary glands represent a unique model of interphase chromosomes. Their giant size allows visualization of replication and transcription processes, investigation of distribution of chromosomal proteins at the cytological level, and gene mapping [1]. The pattern of alternating bands and interbands characteristic for PChs has been used for many years for fine mapping of various cytogenetic markers. Interbands are the least compact regions in PChs with the degree of DNA compaction of (1 : 3)-(1 : 15). Based on morphological features, the bands can be divided into two classes: grey and black. Grey bands are relatively fine decondensed structures with the degree of DNA compaction of (1 : 54)-(1 : 63). Black bands are as a rule thicker and contain the most compacted material; the degree of DNA compaction in them can exceed 1 : 200 [2-5]. These bands are positioned at the beginning of sections and subsections in the classical cytological maps of PChs [6, 7]. The intercalary heterochromatin (IH) bands are the largest among black bands. They replicate very late in PChs and do not finish replication, which results in their distinct morphology that includes breaks, constrictions, and ectopic contacts [8]. It was shown for various Diptera species that the banding patterns are stable and conserved in the polytene chromosomes prepared from different tissues [9-13]. This fact itself suggests that PCh banding reflects general organization of Drosophila genome common for all tissues.
The problem in the validation of this hypothesis was the absence of the data on the exact correlation between PCh morphological structures and the genome map. Several years ago, 32 interbands were localized on the genome map, which allowed the conclusion that interbands display conserved chromatin organization and similar protein composition in various tissues. They consist of open chromatin, and many of them include 5′-ends of housekeeping genes [5, 14-16]. The conserved properties of interbands in different tissues allowed scientist to develop an approach for predicting morphological structures in PChs based on the distribution of chromatin proteins. According to the results of bioinformatic analysis of the distribution of interband proteins, Drosophila genome was divided into four types of chromatin initially termed cyan, blue, green, and magenta [5]. Later these terms were replaced with aquamarine, lazurite, malachite, and ruby, respectively [17]. In cultured cells, aquamarine chromatin is enriched with proteins characteristic for PCh interbands, such as the interband-specific protein CHRIZ/CHROMATOR, chromatin remodeling proteins, and transcription factors [5, 14, 18]. Genes with promoters associated with this type of chromatin usually demonstrate broad expression patterns. More than 90% of ORC2-binding sites in the nuclei of cultured cells and PChs from the salivary glands are located in this chromatin, which indicates its key role in the replication initiation [5]. Lazurite chromatin contains only small amount of CHRIZ but is enriched with proteins involved in transcription elongation. Lazurite chromatin typically encompasses gene bodies [17]. Interband-specific proteins are completely absent from the ruby chromatin which overlaps with the silent types of chromatin identified based on the analysis of protein distribution in cultured cells [19, 20]. Ruby chromatin is enriched with Lamin, SUUR, and proteins involved in Polycomb-mediated repression [5, 17, 21]. Malachite chromatin is enriched with the insulator proteins CP190, SU(HW), and mod 2.2 [21].
The distribution of all four chromatin types is closely related to the PCh morphology. At present, all precisely localized interbands are associated with the aquamarine chromatin. At the same time, not all aquamarine chromatin fragments correspond to interbands [5, 17, 21]. The thin grey bands and some edge zones of the thick compact bands are formed by lazurite chromatin [5, 22]. It was demonstrated that zones of alternating aquamarine and lazurite chromatin in the 10A-B region coincide with the chromosome fragments in which interbands alternate with grey bands [4, 5]. The IH bands and the large black bands consist mostly of ruby chromatin (80% of total length) with insertions of short fragments of malachite chromatin (11%) [21]. It was shown for the 2R chromosome distal segment (cytological map section 58-60) that ruby chromatin could serve as a marker of all black bands including the thin ones [23]. Hence, the model of four color-coded chromatin is a productive tool for associating various PCh morphological structures with the coordinates on the Drosophila genome map (at least, for some particular chromatin regions).
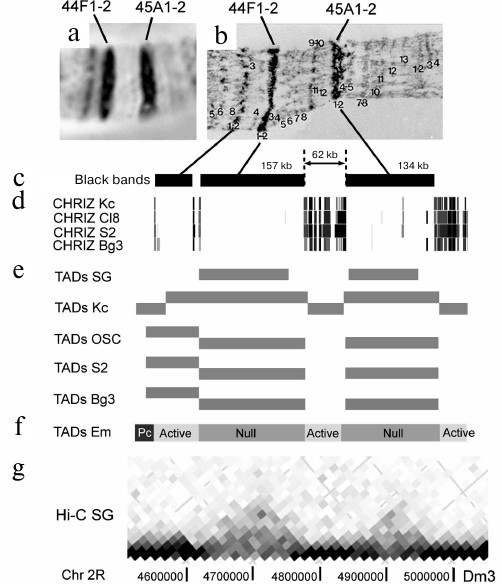
Figure 1 shows a fragment of chromosome 2R with two black bands 44F1-2 and 45A1-2 separated with a decompacted segment in which two fine grey bands could be seen under a light microscope and six very fine grey bands were detected by electron microscopy (EM). Our approach allows to establish the band boundaries and shows that the 44F1-2 band corresponds to the 157-kb DNA fragment, and the 45A1-2 band corresponds to the 134-kb DNA fragment. It is clearly seen that the separating 62-kb fragment is significantly more decompacted and occupies a large portion of the chromosome. It is composed of grey bands alternating with interbands. Considering that grey bands are not clearly distinguished by light microscopy, the terminology in the published literature can be sometimes confusing, when the light decompacted regions between the large black bands are called interbands. According to many years of studies, the “true” interband is a structure with a characteristic size of 0.3-3.8 kb [24, 25]. Hence, the genome section of 62 kb is significantly larger than a single interband. If we use the presence of CHRIZ-binding sites as a criterion for the interbands, the 62-kb fragment should consist of at least six interbands alternating with fine grey bands, which was clearly demonstrated by EM (Fig. 1).
