REVIEW: Role of Nuclear Lamina in Gene Repression and Maintenance of Chromosome Architecture in the Nucleus
Y. Y. Shevelyov1* and S. V. Ulianov2,3
1Institute of Molecular Genetics, Russian Academy of Sciences, 123182 Moscow, Russia; E-mail: shevelev@img.ras.ru2Institute of Gene Biology, Russian Academy of Sciences, 119334 Moscow, Russia
3Lomonosov Moscow State University, Faculty of Biology, 119192 Moscow, Russia
* To whom correspondence should be addressed.
Received November 20, 2017; Revision received December 5, 2017
Nuclear lamina is a protein meshwork composed of lamins and lamin-associated proteins that lines the nuclear envelope from the inside and forms repressive transcription compartment. The review presents current data on the contribution of nuclear lamina to the repression of genes located in this compartment and on the mechanisms of chromatin attachment to the nuclear envelope.
KEY WORDS: nuclear lamina, lamina-associated domain, nuclear envelope, nuclear periphery, repressionDOI: 10.1134/S0006297918040077
Abbreviations: DamID, DNA adenine methyltransferase identification; FISH, fluorescent in situ hybridization; LAD, lamina-associated domain; LBR, lamin B receptor.
NUCLEAR ENVELOPE ORGANIZATION
Nuclear envelope delimits the nucleus from the cytoplasm. It is composed of two lipid bilayer membranes separated by the perinuclear space. The envelope is pierced by nuclear pores – large nucleoporin protein complexes, through which macromolecules are exchanged between the nucleus and the cytoplasm. From the nuclear side, the envelope is lined with a protein meshwork of lamins (type V intermediate filaments classified as A- and B-type lamins depending on whether they are expressed in all cells or only in certain tissues) and lamin-associated proteins anchored in the nuclear membrane [1, 2]. The number of lamin-encoding genes has increased during the evolution. Thus, Caenorhabditis elegans has a single gene coding for lamin (lmn-1), Drosophila has two genes (lamC and Dm0 coding for A- and B-type lamins, respectively), and mammals have three genes (Lmnb1, Lmnb2, and LmnA). Unlike B-type lamins, mammalian A-type lamins (but not those of Drosophila) form a meshwork of filaments not only at the surface, but also inside the nucleus. The nuclear lamina is found in all multicellular animals and has both structural and regulatory functions: it keeps the shape and mechanical stability of the nucleus, provides interactions between the nucleus and the cytoskeleton, and participates in the regulation of transcription, replication, and genome stability. Defects in the nuclear lamina in humans cause disorders with various clinical manifestations collectively named laminopathies [3].
HETEROCHROMATIN ORGANIZATION
Eukaryotic chromosomes are composed of decondensed euchromatin that includes the majority of actively expressed genes and a more tightly packed constitutive heterochromatin that is located in the pericentromeric and subtelomeric regions of chromosomes and contains highly and moderately repetitive DNA sequences. Also, euchromatin chromosome shoulders contain regions of facultative, or intercalary, heterochromatin that encompass mostly silent genes and copies of mobile genetic elements [4, 5]. Microscopy studies showed that in the majority of mammalian cells, constitutive and facultative heterochromatin is located at the nuclear periphery and around the nucleoli [6]. Detection of the so-called inverted nuclear architecture in the retina cells of mammals with nocturnal vision became a sensational discovery. In these cells, multiple chromocenters (constitutive heterochromatin foci) normally located at the nuclear periphery merge in a single chromocenter in the center of the nucleus, while euchromatin resides close to the nuclear envelope [6]. It was shown that the lack of two components of the nuclear lamina – the A-type lamin and the integral protein of a nuclear membrane lamin B receptor (LBR) – is responsible for the conversion of the normal architecture into the inverted one. In presence of any of these proteins, both constitutive and facultative heterochromatin in mammalian cells is located mostly near the nuclear envelope [7].
The idea that heterochromatin is attached to the nuclear envelope and not simply located in its vicinity and that this attachment defines the ordered positioning of chromosomes in the interphase nucleus was proposed a long time ago [8]. Early studies on Drosophila salivary gland cells found a number of polytene chromosome regions to be in a visible contact with the nuclear envelope in the majority of analyzed nuclei [9]. These regions almost completely coincided with the intercalary heterochromatin [10] characterized by late replication, underreplication in the polytene chromosomes, constrictions, frequent breaks, frequent ectopic contacts, and some other properties. The sites of chromosome–envelope contact varied in different types of cells [11], possibly reflecting their dependence on the activity of genes. Fluorescent in situ hybridization (FISH) showed that in cells of early Drosophila embryos, more than 75 chromosomal regions are in frequent contact with the nuclear envelope [12].
During the last decade, the so-called lamina-associated domains (LADs) – extended chromosomal regions contacting and, perhaps, attached to the nuclear lamina – have been identified in Drosophila, mammals, and C. elegans using DamID (DNA adenine methyltransferase identification) [13] and chromatin immunoprecipitation methods [14-26]. LADs occupy ~40% of the genome and contain mainly genes that are silent in this particular type of cells. They are the late-replicating genome regions, poor in acetylated histones. In mammals and C. elegans, LADS are enriched with histone H3 di-/trimethylated at lysine 9 (H3K9me2/3) or trimethylated at lysine 27 (H3K27me3), generally associated with transcription repression [15, 16, 18, 26, 27]. LADs and chromatin domains associated with the Polycomb repressor protein (Pc) supposedly correspond to the intercalary heterochromatin found in Drosophila polytene chromosomes [28].
In addition to multiple sites of contact/attachment to the nuclear lamina, yeast, Drosophila, and mammalian chromosomes are bound to the nuclear pore complexes penetrating the nuclear envelope [29-44]. In the Drosophila genome, thousands short (~2 kb) regions of contact/attachment of chromosomes to the nuclear pores were identified by the DamID method. Moreover, they were found in both active and inactive chromatin [39], which suggests sequence-specific recognition of these regions by yet unidentified components of the nuclear pore complexes. It should be mentioned that in multicellular organisms, nucleoporins (components of the nuclear pore complexes) bind to chromatin not only on the nuclear envelope but also in the nucleoplasm; moreover, in the latter case, they play a role of transcription coactivators [39-41, 43-46].
The data confirming the hypothesis that many chromosomal regions not just contact but attach to the nuclear envelope have been obtained rather recently in experiments on the localization of individual chromosomal loci after depletion of lamin or other components of the nuclear lamina in Drosophila, C. elegans, and mammalian cells using the FISH method. The loci studied lost their preferential peripheral location and moved toward the nuclear interior [47-51]. It should be noted that lamin depletion has an impact on the integrity of the whole nuclear lamina, affecting location of many integral proteins of the nuclear membrane and the nuclear pore complex [52-54]. Therefore, it is unlikely that the lamin is responsible for the attachment of chromatin to the nuclear envelope. Other components of the nuclear lamina or the nuclear pore complexes might be involved in this process as well. For example, after the depletion of one of the nucleoporins (Nup153) in Drosophila SL-2 cells, a number of X-chromosome fragments enriched with nucleoporin association were localized further from the nuclear envelope than in the control cells [40], while in mouse embryonic stem cells, several loci located at the nuclear periphery moved towards the nuclear interior [42], thereby indicating that nuclear pore complexes are involved in the maintenance of chromosome architecture in the nucleus.
