REVIEW: Structure and Functions of the Mediator Complex
E. V. Putlyaev1, A. N. Ibragimov1, L. A. Lebedeva1, P. G. Georgiev1, and Y. V. Shidlovskii1,2*
1Institute of Gene Biology, Russian Academy of Sciences, 119334 Moscow, Russia; E-mail: yul.biogen@gmail.com2Sechenov First Moscow State Medical University, 119048 Moscow, Russia; E-mail: expedition@mma.ru
* To whom correspondence should be addressed.
Received October 31, 2017; Revision received December 4, 2017
Mediator is a key factor in the regulation of expression of RNA polymerase II-transcribed genes. Recent studies have shown that Mediator acts as a coordinator of transcription activation and participates in maintaining chromatin architecture in the cell nucleus. In this review, we present current concepts on the structure and functions of Mediator.
KEY WORDS: Mediator, transcription, RNA polymerase, chromatin, enhancer, promoterDOI: 10.1134/S0006297918040132
Abbreviations: CKM, Cdk8 kinase module; CTD, C-terminal domain of RNA polymerase II Rpb1 subunit; IDR, intrinsically disordered protein regions; ncRNA, noncoding RNA; PIC, preinitiation complex; Pol I, II, III, DNA-dependent RNA polymerase I, II, and III, respectively; UAS, upstream activating sequence.
Mediator is an essential coactivator of transcription by RNA polymerase
II (Pol II) in unicellular and multicellular eukaryotes. Mediator acts
as a scaffold that binds and coordinates most transcription components
[1]. Mediator interacts with general transcription
factors, chromatin-modifying complexes, and numerous gene-specific
activators [2]. It is involved in many signaling
pathways [1] and controls various aspects of cell
metabolism [3]. Mediator plays a key role in
ontogenesis; mutations in the Mediator subunits are associated with the
development of various pathologies [1, 4, 5]. It is not surprising that a
complex with such diverse and broad functions has attracted
considerable interest of researchers. In this review, we present recent
data on the structure and functions of Mediator.
Structure of Mediator complex
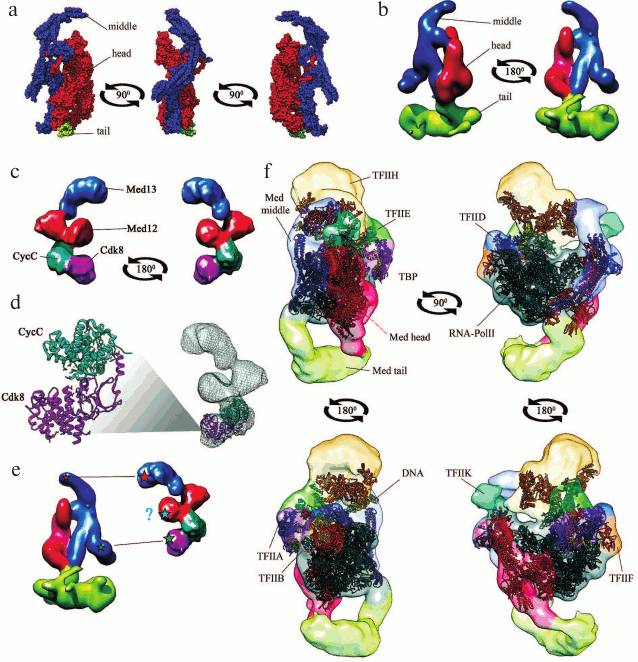
Mediator is a large protein complex with a molecular mass of 1.4 MDa that consists of ~25 subunits in mammals (0.9 MDa and 20 subunits in yeast). The subunits form four major structural modules: head, middle, tail, and a mobile Cdk8 kinase module (CKM) (see table). The subunit composition of the modules and their relative position in the complex have been elucidated after solving the 3D structure of yeast Mediator with high resolution [6, 7] (Fig. 1a). The central role in the organization of Mediator complex belongs to the Med14 subunit, that connects the head, the middle, and the tail modules [8], and to the Med17 subunit, that forms multiple contacts with other subunits [9].
Mediator complex modules and subunits [5] with their
known functions in transcription activation
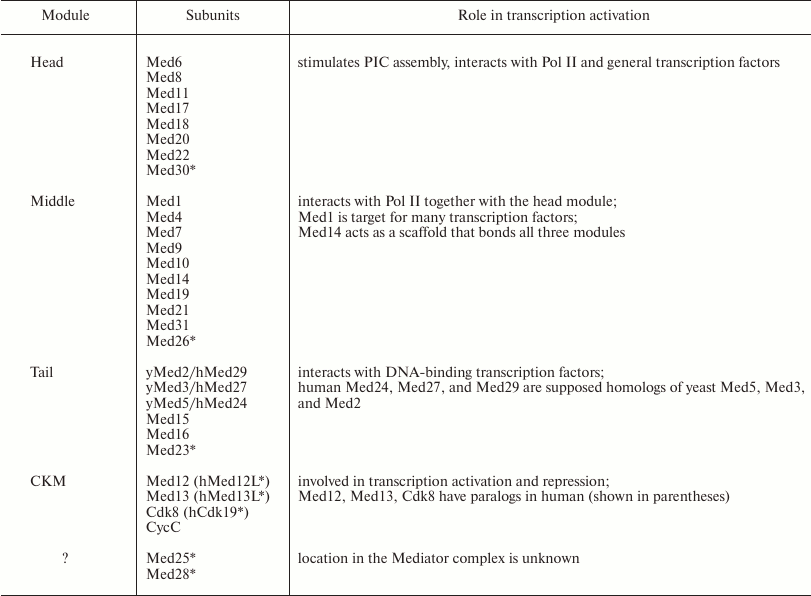
Notes: yMed, yeast Mediator subunits; hMed, human Mediator subunits.