Fig. 1. Correspondence of black condensed bands in PChs to TADs in different cell types exemplified by the bands 44F1-2 and 45A1-2. a) Morphology of a chromosome 2R fragment corresponding to a ~500-kb DNA region including two relatively large condensed bands (aceto orcein staining, phase contrast); b) electron microscopy image of the corresponding DNA fragment of the PCh (adapted from [26]); c) localization of condensed black bands in PCh relative to on the chromosome 2R genome map; band coordinates are predicted with the algorithm described in [23]; d) localization of sites enriched with CHRIZ protein on the genome map (according to ModEncode data [27]); e) localization of TADs on the genome map; TADs are identified using the Hi-C method in salivary glands (SG) [28] and four cell cultures (Kc, OSC, S2, and Bg3) [29]; f) TADs associated with active chromatin (Active) and Polycomb/H3K27me3 chromatin (Pc) or lacking active chromatin markers (Null) as determined by the Hi-C analysis of embryos [30]; g) fragment of the spatial interaction heatmap based on the Hi-C data for PChs of salivary glands [28]. Genome coordinates are presented in accordance with the Dm3 release of the D. melanogaster genome.
Comparison of the data of analysis of selected individual regions of salivary gland PChs and the properties of four chromatin types led to the suggestion that all most compacted black bands associated with the ruby chromatin correspond to the genome sites fragments with the properties similar to those of IH regions, i.e., to the clusters of tissue-specific genes and to chromatin types inactive in the majority of tissues. These fragments also replicate late in most tissues. The regions between them consist of alternating grey bands and interbands and are enriched with housekeeping genes and replication origins, which is characteristic of the permanently open chromatin that replicates early [4, 5, 17, 21, 23].
BLACK BANDS OF POLYTENE CHROMOSOMES CORRESPOND TO TOPOLOGICAL
DOMAINS
The concept on the correspondence between PCh morphology and organization of Drosophila genome has received a new perspective due to the accumulation of data on the chromatin 3D organization [5, 28-32]. Recent studies, especially those using the Hi-C method, showed that partitioning of eukaryotic genome into developmentally stable extended “physical” domains (topologically associating domains, TADs) is very common. Interactions between DNA sections within these domains occur with a high probability and are very dynamic, while interactions between the domains occur at a much lower frequency [33, 34]. These domains are organized hierarchically and can include several levels of smaller contact domains [29, 32, 35].
By now, the results of Drosophila genome Hi-C analysis have been published, and the boundaries of TADs have been determined in cultured cells, embryos, and salivary gland cells [28-30, 32, 36, 37]. Already the earliest studies on the TAD boundaries in Drosophila showed that factors defining these boundaries are to a great extent characteristic of the PCh interbands [29-31, 38]. For example, CHRIZ was found to be the most important marker for TAD boundaries [30]. The functions of this protein are not fully understood; however, it was found to be a typical marker of interbands in PChs [18] that can be used for the prediction of interband location in the genome [4, 5, 39]. It was demonstrated in [38] that the greater is the strength of the boundary between TADs (the value reflecting the drop in the probability of contacts between adjacent DNA regions along the chromosome), the larger is the number of insulator proteins mapped to the respective locus. Another important criterion of the boundary is availability of active promoters. These factors are also essential for the prediction interband location in polytene chromosomes [5, 17].
The majority of TAD boundaries in cultured cells and Drosophila embryos belong to the aquamarine and lazurite types of chromatin, while the ruby chromatin has been predominately detected in the topological domains and is not separated by their boundaries. It was concluded based on these facts that TADs themselves do not correspond to the black bands, and their boundaries lie predominately outside the black bands [5, 17, 29]. In primary spermatocytes, IH bands in PChs were found to colocalize with domains characterized by the presence of H3K27me3, that, in turn, coincided with the prominent topological domains (prominent TADs) in various types of cells [37]. The Hi-C analysis of salivary gland PChs demonstrated that at least 95% of IH bands coincide precisely with TADs [28]. Moreover, the boundaries of TADs identified in PChs allow prediction of boundaries in five out of six black bands in PChs, which was confirmed by in situ hybridization [28]. Despite the fact that the resolution of Hi-C maps (15 kb) does not allow identification of PCh topological domains less than 75 kb long, Eagen et al. hypothesized that all black bands in PChs (including the shorter ones) are direct visualization of TADs [28].
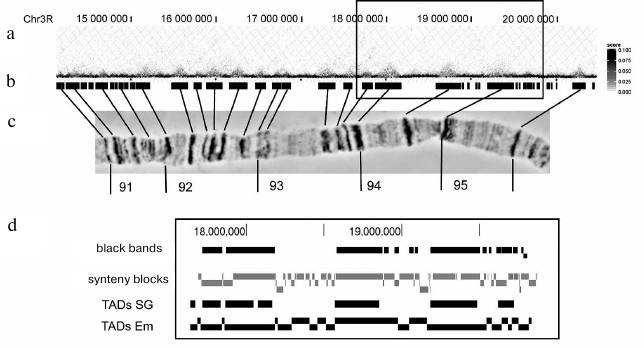
The Hi-C heatmaps of the probability of contacts in PChs presented in [28] clearly show TADs as regions with a high probability of inner interactions located along the diagonal. Many TADs look like almost uniformly stained squares. Such homogenous distribution of color indicates almost identical probability of interactions between a pair of DNA sequences inside the band. Therefore, it can be suggested that PCh band is a relatively randomly packed chromatin globule. Figure 2 shows a fragment of chromosome 3R. Using the approach described in [23], we predicted coordinates of black bands in this region by constructing a heatmap using the matrix of contact probabilities in PCh (available in GEO:GSE72512) [28]. The map is rotated by 45° for the convenience of comparison, and TADs look like intensely stained triangles. It can be seen that all the bands clearly visible under a light microscope correspond to the extended genome fragments and form distinct domains enriched with inner contacts. They are separated by zones with predominance of exclusively short-range contacts. Resolution of the Hi-C analysis and, as a consequence, of the heatmap does not allow elucidation of the chromatin organization in these areas.
Fig. 2. Correspondence of PCh black bands to contact domains in the heatmap constructed using the Hi-C data [28] for a ~5 Mb segment of chromosome 3R (PCh regions 91-95). a) Fragment of the spatial interaction heatmap constructed using the Hi-C data for salivary gland PChs [28]. Genome coordinates are adjusted according to the Dm3 release of D. melanogaster genome; b) location of black compacted bands in PCh relatively to the chromosome 3R genome map; band coordinates are predicted with the algorithm described in [23]; c) morphology of the corresponding 3R chromosome fragment (aceto orcein staining, phase contrast); d) scaled-up chromosome fragment (marked with rectangle above). The distribution of black bands was compared to the distribution of TADs in salivary gland PChs (TAD SG) [28] and embryos (TADs Em) [30], as well as to the distribution of synteny blocks [66].