MECHANISMS OF LAD ATTACHMENT TO THE NUCLEAR ENVELOPE
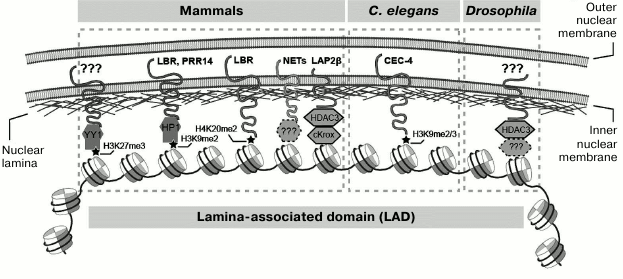
Despite the long history of LAD studies, it still remains unclear how LADs attach to the nuclear envelope. Theoretically, nuclear lamina proteins can bind either DNA or modified histones in LADs (or both). Both types of interactions were found in mammalian cells. For example, the repressor protein cKrox, which binds to (GA)n tracks in DNA, forms a complex with the HDAC3 histone deacetylase and the integral nuclear membrane protein Lap2β that participates in the attachment of several LADs to the nuclear envelope in mouse fibroblasts [49]. Moreover, proteomic analysis revealed the presence of HDAC3 in complexes with lamina proteins in mammalian cells [55, 56]. In mouse embryonic stem cells, HDAC3 (independently of its catalytic activity and together with Lap2β) is involved in the maintenance of peripheral localization of several loci (Fig. 1) [57]. These data illustrate the mechanism of sequence-specific attachment. There are also examples of interactions based on the recognition of a certain type of inactive chromatin. As mammalian LADs are enriched with H3K9me2/3 throughout their entire length [15, 17, 57-59], the presence of this modification being essential for maintaining their peripheral location [59-62]. Besides, the borders of mammalian LADs contain the H3K27me3 mark [15] that participates in the attachment of LADs to the nuclear envelope [62] with the involvement of the DNA-binding protein YY1 (Fig. 1) [61]. Moreover, in C. elegans, H3K9me2/3 histone modification is necessary for the location of the transgene carrying an extended heterochromatin repeat at the nuclear periphery [63, 64].
Fig. 1. Known mechanisms of chromatin attachment to the nuclear envelope in mammals, C. elegans, and Drosophila.
Interestingly, Drosophila LADs, which have been till now mapped only in Kc167 cells [18], are not enriched with either H3K9me2/3 or the main heterochromatin protein HP1a [65]. Therefore, if in other types of Drosophila cells the mechanism of H3K9me2/3-modified chromatin binding with the nuclear envelope exists, it is obviously not the only one. In Kc167 cells, LADs overlap with approximately 40% of Pc domains; hence, these LADs are enriched with H3K27me3 [18]. However, the role of this modification in the chromatin attachment to the nuclear lamina in Drosophila has not been studied. The depletion of HDAC3 in Drosophila S2 cells led to the removal of the 60D1 locus from the nuclear envelope, indicating that the sequence-specific recognition involving HDAC3, similar to that in mammals, also exist in Drosophila [66].
Proteins that presumably introduce repressive histone modifications into LAD chromatin include histone methyltransferase G9a (MET-2 in C. elegans) and histone deacetylase HDAC3, since mutations or knockdown of the corresponding genes result in the loss of interactions between LADs and nuclear envelope in Drosophila [66], mammals [49, 57, 59, 60, 62], and C. elegans [63].
Which proteins of the nuclear lamina are responsible for the chromatin attachment to the nuclear envelope? In vitro experiments showed that lamins are able to bind DNA directly, but they can also bind chromatin and histones H2A and H2B [67-72]. However, this type of interactions does not explain specific binding of inactive chromatin to the nuclear lamina. It is possible that other components of the nuclear lamina also bind chromatin [73, 74]. For example, it was shown that LBR binds HP1 [75] and thus can probably interact with H3K9me2/3-modified histones in mammalian LADs (Fig. 1). However, LBR and HP1 bind each other indirectly, through the histone H3/H4 dimer; acetylation of this histone prevents interaction between these two proteins [76, 77]. Moreover, LBR can bind the H4K20me2 mark of the peripheral heterochromatin both in vitro and in vivo (Fig. 1) [78]. It is important to note that in a number of studies, LBR was shown to be indispensable for the maintenance of the peripheral location of pericentromeric heterochromatin in mammals [7, 79-81]. Taken together, these data suggest that LBR is directly involved in the attachment of the constitutive heterochromatin and, probably, LADs to the nuclear envelope in mammals.
Another lamina-associated protein, Lap2β, which belongs to the LEM-domain protein family, is also involved in the attachment of at least several LADs to the nuclear envelope in mammals [49]. The PRR14 protein associated with the nuclear lamina in mammals was found to bind H3K9-methylated heterochromatin (presumably via interaction between PRR14 and HP1) and to attach it to the nuclear envelope (Fig. 1) [51]. The C. elegans protein CEC-4, that is localized at the nuclear periphery and contains a chromodomain (similarly to HP1), is able to anchor the H3K9-methylated chromatin to the nuclear envelope [64]. Finally, screening of human cells revealed several new nuclear envelope transmembrane proteins, whose expression increased and depletion, to the contrary, reduced the portion of cells with peripheral location of certain chromosomes [82]. Further analysis showed that some of these proteins specifically retain certain gene loci at the periphery of the nucleus, which provides gene repression necessary for tissue-specific cell differentiation [83]. Unfortunately, there are still no data on the factors involved in the attachment of LADs and/or constitutive heterochromatin to the nuclear envelope in Drosophila.
CHROMATIN ATTACHMENT TO THE NUCLEAR ENVELOPE IN INDIVIDUAL
CELLS
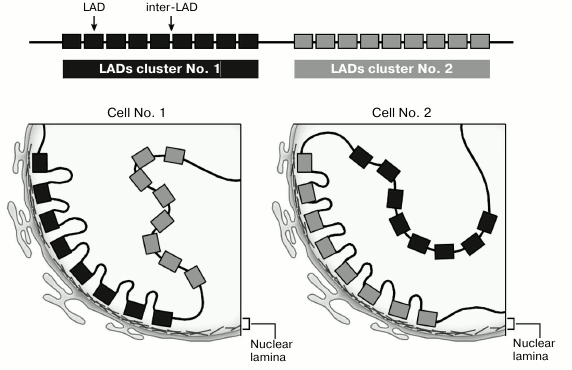
Mapping of LADs by DamID or chromatin immunoprecipitation gives an averaged distribution pattern of the sites of chromosome attachment to the nuclear lamina in a cell population. At the same time, numerous data obtained by FISH showed considerable variability in the positions of loci relative to the nuclear envelope in individual cells of the same homogeneous population. Therefore, studies of the sites of chromatin attachment to the nuclear lamina in individual cells are of considerable interest. Recently, two new approaches have been developed for this purpose. The first one consisted in the binding of the EGFP–DpnI hybrid protein, expressed in human cells, to the chromosome fragments, in which adenine in the GATC sequences was methylated with E. coli Dam methylase fused to B-type lamin. The authors [59] found that in living cells, EGFP-labeled LADs containing methylated adenine were in contact with the nuclear lamina [59]. This gave a possibility to trace nuclear localization of LADs at different stages of the cell cycle. It was found that in individual cells, no more than one third of LADs were located at the nuclear periphery. During mitotic division, LADs were randomly redistributed between the nuclear periphery and interior – more than a half of chromosomal regions bound to the nuclear lamina before mitosis were moved away from it [59]. Similar conclusions were made using the DamID analysis of lamin in individual human cells [84]. Only a small number of LADs mapped as nuclear periphery-associated in cell populations are in contact with the nuclear lamina in every single cell. The authors showed that only ~15% LADs are in contact with lamina in every cell. It was also demonstrated that in each individual cell, long chromosomal regions (6-8 Mb) can be synchronously located either near to or far from the nuclear lamina [84]. Therefore, long regions of chromosome fiber (containing both LADs and inter-LAD sequences) can be localized near the nuclear envelope in a portion of a cell population, while in the other portion of the same population, the same long fragments can be located closer to the center of the nucleus. When a chromosome fiber is located close to the nuclear envelope, its regions of inactive chromatin (LADs) are attached to the lamina, while the regions of active chromatin (inter-LADs) are looping out to the nucleoplasm (Fig. 2) [85].