* Metazoan-specific subunits.
Fig. 1. Three-dimensional structures. a) Mediator complex in free state based on data of X-ray analysis at 4.4 Å resolution [8] (RCSB PDB ID: 5u0p [150]). All subunits are shown except Med1. The structure of the tail module is incomplete because of limitations of the method. b) Yeast Mediator complex in free state based on data of cryo-EM at ~1.8 nm resolution [11] (ePDB EMDB ID: 2634 [151]). Structural modules (head, middle, tail) are shown with different colors. c) CKM in free state based on data of cryo-EM [20] (ePDB EMDB ID: 5588 [151]). d) Superposition of the structures of the complete CKM (cryo-EM) and Cdk8–CycC complex [152] (RCSB PDB ID: 3rgf [150]). e) Contacts between CKM and Mediator (asterisks, presumed areas of contacts; ?, interaction between Med12 and middle module was demonstrated for human Mediator but not for yeast Mediator [20]); f) Mediator-containing PIC, as obtained by superposition of cryo-EM data (resolution, 21.9 Å; ePDB EMDB ID: 8308 [151]) and X-ray analysis data (resolution, 15.3 Å; RCSB PDB ID: 5sva [150]). The figure shows DNA molecules, RNA polymerase II (Pol II), TATA-binding protein (TBP), and transcription initiation factors TFIIA, TFIIB, TFIID, TFIIE, TFIIF, TFIIH, and TFIIK. Structural modules (head, middle, tail) are shown with different colors.
Recently, the structure of the Mediator complex has been resolved by X-ray analysis, cryoelectron microscopy (cryo-EM), and mass spectrometry of cross-linked subunits [9]. Most data have been obtained for yeast Mediator, including the results of cryo-EM that allowed reconstructing the 3D structure of the Mediator complex consisting of its three major structural modules (Fig. 1b). Although the structure of Mediator is conserved in all eukaryotes [10, 11], its details in higher eukaryotes have not been completely elucidated.
The structures of the Mediator core 15-subunit complex with general transcription factors [12] and of the complete Mediator–Pol II preinitiation complex (PIC) on the promoter [13] have been obtained recently. It was found that the Mediator head module forms multiple contacts with RNA polymerase; the most important contact is binding of the head module to the unphosphorylated C-terminal domain (CTD) of the RNA polymerase Rpb1 subunit [13]. The head and the middle modules are responsible for the interaction with general transcription factors on promoters. The tail module protrudes from the PIC toward the upstream DNA, thereby creating a platform for the interaction of PIC with transcriptional activators [14] (Fig. 1f). Taken together, these results corroborate the suggestion on the major functional role of Mediator as a scaffold for PIC assembly and stabilization on a promoter [13].
An important feature of the Mediator complex is that its subunit composition can change depending on the biological context. The most extensively characterized alteration in the Mediator complex is reversible binding of CKM [1]. Mass-spectrometry analysis showed that relative contents of individual subunits in the Mediator complex preparations may differ. Also, the subunit composition of Mediator in differentiated cells becomes less diverse, as it was also described for TFIID complex [15]. The presence of particular subunits in the complex might be tissue-specific, as demonstrated for the Med26 subunit in Drosophila [16]. In yeast, the Mediator complex composition can vary as well. Thus, Med3 and Med15 subunits of the tail module can form amyloid-like aggregates under stress conditions, which changes the subunit composition of the whole complex [17]. These data suggest the existence of Mediator complexes of varying composition. Since different Mediator subunits bind different transcription factors, alternative forms of the complex might be involved in the generation of alternative transcriptional responses in cells.
Mediator is a complex that is dynamic in its subunit composition and structure. Many subunits in this complex (both in humans and yeast) have intrinsically disordered regions (IDRs). It is possible that the presence of many IDRs provides Mediator with the ability to interact with structurally diverse transcriptional factors [18]. Structural rearrangements in the Mediator complex come along with its binding to the CKM and transcription factors or its interaction with PIC [8, 14]. It was suggested that structural rearrangements of Mediator are related to the sequence of processes during gene transcription activation and play an important role in the functioning of Mediator [8].
CYCLIN-DEPENDENT KINASE MODULE OF MEDIATOR
The only enzymatic activity of Mediator is provided by the kinase module that is universally conserved in eukaryotes [4]. Yeast CKM is a protein complex with a molecular mass of ~430 kDa that consists of four subunits: Cdk8, CycC, Med12, and Med13. In mammals, Cdk8, Med12, and Med13 subunits have also paralogs Cdk19, Med12L, and Med13L, respectively. The paralogs are required for the normal course of ontogenesis (in particular, neurogenesis); their presence in the CDK–Mediator complex is mutually exclusive [19]. CKM has an elongated structure; it binds to the Mediator middle module via the Med13 subunit in both mammals and yeast [20, 21] (Fig. 1, c-e).