The current terminology recognizing TADs, inter-TADs, and TAD boundaries has been suggested based on the early papers on the whole-genome analysis of 3D chromatin organization [40]. However, it has become clear that classifying the 3D organization using linear domains oversimplifies the subject. In reality, genome 3D structure is a complex, multilevel system of domains with a high probability of intra-contacts. The hierarchical level TAD is only a matter of convention [29, 32, 34, 35].
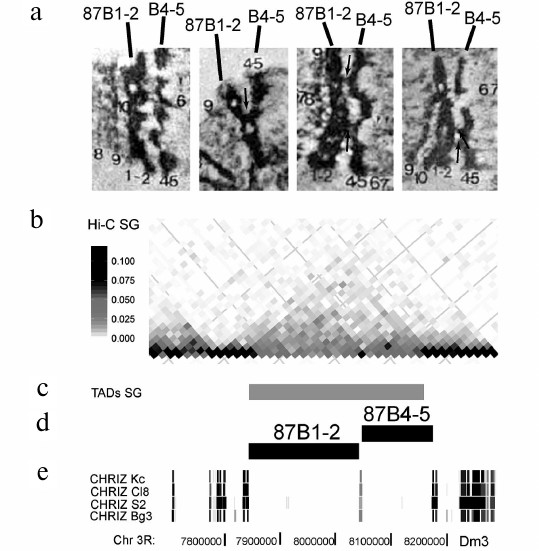
Comparison of the pattern of wide bands in PChs and the heatmap of contacts along the chromosome reveals that clearly defined undivided TADs exists only when the extended bands are separated by lengthy regions of decompacted grey bands and interbands. At the same time, many groups of closely located bands could be associated with a single TAD. However, detailed analysis of the heatmap revealed clearly pronounced subTADs corresponding to individual bands, as shown in Fig. 3 for the 87B region. Two closely located bands, 87B1-2 and 87B4-5, had been identified on the genome map earlier as they are associated with the IH regions [39]. Their boundaries were later refined in [41]. It can be seen in the EM images that these bands are attached to each other at several sites, with chromatin fibers between them. Nevertheless, they are two separate bands. According to Hi-C analysis, these bands form single TAD in the PCh; however, the heatmap clearly demonstrates the existence of individual subdomains separated by the interband (Fig. 3).
Fig. 3. Interband separating two closely located 87B1-2 and 87B4-5 bands divides the TAD into subdomains. a) EM images of four examples of the 87B1-2 and 87B4-5 band pair morphology (adapted from [43]). The bands are clearly separated by an interband; however, there are fibers between them and local contacts (marked with arrows); b) Hi-C heatmap of PCh [28] shows that the chromosome region corresponding to the 87B1-2 and 87B4-5 bands forms a contact domain divided into two subdomains; c) according to [28], this locus contains one TAD that includes both bands and whose boundaries coincide precisely with the boundaries of the bands; d) localization of bands on the genome map according to [41]; e) localization of CHRIZ-enriched sites on the genome maps of four cell lines (according to the data from ModEncode [27]). The CHRIZ-binding site common for all four cell lines coincides precisely with the boundary of subTADs visible in the heatmap.
Comparison of the PCh Hi-C heatmap and distribution of black bands along the entire 2R chromosome showed that virtually all TADs identified in PCh corresponded to the black bands or groups of closely located black bands with each band in these groups corresponding to subTADs [42].
ARE REGIONS BETWEEN BLACK BANDS OF POLYTENE CHROMOSOMES ACTIVE
TADs OR INTER-TADs?
The 15-20-kb resolution of Hi-C maps is not sufficient for identification of TADs less than 75-80 kb in length by statistical methods [28, 29]. The gaps between these large domains are automatically assigned to inter-TADs. Considering that the majority of these extended topological domains are characterized by silent chromatin types, it was suggested that the ability to form TADs is the feature of closed chromatin, its inherent property determined by the propensity of deacetylated histones for short-term association [29]. TADs identified by analysis of 3D chromatin organization in salivary gland PChs correspond to largest black bands that, as shown in other studies, consist predominately of the silent chromatin [5, 39].
Hi-C maps with a resolution of 4-10 kb were obtained by two groups of researchers for Drosophila embryos and Kc cells [30, 36]. TADs identified in these studies harbored 98% of genome euchromatin; hence, the studied genomes lacked inter-TADs. All topological domains in embryos were divided into four groups according to the chromatin specific features: Active, PcG, HP1, and Null domains. The Null domains were not enriched with any known histone modification; instead, they were enriched with histone H1 and lamin [30]. The last three types of domains can be considered silent type domains. Despite the fact that all four types of domains were assigned to TADs, active and silent chromatin domains significantly differ in the manner of chromatin packing [30, 37]. If the probability of interactions between DNA regions in the active domains drops exponentially with the increase in the distance between them, silent type domains demonstrate a high probability of long-range intradomain contacts that does not depend on the distance due probably to the high degree of chromatin compaction [30]. The active domains in Drosophila are usually short. Assignment of these domains to TADs seems paradoxical, it is based on the fact that they contain a large number of binding sites for proteins characteristic of TADs boundaries. Apparently, accepting more rigid criteria for defining the boundaries of topological domains would lead to their fragmentation.
Figure 1 shows that the black bands 44F1-2 and 45A1-2 in a short fragment of chromosome 2R correspond to TADs identified in PCh [28]. Moreover, these bands correspond to TADs in cultured cells [29] and TADs of the Null type found in Drosophila embryos [30]. The gap between the bands was identified as an inter-TAD in PChs and cultured cells, bit as an active TAD in Drosophila embryos. Note that the gap contains multiple interbands in PChs and multiple binding sites for CHRIZ, a marker of interbands and topological domain boundaries. You just have to take a look at the PCh in order to understand that the difference in the flexibility of PChs and chromosomes from diploid cells plays is equally important as the method resolution. If in a diploid cell, closely located small active chromatin domains have a high probability to become spatially close, in the much less flexible PCh, these domains are rigidly separated by interbands.
Recently, the 3D organization of chromatin from Drosophila embryos was analyzed with an unprecedented resolution of 500-1000 bp [32]. Such high resolution allowed identification of contact domains ranging from several kilobases to hundreds kilobases. Using the region 10A-10B, in which all thick and fine bands had been previously mapped in great detail [4], the authors showed that both thick black bands and fine grey bands formed TADs; interbands were located precisely on their boundaries [30].