Fig. 2. LADs can compete for binding to the nuclear envelope.
LADs that move toward the nuclear interior during mitosis no longer experience the repressive influence of the nuclear lamina (see the next section). Therefore, although it is possible that the repressed state of chromatin can be transferred epigenetically from one cell generation to another, one could expect some chromatin derepression during divisions. Indeed, such repression weakening was demonstrated in [59]; however, it is insufficient to trigger the locus into the actively transcribed state.
Why only a portion of inactive chromatin, potentially capable of binding to the nuclear envelope, is located at the nuclear periphery? A possible explanation for this phenomenon was suggested in [84]. Human KBM7 cells can spontaneously pass from the haploid state into the diploid state. The authors performed DamID analysis in individual cells (both haploid and diploid) and found a significant decrease in the Dam-lamin methylation per one chromosome dose in diploid cells compared to the haploid ones. Based on these results, the authors suggested that there might be a competition between different LADs for a limited number of binding sites at the nuclear lamina [84]. If this hypothesis is correct, the limited number of sites for chromatin binding on the nuclear envelope can explain a small portion of LADs forming contacts with lamina in individual cells (Fig. 2). However, this problem requires additional studies.
NUCLER LAMINA INDUCES TRANSCRIPTIONAL REPRESSION OF contacting
genes
The hypothesis that nuclear lamina is a compartment poorly compatible with transcription is supported by numerous experimental data. FISH analysis of multiple loci in different types of cells in different organisms shows strong correlation between the expression or silencing of genes in certain type of cells and their location far from or close to the nuclear envelope, respectively [47, 50, 57, 61, 86-98]. Moreover, in mammals, chromosomal regions attached to the nuclear envelope vary depending on the cell type. During cell differentiation accompanied by activation of tissue-specific genes, DamID-identified contacts between the nuclear lamina and promoter regions of activated genes (and in many cases, the entire genes) are considerably weakened or completely lost [17]. Finally, artificial tethering of the transcriptional activator to the inactive locus results in its relocation from the nuclear envelope towards the nuclear interior [24, 93, 99-101]. Interestingly, such relocation requires chromatin remodeling and decondensation, but not the activation of its transcription [101].
However, the location of silent genes in LADs can either be a consequence of the nuclear lamina ability to suppress transcription or reflects the situation when only inactive chromatin can be attached to the lamina. In order to study relationships between the transcription repression and chromatin attachment to the lamina, several experiments on the artificial tethering of a chromosomal region to the nuclear lamina using the LacI/lacO system were carried out in mammalian and Drosophila cells [102-104]. Suppression of the expression of reporter gene, as well as of several endogenous genes located in the vicinity of the site of chromatin attachment to the lamina, were observed that were accompanied by histone deacetylation. However, when stronger promoter was used, tethering to the nuclear lamina had no effect on the reporter gene expression [105]. Nevertheless, these experiments showed a potential ability of the nuclear lamina to suppress the transcription of genes with low and intermediate levels of expression.
Additional arguments in favor of suppression of gene transcription by the nuclear lamina were obtained by analysis of expression of reporter genes integrated into different genomic regions. Such analysis showed a systematic difference in the levels of reporter gene expression depending on the gene location in either active or inactive chromatin domains [106-108]. For example, in mouse embryonic stem cells, the expression levels of a reporter gene integrated into LADs were on average 5-6 times lower than the expression levels of the same gene integrated into the inter-LADs, with extreme values demonstrating three order of magnitude difference [108].
Studies of gene expression with the knocked-down or knocked-out B-type lamin gene theoretically could reveal the direct effect of the nuclear lamina on the transcription of genes located in LADs. Disruption of the nuclear lamina in C. elegans and Drosophila cells resulted in the derepression of transcription of genes located in LADs [47, 48, 50, 63]. It should be mentioned, however, that genes in LADs are silent not because of their contact with the lamina. Indeed, the ablation of lamin Dm0 in Drosophila S2 cells led to the detachment of these genes from the nuclear envelope, accompanied with only a 1.5- to 3-fold increase of the number of their transcripts still remaining sporadic in the cells [47].
Full transcriptome analysis showed that knock-out of both B-type lamin genes (Lmnb1 and Lmnb2) in mouse embryonic stem cells (ESCs) or trophectoderm cells changed the transcription levels of only a very small number of genes independently of their location in LADs [109, 110]. Therefore, the absence of lamin did not activate transcription of genes in LADs, at least in mouse ESCs. Similarly, depletion of CEC-4 in C. elegans embryos led to the repositioning of H3K9-methylated chromatin from the nuclear periphery towards its interior but was not accompanied by any significant activation of expression of genes located in these regions [64]. It should be kept in mind that transcriptome analysis using microarray or RNA-seq methods does not give full information about the expression levels of all genes in the genome, because these methods are not sensitive enough to detect low-copy transcripts. As a result, about a half of all genes that are not expressed (or poorly expressed) in studied cells [111-113] and are potentially derepressed upon the depletion of lamin are not identified by the researchers.
Another question is whether the actively expressed genes can be localized in LADs and partially repressed because of contacts with the nuclear lamina? DamID analysis showed than the majority of expressed genes [15-18, 23], or at least their promoter regions [22, 114], do not form contacts with the nuclear lamina and, thereby, possibly escape its repressor effect. Even if promoters of actively expressed genes are not bound to the lamina, the bodies of these genes, especially those containing extended introns, can be in contact with the lamina [22, 62, 114]. However, the question whether all the active promoters or only the majority of them avoid contacts with the nuclear lamina is still open. Identification of several promoters of actively expressed genes in LADs can be explained by the errors in delimitation of the LADs borders. We cannot exclude, however, that some promoters of active genes can be located in LADs. The average difference in the expression levels of the reporter gene inserted in LADs and inter-LADs in mouse embryonic stem cells was not altered after the induction of its transcription with doxycycline [108], which indicates that transcription not only of silent, but also of the expressed reporter gene in LAD was partially repressed. This suggests that transcription of genes, whose promoters remain in contact with the nuclear lamina (if such promoters exist), can also be partially repressed, at least, in mammalian cells.
Then why does the knock-out of the Lmnb1 gene in mice cause multiple defects in different organs (in particular, brain) and animal death at early stage of development, as well as knock-downs of the lamin Dm0 gene in Drosophila and the lmn-1 gene in C. elegans lead to embryonic lethality [109, 115-119]? Are these defects caused by the derepression of genes in LADs or by other effects related to the nuclear lamina disruption? For example, conditional knock-out of the Lmnb1 gene in mouse olfactory neurons causes significant changes in the transcriptome and properties of these cells [54]; however, the authors explained these changes not by the disruption of contacts between genes and the nuclear envelope, but by abnormal functioning of the nuclear pores caused by their clustering on the nuclear membrane following lamin depletion [54, 120]. Interpretation of these and other results is also complicated by the fact that LADs in mouse and Drosophila organs and tissues have not been mapped yet.
Summarizing, the available data demonstrate that nuclear lamina does not play a significant role in the transcription repression of genes located in LADs; this repression affects primarily the silent genes (at least, in embryonic cells). Chromatin in LADs is more compact, compared to that in inter-LADs [62, 121, 122]. This can be, at least partially, due to the contacts with lamina, since depletion of lamin leads to chromatin decompaction in Drosophila [66]. The more compact state of chromatin in LADs reduces the probability of nonspecific recruitment of transcription factors to the promoter regions of genes located in LADs, thereby, suppressing their low-level background transcription.