Regulation of CKM binding to Mediator is not completely understood. CKM itself is involved in the interaction with transcription factors in plants, flies, and humans [22-26] and can be recruited to DNA independently, as demonstrated for several genes including yeast heat-shock genes [27, 28]. There are several mechanisms for the dissociation of CKM from the complex. In yeast, Cdk11 phosphorylates subunits Med4 and Med27, which results in CKM dissociation [29]. In mammals, CKM dissociation might be caused by specific degradation of Med13 [30]. PARP-1 regulates CKM dissociation from retinoic acid receptor target genes [31]. In plants, dissociation of CKM from the Mediator complex in the upstream regions of some genes depends on the degradation of the IAA14 transcription repressor [23]. All these data indicate that regulation of CKM binding to Mediator is species- and gene-specific.
Cdk8 is a serine/threonine kinase whose target proteins are SMAD1 and SMAD3 transcription factors [32], Notch ICD [33], SREBP [34], E2F1 [35], STAT1 [36], histone H3 Ser10 [37], and CTD of RNA Pol II [38]. Large-scale search for Cdk8/Cdk19 substrates using cortistatin A allowed to significantly expand this list [39]. Sixty-four most probable targets were identified, most of which were DNA-binding transcription factors, Pol II factors, mRNA-processing factors, and subunits of chromatin-modifying complexes. This kinase also phosphorylates several Mediator subunits including Med12 and Med13. Other targets of CKM are proteins involved in DNA replication and repair and other processes, which indicated that CKM functions in the nucleus are much broader than it was earlier suggested. It was found that in HCT116 cells, phosphorylation does not affect the stability of target proteins except Med13/Med13L. However, in other studies, Cdk8-dependent phosphorylation led to degradation of Notch ICD, SMAD1, SMAD3, and SREBP in higher eukaryotes [32-34] and Ste12p in yeast [40]. It is possible that phosphorylation still plays a role in the stability of the protein, but in a tissue-specific manner.
CKM can either activate or suppress gene expression. However, Cdk8 knockdown in human cells has a very slight effect on gene expression [39]. The functions of Cdk8 in gene expression control are context-dependent – in human cells Cdk8 regulates different genes in response to different factors [41, 42]. The role of cooperation between the module subunits in the regulation of gene expression remains obscure. In yeast, all four subunits control expression of a common set of genes [43]. In mammalian cells, Cdk8 and Med12 sometimes cooperate in the regulation of certain genes; in other cases, Cdk8 and CycC function independently of Med12 [44-46]. Knockdown of Med12/Med13 and CycC/Cdk8 in Drosophila resulted mostly in the opposing effects on gene transcription; though rarely such knockdowns produced similar effects [47]. Two paralogs, Cdk8 and Cdk19, seem to have different gene targets, since knockdown of these subunits affected only slightly overlapping groups of genes in human cells [39].
Therefore, CKM often acts as an independent factor of gene expression regulation that functions in a context-dependent manner. The involvement of CKM in the expression regulation is determined not only by its kinase activity, but also through its interaction with other transcription factors (see below).
ROLE OF MEDIATOR IN ORGANIZATION OF TRANSCRIPTION
Localization of Mediator on chromatin in higher eukaryotes demonstrated that Mediator preferentially binds to enhancers throughout the whole genome [48]. Also, Mediator is typical for large enhancer clusters that initiate tissue-specific transcriptional programs (so-called super-enhancers) [49]. In yeast, Mediator was detected on the majority of the upstream activating sequences (UASs) that serve as the analogs of enhancers in higher located upstream of the promoters [5, 50].
Mediator recruitment to enhancers occurs mostly via interaction of its tail and middle modules with transcriptional activators [2, 51, 52]. Deletion of individual subunits of the tail module results in the inability of Mediator to bind to UASs throughout the entire yeast genome [53, 54].
CKM can play different roles in the recruitment of Mediator to enhancers. In some loci, it acts as a binding antagonist. Thus, removal of the CKM Med13 subunit in yeast increases the amount of Mediator bound to certain UASs [54]. The kinase activity of CKM blocks Mediator-mediated activation of target genes in the EGFR/Ras/ERK signaling pathway in Caenorhabditis elegans [55]. Cdk8 and Cdk19 suppress the activity of super-enhancers in the AML cell line [51]. It is believed that in such cases CKM is required for the fine regulation of recruitment of Mediator to UASs and enhancers [5]. At the same time, CKM can play a role of coactivator for other genes. For example, Cdk8 is required for the activation of the estrogen receptor target genes in humans [56]. CKM is recruited to promoters of target genes during stimulation of the TRL9 receptor; it cooperates with the transcription factors C/EBPβ and NF-κB [57]. Cdk8 and CycC are coactivators of the Drosophila ecdysone receptor [58]. These facts may explain the gene-specific role of CKM in the regulation of gene expression.
Transcription activation in eukaryotes involves physical contacts between an enhancer and a promoter. Using sequential chromatin immunoprecipitation, Petrenko et al. [54] were first to demonstrate the existence of a single Mediator complex that simultaneously associates with an enhancer and a promoter, thereby corroborating the concept that Mediator acts as a dynamic bridge for these two regulatory elements.