Therefore, while black bands of polytene chromosomes have been assigned to TADs in various cell types irrespectively of the resolution of the Hi-C method used, the regions between them can correspond to inter-TADs, active TADs, or consist of alternating short (2-30 kb) domains divided by the boundaries of ~1 kb depending on the method resolution.
POLYTENE CHROMOSOMES AS A MODEL FOR THE STUDIES OF THE MECHANISMS
OF FORMATION OF TAD BOUNDARIES AND THEIR DYNAMICS
Two major mechanisms responsible for the formation of boundaries between TADs are currently debated. The first mechanism involves localization at the domain boundaries of insulator proteins that prevent spatial interactions between the chromatin fibril regions on both sides of the boundary. The alternative mechanism engages active transcription, when the regions of open hyperacetylated chromatin separate the regions of inactive chromatin self-assembling into compact globules [29, 34]. Both mechanisms can be illustrated using PCh morphology as an example.
Loosely compacted interbands in PCh separate compact chromatin globules and coincide with the TAD boundaries established at a very high resolution, which indicates that interbands represent perfect visualization of the TAD boundaries. Interbands in PChs have been attracting attention of researchers for many years. A massive set of cytogenetic tools for their investigation has been developed, in particular, for creation of artificial interbands with the help of ectopic recruitment of interband proteins to the bands [44-46].
Comparison of dynamics of chromosome partition into TADs in different cell types indicates that transcription activation of tissue-specific genes correlates with either the loss of respective TADs or their division into subTADs [34]. If we accept the hypothesis that the morphological structure band reflects local compaction of the genomic material, then the change in PCh morphology following transcription initiation represents perfect illustration of this phenomenon. It is well known that expression of genes in PChs can cause significant changes of their structure. For some genes, it is emergence of huge puffs, for other – slight decompaction and thickening of the band that sometimes can be seen only by EM. Nevertheless, high levels of gene expression are accompanied by changes in the PCh morphology [47-51].
Detailed EM analysis of the development of 18 puffs [48] showed that in the majority of cases, uniform decondensation of the entire band occurred during the puff development. However, there were cases when only a middle part or the edges of the band decondensed. Moreover, sometimes puffs were formed as a result of simultaneous decondensation of two, three, or four bands. These results indicate complex spatial organization of chromatin bands and illustrate unique possibilities of visualization of the chromatin organization dynamics provided by PCh. In the light of newly developed approaches that allow establishment of genomic coordinates of the bands with a high accuracy, it would be extremely interesting to analyze 3D chromatin structure and properties of expression of genes forming different types of puffs.
As a rule, puffs develop during expression of genes with specific functions in PChs. These genes are mostly localized in the ruby chromatin, i.e., in the black bands [5]. Interbands, on the other hand, are enriched with promoters of housekeeping genes. In many cases, when genes have promoters in an interband adjacent to the black band, the gene bodies are located within the black band [52].
Hou et al. compared genome division into TADs based on the analysis of 3D chromatin structure in Kc cells [36], and five types of chromatin identified by analysis of protein distribution in these cells [19]. The authors found that many topological domains were represented by inactive chromatin, but also contained regions of open, predominately YELLOW chromatin (typical for housekeeping genes) at their boundaries. It was found that TAD boundaries corresponded to the gene promoters, while the bodies of these genes, even when transcriptionally active, with within the TADs. This fact once again illustrates the similarity between TADs and bands and demonstrates that active chromatin can be a part of generally inactive contact domain.
The question on how the presence of transcriptionally active gene at the band boundary affects the morphology of this boundary requires special investigation. Detailed cytogenetic analysis of the small locus 61C7-61C8 of chromosome 3L using the high-resolution microscopy was reported in [53]. Cytological structures and genome coordinates in this region were compared with a high accuracy by in situ hybridization using over 20 probes. The authors assigned positions of four identified bands in this region to the color model of chromatin [5] and showed that in three cases (when the boundary of the band predicted by the model did not contain an actively transcribed gene and lazurite chromatin), the model worked very accurately. In the case, when lazurite chromatin was located at the boundary of a predicted band, the boundary of the morphologically decompacted chromatin was shifted (see Fig. 4 in [53]).
These examples demonstrate that the PCh morphology allows visualization of both stable spatial chromatin organization into TADs conserved in different types of cells and its dynamics related to the gene expression.
SPECIFIC PROPERTIES OF PCh SPATIAL ORGANIZATION
In this review, we aimed to demonstrate that PCh morphology represents visualization of the universal organization of an interphase chromosome (see above). At the same time, it is obvious that the properties of PChs differ significantly from those of regular chromosomes. The structure formed as a result of 10 rounds of replication and consisting of more than one thousand DNA strands cannot be as flexible as a single DNA molecule. Moreover, DNA strands in the PCh cannot form contacts with the remote regions of the same chromosome or of other chromosomes. This results in the absence of compartments and long-range interactions, as it was demonstrated directly by the Hi-C analysis of salivary gland PChs [28]. At the same time, long-range interactions still exist in the squash PCh preparations. Thus, it is well known that the IH regions interact with each other forming the so-called ectopic contacts [8]. These regions can be covalently linked due to incomplete replication in a combination with the DNA repair [54, 55]. This is why ectopic contacts can be seen in chromosome preparations fixed with acetic acid (used for common staining). Preparation treatment with formaldehyde results in the larger number of attachments between the regions. However, the frequency of these contacts is negligibly low in comparison with the frequency of contacts between the distant regions in the open or closed chromatin in diploid cells. This raises the question on the importance of long-range interactions and organization of chromatin into compartments in the interphase nucleus for transcription. Many Drosophila larval tissues carry polytene chromosome with a high level of ploidy that are actively transcribed [56].
Classical Hi-C matrices of spatial interactions are constructed based on averaged data from a large number of cells. It is this averaging that allows identification of stable topological domains with sharp boundaries. The packing of individual molecules is stochastic, as revealed by analysis of spatial interaction in individual cells (see review [57]). In PChs, we see bundles of chromatin fibers, in which fragments corresponding to physical domains are aligned and look like single structures; in an individual chromosome, we can see an averaged pattern of chromatin packing.