QUESTIONS TO BE ANSWERED
Even with the use of modern techniques, such as DamID in single cells and super-resolution microscopy, in the studies of chromosome architecture, many questions still remain open. Despite significant progress in the understanding of mechanisms of chromatin attachment to the nuclear envelope, not all factors involved in this process have been identified. It is still unclear, which protein complexes bound to the nuclear lamina are involved in the repression/compaction of chromatin that is in contact with them and why the remodeled chromatin loses this connection. The impact of chromatin binding to the nuclear envelope on the overall chromosome architecture in the nucleus is also poorly understood. It was shown that this binding is important in human cells, since depletion of lamin A/C results in the relocation of the majority of LADs away from the nuclear envelope and in considerable perturbations of chromosomal territories, in particular, in the increased intermingling of inactive and active chromatin [62]. These disturbances can change the number of contacts between active loci, which might explain alterations in the expression of genes located outside LADs. It is especially interesting, as the reasons of why the absence of lamin causes serious problems in certain tissue and organs are still obscure. Finally, it is not known at which stages of cell cycle the repression occurs and how it is transferred through cell generations. Elucidation of these and other questions is necessary for better understanding of fundamental mechanisms of eukaryotic cell functioning, in particular, of aging and development of laminopathies in humans.
Acknowledgments
The work was supported by the Russian Science Foundation (grant No. 16-14-10081).
REFERENCES
1.Schirmer, E. C., and Foisner, R. (2007) Proteins
that associate with lamins: many faces, many functions, Exp.
Cell Res., 313, 2167-2179.
2.Prokocimer, M., Davidovich, M., Nissim-Rafinia, M.,
Wiesel-Motiuk, N., Bar, D. Z., Barkan, R., Meshorer, E., and Gruenbaum,
Y. (2009) Nuclear lamins: key regulators of nuclear structure and
activities, J. Cell Mol. Med., 13,
1059-1085.
3.Worman, H. J., Ostlund, C., and Wang, Y. (2010)
Diseases of the nuclear envelope, Cold Spring Harb.
Perspect. Biol., 2, a000760.
4.Trojer, P., and Reinberg, D. (2007) Facultative
heterochromatin: is there a distinctive molecular signature?
Mol. Cell, 28, 1-13.
5.Cabianca, D. S., and Gasser, S. M. (2016) Spatial
segregation of heterochromatin: uncovering functionality in a
multicellular organism, Nucleus, 7, 301-307.
6.Solovei, I., Kreysing, M., Lanctot, C., Kosem, S.,
Peichl, L., Cremer, T., Guck, J., and Joffe, B. (2009) Nuclear
architecture of rod photoreceptor cells adapts to vision in mammalian
evolution, Cell, 137, 356-368.
7.Solovei, I., Wang, A. S., Thanisch, K., Schmidt, C.
S., Krebs, S., Zwerger, M., Cohen, T. V., Devys, D., Foisner, R.,
Peichl, L., Herrmann, H., Blum, H., Engelkamp, D., Stewart, C. L.,
Leonhardt, H., and Joffe, B. (2013) LBR and lamin A/C sequentially
tether peripheral heterochromatin and inversely regulate
differentiation, Cell, 152, 584-598.
8.Comings, D. E. (1968) The rationale for an ordered
arrangement of chromatin in the interphase nucleus, Am.
J. Hum. Genet., 20, 440-460.
9.Hochstrasser, M., Mathog, D., Gruenbaum, Y.,
Saumweber, H., and Sedat, J. W. (1986) Spatial organization of
chromosomes in the salivary gland nuclei of Drosophila
melanogaster, J. Cell Biol., 102, 112-123.
10.Zhimulev, I. F., Semeshin, V. F., Kulichkov, V.
A., and Belyaeva, E. S. (1982) Intercalary heterochromatin in
Drosophila. I. Localization and general characteristics,
Chromosoma, 87, 197-228.
11.Hochstrasser, M., and Sedat, J. W. (1987)
Three-dimensional organization of Drosophila melanogaster
interphase nuclei. II. Chromosome spatial organization and gene
regulation, J. Cell Biol., 104, 1471-1483.
12.Marshall, W. F., Dernburg, A. F., Harmon, B.,
Agard, D. A., and Sedat, J. W. (1996) Specific interactions of
chromatin with the nuclear envelope: positional determination within
the nucleus in Drosophila melanogaster, Mol. Biol.
Cell, 7, 825-842.
13.Van Steensel, B., and Henikoff, S. (2000)
Identification of in vivo DNA targets of chromatin proteins
using tethered Dam methyltransferase, Nat. Biotechnol.,
18, 424-428.
14.Pickersgill, H., Kalverda, B., de Wit, E.,
Talhout, W., Fornerod, M., and van Steensel, B. (2006) Characterization
of the Drosophila melanogaster genome at the nuclear lamina,
Nat. Genet., 38, 1005-1014.
15.Guelen, L., Pagie, L., Brasset, E., Meuleman, W.,
Faza, M. B., Talhout, W., Eussen, B. H., de Klein, A., Wessels, L., de
Laat, W., and van Steensel, B. (2008) Domain organization of human
chromosomes revealed by mapping of nuclear lamina interactions,
Nature, 453, 948-951.
16.Ikegami, K., Egelhofer, T. A., Strome, S., and
Lieb, J. D. (2010) Caenorhabditis elegans chromosome arms are
anchored to the nuclear membrane via discontinuous association with
LEM-2, Genome Biol., 11, R120.
17.Peric-Hupkes, D., Meuleman, W., Pagie, L.,
Bruggeman, S. W., Solovei, I., Brugman, W., Graf, S., Flicek, P.,
Kerkhoven, R. M., van Lohuizen, M., Reinders, M., Wessels, L., and van
Steensel, B. (2010) Molecular maps of the reorganization of
genome–nuclear lamina interactions during differentiation,
Mol. Cell, 38, 603-613.
18.Van Bemmel, J. G., Pagie, L., Braunschweig, U.,
Brugman, W., Meuleman, W., Kerkhoven, R. M., and van Steensel, B.
(2010) The insulator protein SU(HW) fine-tunes nuclear lamina
interactions of the Drosophila genome, PLoS One,
5, e15013.
19.Handoko, L., Xu, H., Li, G., Ngan, C. Y., Chew,
E., Schnapp, M., Lee, C. W., Ye, C., Ping, J. L., Mulawadi, F., Wong,
E., Sheng, J., Zhang, Y., Poh, T., Chan, C. S., Kunarso, G., Shahab,
A., Bourque, G., Cacheux-Rataboul, V., Sung, W. K., Ruan, Y., and Wei,
C. L. (2011) CTCF-mediated functional chromatin interactome in
pluripotent cells, Nat. Genet., 43, 630-638.
20.Meuleman, W., Peric-Hupkes, D., Kind, J.,
Beaudry, J. B., Pagie, L., Kellis, M., Reinders, M., Wessels, L., and
van Steensel, B. (2013) Constitutive nuclear lamina–genome
interactions are highly conserved and associated with A/T-rich
sequence, Genome Res., 23, 270-280.
21.Sadaie, M., Salama, R., Carroll, T., Tomimatsu,
K., Chandra, T., Young, A. R., Narita, M., Perez-Mancera, P. A.,
Bennett, D. C., Chong, H., Kimura, H., and Narita, M. (2013)
Redistribution of the lamin B1 genomic binding profile affects
rearrangement of heterochromatic domains and SAHF formation during
senescence, Genes Dev., 27, 1800-1808.