The presence of Mediator on yeast promoters is difficult to demonstrate by chromatin immunoprecipitation [59]. The amount of Mediator on a promoter significantly increases when TFIIH kinase is inhibited, what significantly stabilizes PIC [53, 54]. The reason for a weak signal from the promoter is that the interaction of Mediator with PIC is very short-termed (estimated time, 1/8 s) [60]. Despite its short duration, it is this involvement of Mediator in PIC formation that is believed to be its major genome-wide function [5, 61]. In yeast, promoter- and enhancer-bound Mediator complexes have different subunit composition (promoter-bound Mediator lacks the CKM) [53, 54]. The interaction between the enhancer and the promoter is provided by the Mediator in the absence of CKM.
The major target of Mediator complex on a promoter is Pol II CTD, which acts as a scaffold for many transcription factors and serves as a coordinator of the entire transcription process [62]. Besides, Mediator regulates recruitment and activity of other PIC components, such as general transcription factors TFIIA, B, D, E, and F [50, 63-65], and stimulates recruitment and enzymatic activity of TFIIH [66, 67]. Structural analysis revealed that Mediator forms direct contacts with these factors while being a part of the PIC [8, 12, 13] (Fig. 1f). In addition to a purely structural role in PIC assembly, Mediator is essential for coordination of the sequential processes involved in PIC assembly on the promoter [68, 69] – the role of Mediator in this process depends on the promoter architecture [63, 70].
At the initial stages of gene activation, Mediator forms transcriptionally inactive PIC that is then activated by CTD phosphorylation by TFIIH. This stage is stimulated by Mediator [69, 71] that dissociates from the polymerase (as phosphorylated CTD is incapable of Mediator binding [60, 72]) and leaves the promoter [60]. Hence, the interaction between Mediator and TFIIH is important for transcription initiation and Mediator dissociation from the promoter.
Mediator also provides rapid reinitiation of transcription. It was shown for the HIV-1 promoter that rapid Mediator-dependent reinitiation of transcription results in the appearance of polymerase convoys (groups of polymerase molecules that move along the gene one after another) and transcriptional bursting [73].
Beside transcription initiation, Mediator is involved in the regulation of elongation. For instance, mutations in the middle module of yeast Mediator do not affect recruitment of polymerase to the promoter but decrease its concentration in the gene transcribed region and prevent a decrease in the number of nucleosomes along the gene [74]. In vertebrates, inhibition of Cdk8/19 activity results in the suppression of CTD phosphorylation and inhibition of transcription elongation on the NF-κB-controlled genes [38].
After dissociation from the promoter and synthesis of several tens of nucleotides of the transcript, RNA polymerase stops. This RNA polymerase pausing plays an important regulatory role [75]. Mediator is involved in this process via interacting with DSIF [76], cohesin [48], and TFIIS [77, 78]. Cooperation between Mediator and transcription elongation factor TFIIS helps to prevent the negative effect of a nucleosome at the start of the transcribed sequence [79]. Also, Mediator from multicellular organisms can initiate transcription in vitro if RNA polymerase contains the Gdown1/Pol2M subunit [80]. It was found recently that this subunit binds to polymerase after the start of the RNA synthesis and stabilizes the enzyme in the paused state [81]. Possibly Mediator overcomes the negative effect of Gdown1 and facilitates polymerase exit from pausing [5].
P-TEFb, one of the major factors stimulating transition to elongation, could be recruited to promoters through interaction with CKM [41] and Med26 [82]. Perhaps these two mechanisms of P-TEFb recruitment occur in different groups of genes [83]. Cooperative action of Mediator and P-TEFb also takes place on enhancers, as shown for the T cell-specific enhancer (both factors stimulated synthesis of enhancer ncRNA) [84], enhancers of Hippo/YAP signaling pathway target genes [85], and BRD4-controlled regulatory elements in AML cells [86].
Med26 interacts with the EAF subunit of the super elongation complex (SEC) to ensure initiation of elongation on some genes. Med26 knockdown does not impair PIC assembly but suppresses SEC recruitment [87]. Interestingly, Med26 also interacts with TFIID via the same domain that provides its binding to EAF [88]. Interaction of this subunit with alternating partners might be the mechanism for switching between transcription initiation and elongation. Med26 recruits the LEC initiation complex to genes encoding small nuclear RNAs [89]. SEC subunits also interact with the kinase module of Mediator [42]. Cdk8 knockdown inhibits transcription elongation and SEC recruitment to activated promoters [41, 42]. Cooperation between all three factors (Mediator, SEC, and P-TEFb) is essential for the regulation of transcription of genes controlling early embryonic development in Drosophila [90].
Therefore, Mediator is involved in all stages of transcription activation. Based on the existing data, the following mechanism of Mediator functioning was suggested [5]. Initially, the complete Mediator complex is recruited to enhancers via transcription factors; this stage is negatively regulated by the CKM. After CKM dissociation, Mediator is integrated into PIC, which results in PIC stabilization. As a component of PIC, Mediator stimulates kinase activity of TFIIH, thereby causing phosphorylation of Ser5 residues in CTD and polymerase escape from the promoter. Mediator dissociates from the promoter and binds CKM, which stimulates recruitment of factors (e.g., P-TEFb) favoring the polymerase exit from pausing. P-TEFb phosphorylates Ser2 residues in CTD, and polymerase starts elongation.