Probes corresponding to interbands are positioned strictly perpendicular to the chromosome in in situ hybridization [4, 53], which indicates that DNA molecules in these structures are well aligned relatively to each other along the chromosome axis. This is also corroborated by the morphology of the interband itself. How is DNA arranged in the bands? Based on the Hi-C heatmap, where large bands correspond to the regions with high number of inner contacts and each site possesses an ability to interact with any other site, it can be suggested that a PCh band comprises a globule of uniformly mixed material. However, there are also data that distribution of DNA in large bands is not random. The results of in situ hybridization of probes specific for the 54B1-2 band on the sections of polytene chromosomes were analyzed by electron microscopy [58]. Despite the fact that the band structure changes during denaturation required for hybridization, the signal localized perpendicular to the band; the dots corresponding to the probe location on individual DNA strands formed a “cloud” that only partially occupied the band area. The results of in situ hybridization of probes corresponding to the band middle part and boundaries were presented in [28]. It can be seen in the images that the probes did not mix; the area that bound the probe specific to the band middle part was separated by unstained section from the parallel areas stained with the probe specific for the band boundaries. We performed simultaneous immunolocalization of probes corresponding to the boundaries and the middle part of the complex IH band 75C1-4 on chromosomes of Drosophila larvae with mutations in the SuUR gene (with no inderreplication in the IH bands) [59]. In all the cases, signals corresponding to the boundary- and middle-specific probes did not overlap. Overexpression of the SuUR genes causes significant changes in the morphology of the largest bands – they turn into huge “bubbles” from which the majority of proteins, including histones, escape [59, 60]. It is possible that one of the reasons for the “bubble” formation is electrostatic repulsion of DNA molecules lacking histones. The probes corresponding to the middle part and boundaries of the bans hybridized to the significant portion of the “bubble”, but the signals from these probes did not overlap, which once again suggests a certain order in the DNA distribution in the band [59, 60]. Another proof for the DNA organization in at least some individual bands is puff formation in the middle of the band. It separates the band into two parts, and the decondensed fragment is strictly perpendicular to the chromosome axis [48].
Compact bands are very similar in their morphology. They are all associated with the ruby chromatin. However, ruby chromatin combines two fundamentally different types of chromatin: Polycomb chromatin (BLUE chromatin according to Filion et al. [19]) and Null chromatin (BLACK chromatin [19]). The first one is characterized by gene repression mediated by proteins from the Polycomb group. The second one is characterized by the absence of active transcription and active chromatin marks, high levels of histone H1 and lamin, and the absence of specific mechanisms for additional chromatin compaction. Analysis of 3D chromatin organization in Drosophila embryo cells showed [30] that all three types of inactive topological domains differed considerably from the active domains in the type of chromatin packing; however, there was no significant difference between the Null, Polycomb, and HP1 domains. Examination of chromatin in diploid cells using super-resolution microscopy demonstrated that not only active and inactive chromatin domains, but also various types of inactive chromatin domains display different types of inner chromatin packing [61]. The packing corresponding to the model of self-organizing fractal globule was observed in transcriptionally active and transcriptionally inactive (Null) chromatin. Polycomb-repressed domains were the most compactly packed and displayed a complex type of packing. All three types of domains exhibited an exponential dependence of the 3D physical size and the domain length on the genome map; however, the exponents values differed significantly. The authors concluded that transcription actively decompacts domains, the compacting proteins actively compact the domains, and the Null chromatin is packed in accordance with the molecular crowding laws. The question whether the bands formed by the Polycomb and Null types of chromatin in PChs differ in the DNA organization still remains.
BLACK BANDS OF POLYTENE CHROMOSOMES ARE EVOLUTIONARILY CONSERVED
TOPOLOGICAL DOMAINS
Organization of eukaryotic genomes into relatively autonomous physical domains in different types of cells raises the question on functional significance of these domains. It was shown both for mammals and Drosophila that the majority of enhancer–promoter interactions were within the limits of topological domains [34]. Razin and Ulianov suggested in their review [34] that the role of TADs in the ordering of interactions between regulatory domains is more pronounced in mammals than in Drosophila. Genome organization into topological domains in Drosophila is formed passively due to the self-assembly of inactive regions. However, Drosophila has a complex network of interactions between enhancers and promoters [62]. Comparison of the distribution of conserved non-coding genomic elements (many of which correspond to enhancers of developmental genes and form evolutionarily conserved syntenic clusters [63, 64]) with TAD boundaries demonstrated close relationship between them. The majority of clusters colocalize with single TADs; moreover, the boundaries of the cluster and the respective TADs almost coincide [64].
We investigated the degree of conservation of 60 IH regions described in [39] and showed that these regions were genome fragments with a conserved order of genes characterized by low frequency of synteny breaks. It was found that syntenic blocks corresponding to the IH regions were located among the most prolonged blocks in genomes of D. melanogaster and other species. We also demonstrated that the low gene density in the IH regions of D. melanogaster is an evolutionarily conserved feature. We suggested that not only the order of genes but also the repressive status of chromatin have been preserved during evolution, and these blocks remain silent and late-replicating domains in other species of Drosophila genus as well [41]. We also showed that the IH regions are enriched with highly conserved non-coding sequences, thereby suggesting the existence of a complex network of regulatory elements in these regions that defines a high degree of conservation of the gene order in them [65].
Coincidence of IH bands with topological domains allows to extrapolate the conclusion on the conservation of the gene order in the largest black bands onto TADs. The similarity of chromatin organization in IH bands and smaller black bands suggests that the revealed features can be typical for all inactive TADs. Black bands, TADs, and synteny blocks identified in the study of Von Grotthuss and co-authors [66] are compared in Fig. 1. It can be seen that the majority of bands are represented by the combined synteny blocks, and the boundaries of synteny blocks are close to the band boundaries and to the TAD boundaries. Hence, PChs can serve as a useful tool for analyzing the features of Drosophila genome evolution.
In conclusion, the morphology of PCh is a direct visualization of the universal mechanisms of the interphase nucleus spatial organization into topological domains. Compact black bands correspond to extended topological domains consisting predominantly of inactive chromatin. Interbands comprise barriers between the contact domains. We believe that PChs could be a perfect model for investigation of the mechanisms of interphase chromosome spatial organization, dynamics, and evolution not only in the past, but also in the future.
Acknowledgments
Author is grateful to E. I. Volkova, S. A. Demakov, O. V. Demakova, and T. Yu. Zykova for critical remarks and help with the literature search, to F. P. Goncharov for help in visualization of Hi-C data published by other authors, and to V. S. Fishman for his help in interpretation of Hi-C data.
This work was financially supported by the Russian Science Foundation (grant 14-14-00934).