22.Wu, F., and Yao, J. (2013) Spatial
compartmentalization at the nuclear periphery characterized by
genome-wide mapping, BMC Genomics, 14, 591.
23.Gonzalez-Aguilera, C., Ikegami, K., Ayuso, C., de
Luis, A., Iniguez, M., Cabello, J., Lieb, J. D., and Askjaer, P. (2014)
Genome-wide analysis links emerin to neuromuscular junction activity in
Caenorhabditis elegans, Genome Biol., 15, R21.
24.Kind, J., and van Steensel, B. (2014) Stochastic
genome–nuclear lamina interactions: modulating roles of lamin A
and BAF, Nucleus, 5, 124-130.
25.Letourneau, A., Santoni, F. A., Bonilla, X.,
Sailani, M. R., Gonzalez, D., Kind, J., Chevalier, C., Thurman, R.,
Sandstrom, R. S., Hibaoui, Y., Garieri, M., Popadin, K., Falconnet, E.,
Gagnebin, M., Gehrig, C., Vannier, A., Guipponi, M., Farinelli, L.,
Robyr, D., Migliavacca, E., Borel, C., Deutsch, S., Feki, A.,
Stamatoyannopoulos, J. A., Herault, Y., van Steensel, B., Guigo, R.,
and Antonarakis, S. E. (2014) Domains of genome-wide gene expression
dysregulation in Down’s syndrome, Nature, 508,
345-350.
26.Ragoczy, T., Telling, A., Scalzo, D., Kooperberg,
C., and Groudine, M. (2014) Functional redundancy in the nuclear
compartmentalization of the late-replicating genome, Nucleus,
5, 626-635.
27.Ryba, T., Hiratani, I., Lu, J., Itoh, M., Kulik,
M., Zhang, J., Schulz, T. C., Robins, A. J., Dalton, S., and Gilbert,
D. M. (2010) Evolutionarily conserved replication timing profiles
predict long-range chromatin interactions and distinguish closely
related cell types, Genome Res., 20, 761-770.
28.Zhimulev, I. F., Belyaeva, E. S., Vatolina, T.
Y., and Demakov, S. A. (2012) Banding patterns in Drosophila
melanogaster polytene chromosomes correlate with DNA-binding
protein occupancy, Bioessays, 34, 498-508.
29.Brickner, J. H., and Walter, P. (2004) Gene
recruitment of the activated INO1 locus to the nuclear membrane,
PLoS Biol., 2, e342.
30.Casolari, J. M., Brown, C. R., Komili, S., West,
J., Hieronymus, H., and Silver, P. A. (2004) Genome-wide localization
of the nuclear transport machinery couples transcriptional status and
nuclear organization, Cell, 117, 427-439.
31.Cabal, G. G., Genovesio, A., Rodriguez-Navarro,
S., Zimmer, C., Gadal, O., Lesne, A., Buc, H., Feuerbach-Fournier, F.,
Olivo-Marin, J.-C., Hurt, E. C., and Nehrbass, U. (2006) SAGA
interacting factors confine sub-diffusion of transcribed genes to the
nuclear envelope, Nature, 441, 770-773.
32.Dieppois, G., Iglesias, N., and Stutz, F. (2006)
Cotranscriptional recruitment to the mRNA export receptor Mex67p
contributes to nuclear pore anchoring of activated genes, Mol.
Cell. Biol., 26, 7858-7870.
33.Schmid, M., Arib, G., Laemmli, C., Nishikawa, J.,
Durussel, T., and Laemmli, U. K. (2006) Nup-PI: the
nucleopore–promoter interaction of genes in yeast, Mol.
Cell, 21, 379-391.
34.Taddei, A., Houwe, G. V., Hediger, F., Kalck, V.,
Cubizolles, F., Schober, H., and Gasser, S. M. (2006) Nuclear pore
association confers optimal expression levels for an inducible yeast
gene, Nature, 441, 774-778.
35.Brown, C. R., Kennedy, C. J., Delmar, V. A.,
Forbes, D. J., and Silver, P. A. (2008) Global histone acetylation
induces functional genomic reorganization at mammalian nuclear pore
complexes, Genes Dev., 22, 627-639.
36.Kohler, A., Schneider, M., Cabal, G. G.,
Nehrbass, U., and Hurt, E. (2008) Yeast ataxin-7 links histone
deubiquitination with gene gating and mRNA export, Nature Cell
Biol., 10, 707-715.
37.Rougemaille, M., Dieppois, G.,
Kisseleva-Romanova, E., Gudipati, R. K., Lemoine, S., Blugeon, C.,
Boulay, J., Jensen, T. H., Stutz, F., Devaux, F., and Libri, D. (2008)
THO/Sub2p functions to coordinate 3′-end processing with
gene–nuclear pore association, Cell, 135,
308-321.
38.Ahmed, S., Brickner, D. G., Light, W. H.,
Cajigas, I., McDonough, M., Froyshteter, A. B., Volpe, T., and
Brickner, J. H. (2010) DNA zip codes control an ancient mechanism for
gene targeting to the nuclear periphery, Nat. Cell Biol.,
12, 111-118.
39.Kalverda, B., Pickersgill, H., Shloma, V. V., and
Fornerod, M. (2010) Nucleoporins directly stimulate expression of
developmental and cell-cycle genes inside the nucleoplasm, Cell,
140, 360-371.
40.Vaquerizas, J. M., Suyama, R., Kind, J., Miura,
K., Luscombe, N. M., and Akhtar, A. (2010) Nuclear pore proteins Nup153
and Megator define transcriptionally active regions in the
Drosophila genome, PLoS Genet., 6, e1000846.
41.Liang, Y., Franks, T. M., Marchetto, M. C., Gage,
F. H., and Hetzer, M. W. (2013) Dynamic association of NUP98 with the
human genome, PLoS Genet., 9, e1003308.
42.Jacinto, F. V., Benner, C., and Hetzer, M. W.
(2015) The nucleoporin Nup153 regulates embryonic stem cell
pluripotency through gene silencing, Genes Dev., 29,
1224-1238.
43.Ibarra, A., Benner, C., Tyagi, S., Cool, J., and
Hetzer, M. W. (2016) Nucleoporin-mediated regulation of cell identity
genes, Genes Dev., 30, 2253-2258.
44.Pascual-Garcia, P., Debo, B., Aleman, J. R.,
Talamas, J. A., Lan, Y., Nguyen, N. H., Won, K. J., and Capelson, M.
(2017) Metazoan nuclear pores provide a scaffold for poised genes and
mediate induced enhancer–promoter contacts, Mol.
Cell, 66, 63-76.
45.Capelson, M., Liang, Y., Schulte, R., Mair, W.,
Wagner, U., and Hetzer, M. W. (2010) Chromatin-bound nuclear pore
components regulate gene expression in higher eukaryotes, Cell,
140, 372-383.
46.Pascual-Garcia, P., Jeong, J., and Capelson, M.
(2014) Nucleoporin Nup98 associates with Trx/MLL and NSL
histone-modifying complexes and regulates Hox gene expression,
Cell Rep., 9, 433-442.
47.Shevelyov, Y. Y., Lavrov, S. A., Mikhaylova, L.
M., Nurminsky, I. D., Kulathinal, R. J., Egorova, K. S., Rozovsky, Y.
M., and Nurminsky, D. I. (2009) The B-type lamin is required for
somatic repression of testis-specific gene clusters, Proc.
Natl. Acad. Sci. USA, 106,
3282-3287.
48.Mattout, A., Pike, B. L., Towbin, B. D., Bank, E.