CKM plays a dual role in this model. Most likely, its repressor function is not related to the enzymatic activity but results from the binding of CKM to Mediator, since Mediator can only bind either CKM or RNA polymerase [20]. The activator function of CKM is related to its ability to directly bind to some transcriptional activators and coactivators.
In the yeast nucleus, ~15% of all Mediator complexes interact with RNA Pol II, 15% are bound to CKM, and therefore excluded from this interaction, and the remaining 70% are in a free state [91].
It is important to emphasize that Mediator is not an obligatory component of PIC, however, it stimulates consequent stages of PIC maturation on the promoter. Also, some Mediator subunits can be functionally duplicated by other factors. Thus, deletion of individual Mediator subunits produces very moderate effect on yeast transcription, and only impairments in all three modules – head, middle, and tail – significantly disturb cell translation and viability [70].
Mediator is also involved in posttranscriptional events. Med23 interacts with mRNA-processing factors and regulates alternative splicing and alternative mRNA polyadenylation [92]. Mediator directly binds various factors regulating mRNA 3′-end processing and mRNA degradation [93]. The Med31/Med7N module interacts with the mRNA nuclear export factor TREX-2. Interestingly, the latter is required for the binding of CKM to Mediator and regulates Mediator binding to polymerase [94].
ROLE OF MEDIATOR IN REGULATION OF CHROMATIN STRUCTURE
In addition to direct interaction with transcription machinery components, Mediator controls gene activity by regulating chromatin structure. Mediator binds to chromatin through association with histone H3 and H4 tails [95, 96]. It also interacts with chromatin-remodeling and chromatin-modifying factors.
Mediator participates in keeping promoter regions of active genes in a nucleosome-free state. It was found to interact with the chromatin-remodeling complex SWI/SNF in yeast [97, 98]. In humans, CKM interacts with Brg1 [99]. The yeast Med15 subunit binds to the Hrp1 remodeling factor of the CHD1 family [100]; Med15 interacts with CHD1 in mouse cells, and the resulting complex is recruited by active genes [101]. Binding of Mediator to the yeast HO gene requires recruitment of the SWI/SNF complex [102]. For some genes, on the contrary, recruitment of the SWI/SNF complex happens after binding of Mediator to the genes [97, 98, 103]. Therefore, cooperation between Mediator and remodeling factors is tissue-specific. Moreover, the presence of Mediator in the content of PIC on a promoter is important for DNA dissociation from nucleosomes irrespectively of the chromatin remodeling factors [104].
Mediator regulates epigenetic events. For instance, mediator and histone acetyltransferase p300 function synergistically during estrogen receptor transcription in human cells [105] and on active enhancers of mouse hemopoietic cells [106]. Mediator can be recruited to target genes of frog androgen and thyroid hormone receptors directly or via interaction with p300 [107]. In human cells, Mediator binds to the histone acetyltransferase Gcn5 [108]. The two proteins function cooperatively: Gcn5 together with Cdk8 perform tandem modification of histone H3 [109]. Yeast have promoters that recruit the histone acetyltransferase complex SAGA and Mediator in a coordinated manner [110, 111]. However, yeast cells also have promoters to which these complexes bind independently of each other [112, 113]. In higher eukaryotes, Mediator is recruited cooperatively with the histone acetyltransferase complexes STAGA [114, 115] and ATAC [116].
Mediator also plays a role in H2B ubiquitination. In human cells, Med23 recruits the corresponding RNF20/40 enzyme to chromatin [117].
Therefore, Mediator is involved in the recruitment of complexes that establish active chromatin markers. However, Mediator also participates in epigenetic repression, mostly via its kinase module. Gene-specific suppression of trimethylation of the H3K4 residue by CKM [118] was found in yeast. It was demonstrated for one of these genes that CKM blocks the binding of the chromatin-modifying Set1p/COMPASS activator complex [40]. In mammals, downregulation of neuronal genes outside of the nervous system is mediated by the Mediator-dependent recruitment of histone methyltransferase G9a, and this function of Mediator depends on CKM [119]. Repression of immune response genes in humans depends on the Mediator-mediated recruitment of arginine methyltransferase PRMT5; in this case, binding occurs also via the kinase module [120].
Mediator is involved in epigenetic transcriptional memory. In yeast, CKM binds to the INO1 gene in the memory state (the gene is switched off but can be easily reactivated), which is important for polymerase recruitment and pausing for further transcription initiation. This mechanism is evolutionarily conserved – it was also found for the IFNγ-induced genes in human cells [121].
Mediator binds the Polycomb group complexes that repress certain genes in ontogenesis. Cdk8 knockdown in a mouse cancer model resulted in reduction in histone H3K27 trimethylation and derepression of the Polycomb-regulated genes [122]. CKM binds the EZH2 and SUZ12 subunits of the PRC2 complex in humans; this interaction is essential for the timely activation of neuronal genes during development [123]. The CKM subunit Med12 operates together with PRC1 to silence key developmental genes in mouse pluripotent cells. During cell differentiation, Med12 dissociates from PRC1, and Mediator converts from a transcriptional repressor to a transcriptional enhancer [124]. There are also some evidences indicating that Mediator can counteract the Polycomb-dependent repression – its subunit Med25 blocks the binding of PRC2 to the gene targets of HFN4α [125].