REFERENCES
1.Zhimulev, I. F. (1999) Genetic organization of
polytene chromosomes, Adv. Genet., 39, 1-589.
2.Spierer, A., and Spierer, P. (1984) Similar level
of polyteny in bands and interbands of Drosophila giant
chromosomes, Nature, 307, 176-178.
3.Kozlova, T., Semeshin, V. F., Tretyakova, I. V.,
Kokoza, E. B., Pirrotta, V., Grafodatskaya, V. E., Belyaeva, E. S., and
Zhimulev, I. F. (1994) Molecular and cytogenetical characterization of
the 10A1-2 band and adjoining region in the Drosophila
melanogaster polytene X chromosome, Genetics, 136,
1063-1073.
4.Vatolina, T. Y., Boldyreva, L. V., Demakova, O. V.,
Demakov, S. A., Kokoza, E. B., Semeshin, V. F., Babenko, V. N.,
Goncharov, F. P., Belyaeva, E. S., and Zhimulev, I. F. (2011) Identical
functional organization of nonpolytene and polytene chromosomes in
Drosophila melanogaster, PLoS One, 6, e25960.
5.Zhimulev, I. F., Zykova, T. Y., Goncharov, F. P.,
Khoroshko, V. A., Demakova, O. V., Semeshin, V. F., Pokholkova, G. V.,
Boldyreva, L. V., Demidova, D. S., Babenko, V. N., Demakov, S. A., and
Belyaeva, E. S. (2014) Genetic organization of interphase chromosome
bands and interbands in Drosophila melanogaster, PLoS
One, 9, e101631.
6.Bridges, C. B. (1935) Salivary chromosome maps with
a key to the banding of the chromosomes of Drosophila
melanogaster, J. Hered., 26, 60-64.
7.Lefevre, E. G. (1976) A Photographic
Representation and Interpretation of the Polytene Chromosomes of
Drosophila melanogaster Salivary Glands (Ashburner, M., ed.)
Academic Press, London-New York.
8.Zhimulev, I. F., Semeshin, V. F., Kulichkov, V. A.,
and Belyaeva, E. S. (1982) Intercalary heterochromatin in
Drosophila. I. Localization and general characteristics,
Chromosoma, 87, 197-228.
9.Beermann, W. (1952) Chromomerenkonstanz und
specifische modifikationen der chromosomenstruktur in der entwicklung
und organdifferenzierung von Chironomus tentans,
Chromosoma, 5, 139-198.
10.Beermann, W. (1965) Structure and Function of
Interphase Chromosomes (Gerts, S. J., ed.) Pergamon Press,
Oxford-London-Edinburgh-NewYork-Paris-Frankfurt, pp. 375-384.
11.Sinha, P., Mishra, A., and Lakhotia, S. C. (1987)
Chromosomal organization of Drosophila tumors. I. Polytene
chromosome organization and DNA synthesis in ovarian pseudonurse cells
in otu mutants of D. melanogaster, Chromosoma,
95, 108-116.
12.Mal’ceva, N. I., Gyurkovics, H., and
Zhimulev, I. F. (1995) General characteristics of the polytene
chromosome from ovarian pseudonurse cells of the Drosophila
melanogaster otu11 and fs(2)B mutants, Chromosome
Res., 3, 191-200.
13.Heino, T. I. (1994) Polytene chromosomes from
ovarian pseudonurse cells of the Drosophila melanogaster out
mutant. II. Photographic map of the X chromosome, Chromosoma,
103, 4-15.
14.Demakov, S. A., Vatolina, T. Y., Babenko, V. N.,
Semeshin, V. F., Belyaeva, E. S., and Zhimulev, I. F. (2011) Protein
composition of interband regions in polytene and cell line chromosomes
of Drosophila melanogaster, BMC Genomics, 12,
566.
15.Vatolina, T., Demakov, S. A., Semeshin, V. F.,
Makunin, I. V., Babenko, V. N., Beliaeva, E. S., and Zhimulev, I. F.
(2011) Identification and molecular genetic characterization of the
polytene chromosome interbands in Drosophila melanogaster,
Russ. J. Genet., 47, 521-532.
16.Zhimulev, I. F., Belyaeva, E. S., Vatolina, T.
Y., and Demakov, S. A. (2012) Banding patterns in Drosophila
melanogaster polytene chromosomes correlate with DNA-binding
protein occupancy, Bioessays, 34, 498-508.
17.Boldyreva, L. V., Goncharov, F. P., Demakova, O.
V., Zykova, T. Y., Levitsky, V. G., Kolesnikov, N. N., Pindyurin, A.
V., Semeshin, V. F., and Zhimulev, I. F. (2017) Protein and genetic
composition of four chromatin types in Drosophila melanogaster
cell lines, Curr. Genomics, 18, 214-226.
18.Gortchakov, A. A., Eggert, H., Gan, M., Mattow,
J., Zhimulev, I. F., and Saumweber, H. (2005) CHRIZ, a chromodomain
protein specific for the interbands of Drosophila melanogaster
polytene chromosomes, Chromosoma, 114, 54-66.
19.Filion, G. J., van Bemmel, J. G., Braunschweig,
U., Talhout, W., Kind, J., Ward, L. D., Brugman, W., de Castro, I. J.,
Kerkhoven, R. M., Bussemaker, H. J., and van Steensel, B. (2010)
Systematic protein location mapping reveals five principal chromatin
types in Drosophila cells, Cell, 143, 212-224.
20.Kharchenko, P. V., Alekseyenko, A. A., Schwartz,
Y. B., Minoda, A., Riddle, N. C., Ernst, J., Sabo, P. J., Larschan, E.,
Gorchakov, A. A., Gu, T., Linder-Basso, D., Plachetka, A., Shanower,
G., Tolstorukov, M. Y., Luquette, L. J., Xi, R., Jung, Y. L., Park, R.
W., Bishop, E. P., Canfield, T. K., Sandstrom, R., Thurman, R. E.,
MacAlpine, D. M., Stamatoyannopoulos, J. A., Kellis, M., Elgin, S. C.,
Kuroda, M. I., Pirrotta, V., Karpen, G. H., and Park, P. J. (2011)
Comprehensive analysis of the chromatin landscape in Drosophila
melanogaster, Nature, 471, 480-485.