M., Gonzalez-Sandoval, A., Stadler, M. B., Meister, P., Gruenbaum, Y.,
and Gasser, S. M. (2011) An EDMD mutation in C. elegans lamin
blocks muscle-specific gene relocation and compromises muscle
integrity, Curr. Biol., 21, 1603-1614.
49.Zullo, J. M., Demarco, I. A., Pique-Regi, R.,
Gaffney, D. J., Epstein, C. B., Spooner, C. J., Luperchio, T. R.,
Bernstein, B. E., Pritchard, J. K., Reddy, K. L., and Singh, H. (2012)
DNA sequence-dependent compartmentalization and silencing of chromatin
at the nuclear lamina, Cell, 149, 1474-1487.
50.Kohwi, M., Lupton, J. R., Lai, S. L., Miller, M.
R., and Doe, C. Q. (2013) Developmentally regulated subnuclear genome
reorganization restricts neural progenitor competence in
Drosophila, Cell, 152, 97-108.
51.Poleshko, A., Mansfield, K. M., Burlingame, C.
C., Andrake, M. D., Shah, N. R., and Katz, R. A. (2013) The human
protein PRR14 tethers heterochromatin to the nuclear lamina during
interphase and mitotic exit, Cell Rep., 5, 292-301.
52.Wagner, N., Schmitt, J., and Krohne, G. (2004).
Two novel LEM-domain proteins are splice products of the annotated
Drosophila melanogaster gene CG9424 (Bocksbeutel),
Eur. J. Cell Biol., 82, 605-616.
53.Wagner, N., Kagermeier, B., Loserth, S., and
Krohne, G. (2006) The Drosophila melanogaster LEM-domain
protein MAN1, Eur. J. Cell Biol., 85,
91-105.
54.Gigante, C. M., Dibattista, M., Dong, F. N.,
Zheng, X., Yue, S., Young, S. G., Reisert, J., Zheng, Y., and Zhao, H.
(2017) Lamin B1 is required for mature neuron-specific gene expression
during olfactory sensory neuron differentiation, Nat.
Commun., 8, 15098.
55.Somech, R., Shaklai, S., Geller, O., Amariglio,
N., Simon, A. J., Rechavi, G., and Gal-Yam, E. N. (2005) The
nuclear-envelope protein and transcriptional repressor LAP2β
interacts with HDAC3 at the nuclear periphery and induces histone H4
deacetylation, J. Cell Sci., 118, 4017-4025.
56.Holaska, J. M., and Wilson, K. L. (2007) An
emerin “proteome”: purification of distinct
emerin-containing complexes from HeLa cells suggests molecular basis
for diverse roles including gene regulation, mRNA splicing, signaling,
mechanosensing, and nuclear architecture, Biochemistry,
46, 8897-8908.
57.Poleshko, A., Shah, P. P., Gupta, M., Babu, A.,
Morley, M. P., Manderfield, L. J., Ifkovits, J. L., Calderon, D.,
Aghajanian, H., Sierra-Pagan, J. E., Sun, Z., Wang, Q., Li, L., Dubois,
N. C., Morrisey, E. E., Lazar, M. A., Smith, C. L., Epstein, J. A., and
Jain, R. (2017) Genome–nuclear lamina interactions regulate
cardiac stem cell lineage restriction, Cell, 171,
573-587.
58.Wen, B., Wu, H., Shinkai, Y., Irizarry, R. A.,
and Feinberg, A. P. (2009) Large histone H3 lysine 9 dimethylated
chromatin blocks distinguish differentiated from embryonic stem cells,
Nat. Genet., 41, 246-250.
59.Kind, J., Pagie, L., Ortabozkoyun, H., Boyle, S.,
de Vries, S. S., Janssen, H., Amendola, M., Nolen, L. D., Bickmore, W.
A., and van Steensel, B. (2013) Single-cell dynamics of
genome–nuclear lamina interactions, Cell, 153,
178-192.
60.Bian, Q., Khanna, N., Alvikas, J., and Belmont,
A. S. (2013) β-Globin cis-elements determine differential
nuclear targeting through epigenetic modifications, J. Cell
Biol., 203, 767-783.
61.Harr, J. C., Luperchio, T. R., Wong, X., Cohen,
E., Wheelan, S. J., and Reddy, K. L. (2015) Directed targeting of
chromatin to the nuclear lamina is mediated by chromatin state and
A-type lamins, J. Cell Biol., 208,
33-52.
62.Luperchio, T. R., Sauria, M. E. G., Wong, X.,
Gaillard, M.-C., Tsang, P., Pekrun, K., Ach, R. A., Yamada, N. A.,
Taylor, J., and Reddy, K. L. (2017) Chromosome conformation paints
reveal the role of lamina association in genome organization and
regulation, bioRxiv preprint first posted online March 30; doi:
https://doi.org/10.1101/122226.
63.Towbin, B. D., Meister, P., Pike, B. L., and
Gasser, S. M. (2011) Repetitive transgenes in C. elegans
accumulate heterochromatic marks and are sequestered at the nuclear
envelope in a copy-number- and lamin-dependent manner, Cold Spring
Harb. Symp. Quant. Biol., 75,
555-565.
64.Gonzalez-Sandoval, A., Towbin, B. D., Kalck, V.,
Cabianca, D. S., Gaidatzis, D., Hauer, M. H., Geng, L., Wang, L., Yang,
T., Wang, X., Zhao, K., and Gasser, S. M. (2015) Perinuclear anchoring
of H3K9-methylated chromatin stabilizes induced cell fate in C.
elegans embryos, Cell, 163, 1333-1347.
65.Filion, G. J., van Bemmel, J. G., Braunschweig,
U., Talhout, W., Kind, J., Ward, L. D., Brugman, W., de Castro, I. J.,
Kerkhoven, R. M., Bussemaker, H. J., and van Steensel, B. (2010)
Systematic protein location mapping reveals five principal chromatin
types in Drosophila cells, Cell, 143, 212-224.
66.Milon, B. C., Cheng, H., Tselebrovsky, M. V.,
Lavrov, S. A., Nenasheva, V. V., Mikhaleva, E. A., Shevelyov, Y. Y.,
and Nurminsky, D. I. (2012) Role of histone deacetylases in gene
regulation at nuclear lamina, PLoS One, 7, e49692.
67.Hoger, T. H., Krohne, G., and Kleinschmidt, J. A.
(1991) Interaction of Xenopus lamins A and LII with chromatin
in vitro mediated by a sequence element in the carboxyterminal
domain, Exp. Cell Res., 197, 280-289.
68.Glass, C. A., Glass, J. R., Taniura, H., Hasel,
K. W., Blevitt, J. M., and Gerace, L. (1993) The α-helical rod
domain of human lamins A and C contains a chromatin binding site,
EMBO J., 12, 4413-4424.
69.Luderus, M. E., den Blaauwen, J. L., de Smit, O.
J., Compton, D. A., and van Driel, R. (1994) Binding of matrix
attachment regions to lamin polymers involves single-stranded regions
and the minor groove, Mol. Cell. Biol., 14,
6297-6305.
70.Goldberg, M., Harel, A., Brandeis, M.,
Rechsteiner, T., Richmond, T. J., Weiss, A. M., and Gruenbaum, Y.
(1999) The tail domain of lamin Dm0 binds histones H2A and H2B,
Proc. Natl. Acad. Sci. USA,
96, 2852-2857.
71.Stierle, V., Couprie, J., Ostlund, C., Krimm, I.,
Zinn-Justin, S., Hossenlopp, P., Worman, H. J., Courvalin, J. C., and
Duband-Goulet, I. (2003) The carboxyl-terminal region common to lamins
A and C contains a DNA binding domain, Biochemistry, 42,
4819-4828.