Finally, Mediator is involved in the formation and maintenance of the structure of pericentric and telomeric heterochromatin [1] and in the establishment of borders between active and inactive chromatin in yeast [126]. In accordance with it, Med26 (but not other subunits) was found in the pericentric heterochromatin in Drosophila [16]. Perhaps this subunit has a specific function that differs from the functions of the complex. Mediator is also required for the synthesis and processing of centromeric ncRNAs involved in heterochromatin formation in yeast and plants [127-129]. In telomeric heterochromatin of yeast, Mediator operates together with histone deacetylase Sir2 [130].
Therefore, due to many interactions with enzymes that modify chromatin, Mediator acts as an important factor in epigenetic events in all genome regions in a broad range of living organisms.
ROLE OF MEDIATOR IN CHROMATIN ARCHITECTURE FORMATION
The presence of Mediator at a certain locus does not always correlate with the transcriptional activity of this locus. In both yeast and higher eukaryotes, Mediator has been found on many regulatory elements along the entire genome regardloss of their activity [48, 53]. It was suggested that besides regulating gene activity, Mediator is important for maintaining general chromatin architecture [50]. Indeed, Mediator plays an essential role in the formation of contacts between gene regulatory elements in higher eukaryotes [131-134]. Such contacts are a structural feature of DNA packing in higher eukaryotes. Interactions between enhancer-anchored Mediator and promoter-bound PIC are short-termed, as mentioned above, and most probably cannot serve as a basis for the formation of stable contacts in chromatin.
One of the proteins involved in the formation of long-range contacts in chromatin is cohesin. In mouse cells, Mediator and cohesin interact, and the resulting complex binds to the factor Nipbl that provides cohesin docking on DNA. The loci that bind Mediator and cohesin form long-range contacts in a process that depends on Med12; the pattern of such contacts is tissue-specific [48, 108]. Synergistic action of Med12 and Nipbl was also shown in the Danio rerio zebrafish [135]. In human cells, Mediator and cohesin act synergistically to maintain cell type [136]. Analysis of chromatin packing hierarchy in mammalian cells showed that Mediator and cohesin are required for the formation of short-range (up to 100 kb) tissue-specific contacts between enhancers and promoters, while CTCF and cohesin are important for the establishment of stable long-range (over 1000 kb) contacts. Formation of short-range contacts depends on Med12. There are also small loops (600-1000 bp), the formation of which is regulated by Mediator, but not by cohesin [137]. At the same time, Mediator is not an obligatory component of the enhancer–promoter contacts in the genome – both Mediator-independent and cohesin-independent contacts were described for the beta-globin locus [138].
Although gene regulation in yeast does not proceed via formation of long-range contacts [139], in these organisms Mediator determines the distance over which UASs can activate transcription [140]. In yeast, Mediator is also involved in the formation of contacts between the 5′- and 3′-ends of genes [141], general packing of chromosomes, and formation of chromatin domains (lack of Mediator results in chromatin decompaction in the nucleus) [142]. Analysis of the distribution of Mediator in the yeast genome showed that it is more abundant at the boundaries of the so-called chromosomal interacting domain (CIDs), which are analogs of TADs (topologically associating domains) in higher organisms. Mediator is also associated with various factors involved in the organization of general chromatin structure in the yeast nucleus [93]. Therefore, Mediator has an evolutionarily conserved function involving regulation of distant genomic elements and control of chromatin packing in the nucleus of all eukaryotic organisms.
Other molecules required for the formation of long-range interactions are ncRNAs. It was found that ncRNA-a (one of the classes of mammalian ncRNAs involved in gene activation) interacts with Med12. Mediator is recruited to the sites of synthesis of this RNA; ncRNA-a and Mediator operate together to promote long-range interactions and to activate target genes [143]. In a similar manner, Mediator is required for the activity of enhancer RNAs synthesized on active enhancers, as it was shown for the target genes of androgen receptor [144] and PPARγ receptor [145].
Therefore, Mediator plays an important role not only in transcription, but also in the formation of contacts between distant genome loci. Perhaps such broad activity of Mediator is related to its ability to interact with different types of molecules, including ncRNAs.