21.Khoroshko, V. A., Levitsky, V. G., Zykova, T. Y.,
Antonenko, O. V., Belyaeva, E. S., and Zhimulev, I. F. (2016) Chromatin
heterogeneity and distribution of regulatory elements in the
late-replicating intercalary heterochromatin domains of Drosophila
melanogaster chromosomes, PLoS One, 11, e0157147.
22.Demakova, O. V., Boldyreva, L. V., Demakov, S.
A., Goncharov, F. P., Antonenko, O. V., and Zhimulev, I. F. (2016)
Characteristic of the chromatin type corresponding to thin
“grey” bands in polythene chromosomes of Drosophila
melanogaster, Tsitologiya, 58, 248-252.
23.Kolesnikova, T. D., and Zhimulev, I. F. (2016)
Comprehensive approach to mapping late-replicating bands in polytene
chromosomes of Drosophila melanogaster, Tsitologiya,
58, 262-266.
24.Beermann, W. (1972) Chromomeres and Genes
in Results and Problems in Cell Differentiation (Beermann, W.,
ed.) Springer, Berlin-Heidelberg-New York, pp. 1-33.
25.Sorsa, V. (1984) Electron microscopic mapping and
ultrastructure of Drosophila polytene chromosomes, in Insect
Ultrastructure (King, R. C., and Akai, H., eds.) Plenum Press, New
York, pp. 75-107.
26.Saura, A. (1986) Electron Microscopic Mapping
of the Second Polytene Chromosome of Drosophila
melanogaster, Ph. D. Thesis, University of Helsinki, Dept. of
Genetics., p. 58.
27.Celniker, S. E., Dillon, L. A., Gerstein, M. B.,
Gunsalus, K. C., Henikoff, S., Karpen, G. H., Kellis, M., Lai, E. C.,
Lieb, J. D., MacAlpine, D. M., Micklem, G., Piano, F., Snyder, M.,
Stein, L., White, K. P., Waterston, R. H., and modENCODE Consortium
(2009) Unlocking the secrets of the genome, Nature, 459,
927-930.
28.Eagen, K. P., Hartl, T. A., and Kornberg, R. D.
(2015) Stable chromosome condensation revealed by chromosome
conformation capture, Cell, 163, 934-946.
29.Ulianov, S. V., Khrameeva, E. E., Gavrilov, A.
A., Flyamer, I. M., Kos, P., Mikhaleva, E. A., Penin, A. A., Logacheva,
M. D., Imakaev, M. V., Chertovich, A., Gelfand, M. S., Shevelyov, Y.
Y., and Razin, S. V. (2016) Active chromatin and transcription play a
key role in chromosome partitioning into topologically associating
domains, Genome Res., 26, 70-84.
30.Sexton, T., Yaffe, E., Kenigsberg, E.,
Bantignies, F., Leblanc, B., Hoichman, M., Parrinello, H., Tanay, A.,
and Cavalli, G. (2012) Three-dimensional folding and functional
organization principles of the Drosophila genome, Cell,
148, 458-472.
31.White, R. (2012) Packaging the fly genome:
domains and dynamics, Brief. Funct. Genomics, 11,
347-355.
32.Stadler, M. R., Haines, J. E., and Eisen, M. B.
(2017) Convergence of topological domain boundaries, insulators, and
polytene interbands revealed by high-resolution mapping of chromatin
contacts in the early Drosophila melanogaster embryo,
Elife, 6, e29550.
33.Ea, V., Baudement, M. O., Lesne, A., and Forne,
T. (2015) Contribution of topological domains and loop formation to 3D
chromatin organization, Genes (Basel.), 6, 734-750.
34.Razin, S. V., and Ulianov, S. V. (2017) Gene
functioning and storage within a folded genome, Cell. Mol. Biol.
Lett., 22, 18.
35.Weinreb, C., and Raphael, B. J. (2016)
Identification of hierarchical chromatin domains,
Bioinformatics, 32, 1601-1609.
36.Hou, C., Li, L., Qin, Z. S., and Corces, V. G.
(2012) Gene density, transcription, and insulators contribute to the
partition of the Drosophila genome into physical domains,
Mol. Cell, 48, 471-484.
37.El-Sharnouby, S., Fischer, B., Magbanua, J. P.,
Umans, B., Flower, R., Choo, S. W., Russell, S., and White, R. (2017)
Regions of very low H3K27me3 partition the Drosophila genome
into topological domains, PLoS One, 12, e0172725.
38.Van Bortle, K., Nichols, M. H., Li, L., Ong, C.
T., Takenaka, N., Qin, Z. S., and Corces, V. G. (2014) Insulator
function and topological domain border strength scale with
architectural protein occupancy, Genome Biol., 15,
R82.
39.Belyaeva, E. S., Goncharov, F. P., Demakova, O.
V., Kolesnikova, T. D., Boldyreva, L. V., Semeshin, V. F., and
Zhimulev, I. F. (2012) Late replication domains in polytene and
non-polytene cells of Drosophila melanogaster, PLoS One,
7, e30035.
40.Dixon, J. R., Selvaraj, S., Yue, F., Kim, A., Li,
Y., Shen, Y., Hu, M., Liu, J. S., and Ren, B. (2012) Topological
domains in mammalian genomes identified by analysis of chromatin
interactions, Nature, 485, 376-380.
41.Andreyenkova, N. G., Kolesnikova, T. D., Makunin,
I. V., Pokholkova, G. V., Boldyreva, L. V., Zykova, T. Y., Zhimulev, I.
F., and Belyaeva, E. S. (2013) Late replication domains are
evolutionary conserved in the Drosophila genome, PLoS
One, 8, e83319.
42.Kolesnikova, T. D., Goncharov, F. P., and
Zhimulev, I. F. (2018) Similarity in replication timing between
polytene and diploid cells is associated with the organization of the
Drosophila genome, PLoS One, in press.
43.Saura, A. O., Heino, T. I., and Sorsa, V. (1994)
Electron micrograph map of the Drosophila melanogaster polytene
chromosome 3R divisions 81 through 90, Hereditas, 121,
1-20.
44.Semeshin, V. F., Demakov, S. A., Shloma, V. V.,
Vatolina, T. Y., Gorchakov, A. A., and Zhimulev, I. F. (2008)
Interbands behave as decompacted autonomous units in Drosophila
melanogaster polytene chromosomes, Genetica, 132,
267-279.
45.Zhimulev, I. F., Belyaeva, E. S., Semeshin, V.
F., Koryakov, D. E., Demakov, S. A., Demakova, O. V., Pokholkova, G.