72.Mattout, A., Goldberg, M., Tzur, Y., Margalit,
A., and Gruenbaum, Y. (2007) Specific and conserved sequences in D.
melanogaster and C. elegans lamins and histone H2A mediate
the attachment of lamins to chromosomes, J. Cell Sci.,
120 (Pt. 1), 77-85.
73.Foisner, R., and Gerace, L. (1993) Integral
membrane proteins of the nuclear envelope interact with lamins and
chromosomes, and binding is modulated by mitotic phosphorylation,
Cell, 73, 1267-1279.
74.Furukawa, K., Glass, C., and Kondo, T. (1997)
Characterization of the chromatin binding activity of lamina-associated
polypeptide (LAP) 2, Biochem. Biophys. Res.
Commun., 238, 240-246.
75.Ye, Q., and Worman, H. J. (1996) Interaction
between an integral protein of the nuclear envelope inner membrane and
human chromodomain proteins homologous to Drosophila HP1,
J. Biol. Chem., 271, 14653-14656.
76.Polioudaki, H., Kourmouli, N., Drosou, V., Bakou,
A., Theodoropoulos, P. A., Singh, P. B., Giannakouros, T., and
Georgatos, S. D. (2001) Histones H3/H4 form a tight complex with the
inner nuclear membrane protein LBR and heterochromatin protein 1,
EMBO Rep., 2, 920-925.
77.Makatsori, D., Kourmouli, N., Polioudaki, H.,
Shultz, L. D., McLean, K., Theodoropoulos, P. A., Singh, P. B., and
Georgatos, S. D. (2004) The inner nuclear membrane protein lamin B
receptor forms distinct microdomains and links epigenetically marked
chromatin to the nuclear envelope, J. Biol. Chem., 279,
25567-25573.
78.Hirano, Y., Hizume, K., Kimura, H., Takeyasu, K.,
Haraguchi, T., and Hiraoka, Y. (2012) Lamin B receptor recognizes
specific modifications of histone H4 in heterochromatin formation,
J. Biol. Chem., 287, 42654-42663.
79.Clowney, E. J., LeGros, M. A., Mosley, C. P.,
Clowney, F. G., Markenskoff-Papadimitriou, E. C., Myllys, M., Barnea,
G., Larabell, C. A., and Lomvardas, S. (2012) Nuclear aggregation of
olfactory receptor genes governs their monogenic expression,
Cell, 151, 724-737.
80.Lukasova, E., Kovarik, A., Bacikova, A., Falk,
M., and Kozubek, S. (2017) Loss of lamin B receptor is necessary to
induce cellular senescence, Biochem. J., 474,
281-300.
81.Zhu, Y., Gong, K., Denholtz, M., Chandra, V.,
Kamps, M. P., Alber, F., and Murre, C. (2017) Comprehensive
characterization of neutrophil genome topology, Genes Dev.,
31, 141-153.
82.Zuleger, N., Boyle, S., Kelly, D. A., de las
Heras, J. I., Lazou, V., Korfali, N., Batrakou, D. G., Randles, K. N.,
Morris, G. E., Harrison, D. J., Bickmore, W. A., and Schirmer, E. C.
(2013) Specific nuclear envelope transmembrane proteins can promote the
location of chromosomes to and from the nuclear periphery, Genome
Biol., 14, R14.
83.Robson, M. I., de Las Heras, J. I., Czapiewski,
R., Le Thanh, P., Booth, D. G., Kelly, D. A., Webb, S., Kerr, A. R. W.,
and Schirmer, E. C. (2016) Tissue-specific gene repositioning by muscle
nuclear membrane proteins enhances repression of critical developmental
genes during myogenesis, Mol. Cell, 62,
834-847.
84.Kind, J., Pagie, L., de Vries, S. S., Nahidiazar,
L., Dey, S. S., Bienko, M., Zhan, Y., Lajoie, B., de Graaf, C. A.,
Amendola, M., Fudenberg, G., Imakaev, M., Mirny, L. A., Jalink, K.,
Dekker, J., van Oudenaarden, A., and van Steensel, B. (2015)
Genome-wide maps of nuclear lamina interactions in single human cells,
Cell, 163, 134-147.
85.Shevelyov, Y. Y., and Nurminsky, D. I. (2012) The
nuclear lamina as a gene-silencing hub, Curr. Issues Mol.
Biol., 14, 27-38.
86.Kosak, S. T., Skok, J. A., Medina, K. L., Riblet,
R., Le Beau, M. M., Fisher, A. G., and Singh, H. (2002) Subnuclear
compartmentalization of immunoglobulin loci during lymphocyte
development, Science, 296, 158-162.
87.Hewitt, S. L., High, F. A., Reiner, S. L.,
Fisher, A. G., and Merkenschlager, M. (2004) Nuclear repositioning
marks the selective exclusion of lineage-inappropriate transcription
factor loci during T helper cell differentiation, Eur. J.
Immunol., 34, 3604-3613.
88.Zink, D., Amaral, M. D., Englmann, A., Lang, S.,
Clarke, L. A., Rudolph, C., Alt, F., Luther, K., Braz, C., Sadoni, N.,
Rosenecker, J., and Schindelhauer, D. (2004) Transcription-dependent
spatial arrangements of CFTR and adjacent genes in human cell
nuclei, J. Cell Biol., 166, 815-825.
89.Ragoczy, T., Bender, M. A., Telling, A., Byron,
R., and Groudine, M. (2006) The locus control region is required for
association of the murine β-globin locus with engaged
transcription factories during erythroid maturation, Genes Dev.,
20, 1447-1457.
90.Williams, R. R., Azuara, V., Perry, P., Sauer,
S., Dvorkina, M., Jorgensen, H., Roix, J., McQueen, P., Misteli, T.,
Merkenschlager, M., and Fisher, A. G. (2006) Neural induction promotes
large-scale chromatin reorganization of the Mash1 locus,
J. Cell Sci., 119, 132-140.
91.Ballester, M., Kress, C., Hue-Beauvais, C., Kieu,
K., Lehmann, G., Adenot, P., and Devinoy, E. (2008) The nuclear
localization of WAP and CSN genes is modified by
lactogenic hormones in HC11 cells, J. Cell Biochem.,
105, 262-270.
92.Szczerbal, I., Foster, H. A., and Bridger, J. M.
(2009) The spatial repositioning of adipogenesis genes is correlated
with their expression status in a porcine mesenchymal stem cell
adipogenesis model system, Chromosoma, 118, 647-663.
93.Meister, P., Towbin, B. D., Pike, B. L., Ponti,
A., and Gasser, S. M. (2010) The spatial dynamics of tissue-specific
promoters during C. elegans development, Genes Dev.,
24, 766-782.
94.Kress, C., Kieu, K., Droineau, S., Galio, L., and
Devinoy, E. (2011) Specific positioning of the casein gene cluster in
active nuclear domains in luminal mammary epithelial cells,
Chromosome Res., 19, 979-997.
95.Lee, H. Y., Johnson, K. D., Boyer, M. E., and
Bresnick, E. H. (2011) Relocalizing genetic loci into specific
subnuclear neighborhoods, J. Biol. Chem.,
286, 18834-18844.
96.Yao, J., Fetter, R. D., Hu, P., Betzig, E., and
Tjian, R. (2011) Subnuclear segregation of genes and core promoter
factors in myogenesis, Genes Dev., 25, 569-580.
97.Demmerle, J., Koch, A. J., and Holaska, J. M.
(2013) Emerin and histone deacetylase 3 (HDAC3) cooperatively regulate
expression and nuclear positions of MyoD, Myf5, and
Pax7 genes during myogenesis, Chromosome Res., 21,
765-779.