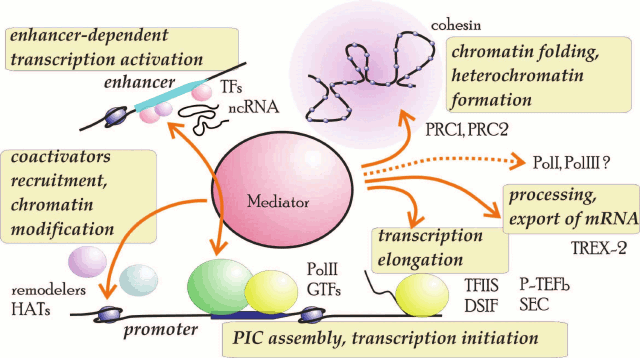
Mediator is believed to be a structural integrating complex that participates in the coordinated recruitment of various activities toward genomic loci. Recent description of the Mediator interactome in yeast corroborated and extended this model [91]. More than 400 Mediator-interacting proteins with different functions were identified. In addition to the earlier described partners, Mediator was found to interact with FACT chaperones Spt16 and Pob3, chromatin-remodeling factors ISWI and INO80, acetylated and trimethylated histones, and proteins involved in mRNA processing such as polyA-binding protein (PABP), proteins of RNA splicing apparatus, decapping proteins, and Xrn1, which is a protein participating in mRNA degradation. Interestingly, Mediator copurifies with Pol I and III; it interacts with all 14 subunits of Pol I, as well as with the initiator complex proteins and factors of RNA processing and ribosome biogenesis. In Pol III, Mediator interacted with its two large subunits and two components of the initiator complex – TFIIIB and TFIIIC. These data indicate that the role of Mediator in transcription is much broader than it had been believed before (Fig. 2).
Fig. 2. Major functions and interaction partners of Mediator complex: GTFs, general transcriptions factors; HATs, histone acyltransferases; ncRNA, noncoding mRNA; remodelers, chromatin-remodeling factors; TFs, transcription factors. Putative involvement of Pol I and Pol III is indicated with a dashed line.
Despite a considerable progress in understanding of the Mediator’s functional role, many mechanisms of its action still require further investigation. Thus, the structure of Mediator complex in higher eukaryotes and contribution of subunits specific for the multicellular organisms remain poorly studied. It is still unclear if the complex has varying subunit composition. Among other unresolved problems are structural rearrangements that occur during binding of Mediator to activators and PIC and the role of these rearrangements in the activity of the complex. The mechanism of CKM binding to Mediator and the role of kinase in the regulation of gene activity are to be studied as well. The structural basis of interactions of Mediator with other factors often remains obscure. Thus, almost nothing is known about interactions of Mediator with ncRNAs. It remains unclear if Mediator’s binding to RNA is sequence-specific, and how such binding affects the structure and functions of Mediator.
The involvement of Mediator in the formation of chromatin architecture is apparently not limited to cohesin binding. It was shown recently that formation of chromatin loops requires participation of many different factors [146]. It seems reasonable to suggest that Mediator acts as a recruiter for these factors. According to another hypothesis, interaction of Mediator with CTD stabilizes long-range contacts in the genome. Since CTDs of higher eukaryotes are long, perhaps they can stimulate formation of long-range enhancer–promoter contacts [1].
Another interesting hypothesis is that the Mediator could be involved in formation of transcription factories. The presence of IDR domains in subunits of the complex might indicate its ability to form a separate phase in the nucleus [1]. IDR domains are common in transcription factors [62, 147, 148], which might be the basis for the concentration of RNA polymerase and general transcription factors into discrete sites inside the regions of transcription. The role of separate protein phase formation in the nucleus has been demonstrated recently for the heterochromatin protein HP1α [149].
Acknowledgments
This work was supported by the Ministry of Education and Science of the Russian Federation (project No. 14.B25.31.0022) and the Russian Science Foundation (project No. 16-14-10346).
REFERENCES
1.Allen, B. L., and Taatjes, D. J. (2015) The
Mediator complex: a central integrator of transcription, Nat. Rev.
Mol. Cell Biol., 16, 155-166.
2.Borggrefe, T., and Yue, X. (2011) Interactions
between subunits of the Mediator complex with gene-specific
transcription factors, Semin. Cell Devel. Biol., 22,
759-768.
3.Youn, D. Y., Xiaoli, A. M., Pessin, J. E., and
Yang, F. (2016) Regulation of metabolism by the Mediator complex,
Biophys. Rep., 2, 69-77.
4.Clark, A. D., Oldenbroek, M., and Boyer, T. G.
(2015) Mediator kinase module and human tumorigenesis, Crit. Rev.
Biochem. Mol. Biol., 5BB0, 393-426.
5.Jeronimo, C., and Robert, F. (2017) The Mediator
complex: at the nexus of RNA polymerase II transcription, Trends
Cell Biol., 27, 765-783.
6.Tsai, K. L., Tomomori-Sato, C., Sato, S., Conaway,
R. C., Conaway, J. W., and Asturias, F. J. (2014) Subunit architecture
and functional modular rearrangements of the transcriptional mediator
complex, Cell, 158, 463.
7.Wang, X., Sun, Q., Ding, Z., Ji, J., Wang, J.,
Kong, X., Yang, J., and Cai, G. (2014) Redefining the modular
organization of the core Mediator complex, Cell Res., 24,
796-808.
8.Tsai, K. L., Yu, X., Gopalan, S., Chao, T. C.,
Zhang, Y., Florens, L., Washburn, M. P., Murakami, K., Conaway, R. C.,
Conaway, J. W., and Asturias, F. J. (2017) Mediator structure and
rearrangements required for holoenzyme formation, Nature,
544, 196-201.
9.Harper, T. M., and Taatjes, D. J. (2017) The
complex structure and function of Mediator, J. Biol. Chem., in
press.