V., and Andreyeva, E. N. (2004) Polytene chromosomes: 70 years of
genetic research, Int. Rev. Cytol., 241, 203-275.
46.Andreenkov, O. V., Andreenkova, N. G., Volkova,
E. I., Georgiev, P. G., Goncharova, A. A., Pokholkova, G. V., and
Demakov, S. A. (2016) Ectopic attraction of the Chromator protein in
the UAS>DBD(GAL4) system as a method for investigation of insulator
proteins in polytene chromosomes from Drosophila melanogaster,
Tsitologiya, 58, 493-496.
47.Zhimulev, I. F., and Belyaeva, E. S. (1999)
Detailed description of puffing patterns in the salivary gland
chromosomes of normally developing larvae and prepupae of Drosophila
melanogaster, Dros. Inf. Serv., 82, 9-20.
48.Semeshin, V. F., Baricheva, E. M., Belyaeva, E.
S., and Zhimulev, I. F. (1985) Electron microscopical analysis of
Drosophila polytene chromosomes. II. Development of complex
puffs, Chromosoma, 91, 210-233.
49.Semeshin, V. F., Zhimulev, I. F., and Belyaeva,
E. S. (1979) Electron microscope autoradiographic study on
transcriptional activity of Drosophila melanogaster polytene
chromosomes, Chromosoma, 73, 163-177.
50.Vlassova, I. E., Umbetova, G. H., Zimmermann, V.
H., Alonso, C., Belyaeva, E. S., and Zhimulev, I. F. (1985)
Immunofluorescence localization of DNA:RNA hybrids in Drosophila
melanogaster polytene chromosomes, Chromosoma, 91,
251-258.
51.Semeshin, V. F., Shloma, V. V., and Zhimulev, I.
F. (2001) Formation and morphology of dark puffs in Drosophila
melanogaster polytene chromosomes, Hereditas, 134,
15-22.
52.Khoroshko, V. A., Zykova, T. Yu., Popova, O. O.,
and Zhimulev, I. F. (2018) Boundary structure of intercalary
heterochromatin bands in Drosophila melanogaster polytene
chromosomes, Dokl. Akad. Nauk, 5,1-4.
53.Zielke. T., Glotov, A., and Saumweber, H. (2015)
High-resolution in situ hybridization analysis on the
chromosomal interval 61C7-61C8 of Drosophila melanogaster
reveals interbands as open chromatin domains, Chromosoma,
125, 423-435.
54.Belyaeva, E. S., Demakov, S. A., Pokholkova, G.
V., Alekseyenko, A. A., Kolesnikova, T. D., and Zhimulev, I. F. (2006)
DNA underreplication in intercalary heterochromatin regions in polytene
chromosomes of Drosophila melanogaster correlates with the
formation of partial chromosomal aberrations and ectopic pairing,
Chromosoma, 115, 355-366.
55.Yarosh, W., and Spradling, A. C. (2014)
Incomplete replication generates somatic DNA alterations within
Drosophila polytene salivary gland cells, Genes Dev.,
28, 1840-1855.
56.Maqbool, S. B., Mehrotra, S., Kolpakas, A.,
Durden, C., Zhang, B., Zhong, H., and Calvi, B. R. (2010) Dampened
activity of E2F1-DP and Myb-MuvB transcription factors in
Drosophila endocycling cells, J. Cell Sci., 123,
4095-4106.
57.Ulianov, S. V., Tachibana-Konwalski, K., and
Razin, S. V. (2017) Single-cell Hi-C bridges microscopy and genome-wide
sequencing approaches to study 3D chromatin organization,
Bioessays, 39, doi: 10.1002/bies.201700104.
58.Semeshin, V. F., Artero, R., Perez-Alonso, M.,
and Shloma, V. V. (1998) Electron microscopic in situ
hybridization of digoxigenin-dUTP-labelled DNA probes with
Drosophila melanogaster polytene chromosomes, Chromosome
Res., 6, 405-410.
59.Kolesnikova, T. D., Semeshin, V. F., Andreyeva,
E. N., Zykov, I. A., Kokoza, E. B., Kalashnikova, D. A., Belyaeva, E.
S., and Zhimulev, I. F. (2011) Induced decondensation of
heterochromatin in Drosophila melanogaster polytene chromosomes
under condition of ectopic expression of the Suppressor of
underreplication gene, Fly (Austin), 5,
181-190.
60.Zhimulev, I. F., Belyaeva, E. S., Semeshin, V.
F., Shloma, V. V., Makunin, I. V., and Volkova, E. I. (2003)
Overexpression of the SuUR gene induces reversible modifications
at pericentric, telomeric and intercalary heterochromatin of
Drosophila melanogaster polytene chromosomes, J. Cell
Sci., 116, 169-176.
61.Boettiger, A. N., Bintu, B., Moffitt, J. R.,
Wang, S., Beliveau, B. J., Fudenberg, G., Imakaev, M., Mirny, L. A.,
Wu, C. T., and Zhuang, X. (2016) Super-resolution imaging reveals
distinct chromatin folding for different epigenetic states,
Nature, 529, 418-422.
62.Ghavi-Helm, Y., Klein, F. A., Pakozdi, T.,
Ciglar, L., Noordermeer, D., Huber, W., and Furlong, E. E. (2014)
Enhancer loops appear stable during development and are associated with
paused polymerase, Nature, 512, 96-100.
63.Engstrom, P. G., Ho Sui, S. J., Drivenes, O.,
Becker, T. S., and Lenhard, B. (2007) Genomic regulatory blocks
underlie extensive microsynteny conservation in insects, Genome
Res., 17, 1898-1908.
64.Harmston, N., Ing-Simmons, E., Tan, G., Perry,
M., Merkenschlager, M., and Lenhard, B. (2017) Topologically
associating domains are ancient features that coincide with Metazoan
clusters of extreme noncoding conservation, Nat. Commun.,
8, 441.
65.Makunin, I. V., Kolesnikova, T. D., and
Andreyenkova, N. G. (2014) Underreplicated regions in Drosophila
melanogaster are enriched with fast-evolving genes and highly
conserved noncoding sequences, Genome Biol. Evol., 6,
2050-2060.
66.Von Grotthuss, M., Ashburner, M., and Ranz, J. M.
(2010) Fragile regions and not functional constraints predominate in
shaping gene organization in the genus Drosophila, Genome
Res., 20, 1084-1096.