98.Zhao, H., Sifakis, E. G., Sumida, N.,
Millan-Arino, L., Scholz, B. A., Svensson, J. P., Chen, X., Ronnegren,
A. L., Mallet de Lima, C. D., Varnoosfaderani, F. S., Shi, C., Loseva,
O., Yammine, S., Israelsson, M., Rathje, L. S., Nemeti, B., Fredlund,
E., Helleday, T., Imreh, M. P., and Gondor, A. (2015) PARP1- and
CTCF-mediated interactions between active and repressed chromatin at
the lamina promote oscillating transcription, Mol. Cell,
59, 984-997.
99.Tumbar, T., and Belmont, A. S. (2001) Interphase
movements of a DNA chromosome region modulated by VP16 transcriptional
activator, Nat. Cell Biol., 3, 134-139.
100.Chuang, C. H., Carpenter, A. E., Fuchsova, B.,
Johnson, T., de Lanerolle, P., and Belmont, A. S. (2006) Long-range
directional movement of an interphase chromosome site, Curr.
Biol., 16, 825-831.
101.Therizols, P., Illingworth, R. S., Courilleau,
C., Boyle, S., Wood, A. J., and Bickmore, W. A. (2014) Chromatin
decondensation is sufficient to alter nuclear organization in embryonic
stem cells, Science, 346, 1238-1242.
102.Finlan, L. E., Sproul, D., Thomson, I., Boyle,
S., Kerr, E., Perry, P., Ylstra, B., Chubb, J. R., and Bickmore, W. A.
(2008) Recruitment to the nuclear periphery can alter expression of
genes in human cells, PLoS Genet., 4, e1000039.
103.Reddy, K. L., Zullo, J. M., Bertolino, E., and
Singh, H. (2008) Transcriptional repression mediated by repositioning
of genes to the nuclear lamina, Nature, 452, 243-247.
104.Dialynas, G., Speese, S., Budnik, V., Geyer, P.
K., and Wallrath, L. L. (2010) The role of Drosophila lamin C in
muscle function and gene expression, Development, 137,
3067-3077.
105.Kumaran, R. I., and Spector, D. L. (2008) A
genetic locus targeted to the nuclear periphery in living cells
maintains its transcriptional competence, J. Cell Biol.,
180, 51-65.
106.Gierman, H. J., Indemans, M. H. G., Koster, J.,
Goetze, S., Seppen, J., Geerts, D., van Driel, R., and Versteeg, R.
(2007) Domain-wide regulation of gene expression in the human genome,
Genome Res., 17, 1286-1295.
107.Babenko, V. N., Makunin, I. V., Brusentsova, I.
V., Belyaeva, E. S., Maksimov, D. A., Belyakin, S. N., Maroy, P.,
Vasil’eva, L. A., and Zhimulev, I. F. (2010) Paucity and
preferential suppression of transgenes in late replication domains of
the D. melanogaster genome, BMC Genomics, 11,
318.
108.Akhtar, W., de Jong, J., Pindyurin, A. V.,
Pagie, L., Meuleman, W., de Ridder, J., Berns, A., Wessels, L. F., van
Lohuizen, M., and van Steensel, B. (2013) Chromatin position effects
assayed by thousands of reporters integrated in parallel, Cell,
154, 914-927.
109.Kim, Y., Sharov, A. A., McDole, K., Cheng, M.,
Hao, H., Fan, C. M., Gaiano, N., Ko, M. S., and Zheng, Y. (2011) Mouse
B-type lamins are required for proper organogenesis but not by
embryonic stem cells, Science, 334, 1706-1710.
110.Amendola, M., and van Steensel, B. (2015)
Nuclear lamins are not required for lamina-associated domain
organization in mouse embryonic stem cells, EMBO Rep.,
16, 610-617.
111.Chintapalli, V. R., Wang, J., and Dow, J. A.
(2007) Using FlyAtlas to identify better Drosophila melanogaster
models of human disease, Nat. Genet., 39,
715-720.
112.Chang, C. W., Cheng, W. C., Chen, C. R., Shu,
W. Y., Tsai, M. L., Huang, C. L., and Hsu, I. C. (2011) Identification
of human housekeeping genes and tissue-selective genes by microarray
meta-analysis, PLoS One, 6, e22859.
113.Graveley, B. R., Brooks, A. N., Carlson, J. W.,
Duff, M. O., Landolin, J. M., Yang, L., Artieri, C. G., van Baren, M.
J., Boley, N., Booth, B. W., Brown, J. B., Cherbas, L., Davis, C. A.,
Dobin, A., Li, R., Lin, W., Malone, J. H., Mattiuzzo, N. R., Miller,
D., Sturgill, D., Tuch, B. B., Zaleski, C., Zhang, D., Blanchette, M.,
Dudoit, S., Eads, B., Green, R. E., Hammonds, A., Jiang, L., Kapranov,
P., Langton, L., Perrimon, N., Sandler, J. E., Wan, K. H., Willingham,
A., Zhang, Y., Zou, Y., Andrews, J., Bickel, P. J., Brenner, S. E.,
Brent, M. R., Cherbas, P., Gingeras, T. R., Hoskins, R. A., Kaufman, T.
C., Oliver, B., and Celniker, S. E. (2011) The developmental
transcriptome of Drosophila melanogaster, Nature,
471, 473-479.
114.Wu, F., and Yao, J. (2017) Identifying novel
transcriptional and epigenetic features of nuclear lamina-associated
genes, Sci. Rep., 7, 100.
115.Liu, J., Rolef Ben-Shahar, T., Riemer, D.,
Treinin, M., Spann, P., Weber, K., Fire, A., and Gruenbaum, Y. (2000)
Essential roles for Caenorhabditis elegans lamin gene in nuclear
organization, cell cycle progression, and spatial organization of
nuclear pore complexes, Mol. Biol. Cell,
11, 3937-3947.
116.Vergnes, L., Peterfy, M., Bergo, M. O., Young,
S. G., and Reue, K. (2004) Lamin B1 is required for mouse development
and nuclear integrity, Proc. Natl. Acad.
Sci. USA, 101, 10428-10433.
117.Wagner, N., Weber, D., Seitz, S., and Krohne,
G. (2004). The lamin B receptor of Drosophila melanogaster,
J. Cell Sci., 117 (Pt. 10), 2015-2028.
118.Coffinier, C., Jung, H. J., Nobumori, C.,
Chang, S., Tu, Y., Barnes, R. H., 2nd, Yoshinaga, Y., de Jong, P. J.,
Vergnes, L., Reue, K., Fong, L. G., and Young, S. G. (2011)
Deficiencies in lamin B1 and lamin B2 cause neurodevelopmental defects
and distinct nuclear shape abnormalities in neurons, Mol.
Biol. Cell, 22, 4683-4693.
119.Kim, Y., McDole, K., and Zheng, Y. (2012) The
function of lamins in the context of tissue building and maintenance,
Nucleus, 3, 256-262.
120.Guo, Y., and Zheng, Y. (2015) Lamins position
the nuclear pores and centrosomes by modulating dynein, Mol.
Biol. Cell, 26, 3379-3389.
121.Milon, B., Sun, Y., Chang, W., Creasy, T.,
Mahurkar, A., Shetty, A., Nurminsky, D., and Nurminskaya, M. (2014) Map
of open and closed chromatin domains in Drosophila genome,
BMC Genomics, 15, 988.
122.Boettiger, A. N., Bintu, B., Moffitt, J. R.,
Wang, S., Beliveau, B. J., Fudenberg, G., Imakaev, M., Mirny, L. A.,
Wu, C. T., and Zhuang, X. (2016) Super-resolution imaging reveals
distinct chromatin folding for different epigenetic states,
Nature, 529, 418-422.