10.Cevher, M. A., Shi, Y., Li, D., Chait, B. T.,
Malik, S., and Roeder, R. G. (2014) Reconstitution of active human core
Mediator complex reveals a critical role of the MED14 subunit, Nat.
Struct. Mol. Biol., 21, 1028-1034.
11.Tsai, K. L., Tomomori-Sato, C., Sato, S.,
Conaway, R. C., Conaway, J. W., and Asturias, F. J. (2014) Subunit
architecture and functional modular rearrangements of the
transcriptional mediator complex, Cell, 157,
1430-1444.
12.Plaschka, C., Lariviere, L., Wenzeck, L., Seizl,
M., Hemann, M., Tegunov, D., Petrotchenko, E. V., Borchers, C. H.,
Baumeister, W., Herzog, F., Villa, E., and Cramer, P. (2015)
Architecture of the RNA polymerase II–Mediator core initiation
complex, Nature, 518, 376-380.
13.Robinson, P. J., Trnka, M. J., Bushnell, D. A.,
Davis, R. E., Mattei, P. J., Burlingame, A. L., and Kornberg, R. D.
(2016) Structure of a complete Mediator–RNA polymerase II
pre-initiation complex, Cell, 166, 1411-1422.
14.Nozawa, K., Schneider, T. R., and Cramer, P.
(2017) Core Mediator structure at 3.4 Å extends model of
transcription initiation complex, Nature, 545,
248-251.
15.D’Alessio, J. A., Ng, R., Willenbring, H.,
and Tjian, R. (2011) Core promoter recognition complex changes
accompany liver development, Proc. Natl. Acad. Sci. USA,
108, 3906-3911.
16.Marr, S. K., Lis, J. T., Treisman, J. E., and
Marr, M. T., 2nd (2014) The metazoan-specific mediator subunit 26
(Med26) is essential for viability and is found at both active genes
and pericentric heterochromatin in Drosophila melanogaster,
Mol. Cell. Biol., 34, 2710-2720.
17.Zhu, X., Chen, L., Carlsten, J. O., Liu, Q.,
Yang, J., Liu, B., and Gustafsson, C. M. (2015) Mediator tail subunits
can form amyloid-like aggregates in vivo and affect stress
response in yeast, Nucleic Acids Res., 43, 7306-7314.
18.Nagulapalli, M., Maji, S., Dwivedi, N., Dahiya,
P., and Thakur, J. K. (2016) Evolution of disorder in Mediator complex
and its functional relevance, Nucleic Acids Res., 44,
1591-1612.
19.Daniels, D., Ford, M., Schwinn, M., Benink, H.,
Galbraith, M., Amunugama, R., Jones, R., Allen, D., Okazaki, N.,
Yamakawa, H., Miki, F., Nagase, T., Espinosa, J., and Urh, M. (2013)
Mutual exclusivity of MED12/MED12L, MED13/13L, and CDK8/19 paralogs
revealed within the CDK–Mediator kinase module, J. Proteom.
Bioinform., S2, 004.
20.Tsai, K. L., Sato, S., Tomomori-Sato, C.,
Conaway, R. C., Conaway, J. W., and Asturias, F. J. (2013) A conserved
Mediator–CDK8 kinase module association regulates
Mediator–RNA polymerase II interaction, Nat. Struct. Mol.
Biol., 20, 611-619.
21.Wang, X., Wang, J., Ding, Z., Ji, J., Sun, Q.,
and Cai, G. (2013) Structural flexibility and functional interaction of
Mediator Cdk8 module, Protein Cell, 4, 911-920.
22.Carrera, I., Janody, F., Leeds, N., Duveau, F.,
and Treisman, J. E. (2008) Pygopus activates Wingless target gene
transcription through the mediator complex subunits Med12 and Med13,
Proc. Natl. Acad. Sci. USA, 105, 6644-6649.
23.Ito, J., Fukaki, H., Onoda, M., Li, L., Li, C.,
Tasaka, M., and Furutani, M. (2016) Auxin-dependent compositional
change in Mediator in ARF7- and ARF19-mediated transcription, Proc.
Natl. Acad. Sci. USA, 113, 6562-6567.
24.Kim, S., Xu, X., Hecht, A., and Boyer, T. G.
(2006) Mediator is a transducer of Wnt/beta-catenin signaling, J.
Biol. Chem., 281, 14066-14075.
25.Tutter, A. V., Kowalski, M. P., Baltus, G. A.,
Iourgenko, V., Labow, M., Li, E., and Kadam, S. (2009) Role for Med12
in regulation of Nanog and Nanog target genes, J. Biol. Chem.,
284, 3709-3718.
26.Zhu, Y., Schluttenhoffer, C. M., Wang, P., Fu,
F., Thimmapuram, J., Zhu, J. K., Lee, S. Y., Yun, D. J., and Mengiste,
T. (2014) Cyclin-dependent kinases differentially regulates plant
immunity to fungal pathogens through kinase-dependent and -independent
functions in Arabidopsis, Plant Cell, 26,
4149-4170.
27.Anandhakumar, J., Moustafa, Y. W., Chowdhary, S.,
Kainth, A. S., and Gross, D. S. (2016) Evidence for multiple Mediator
complexes in yeast independently recruited by activated heat shock
factor, Mol. Cell. Biol., 36, 1943-1960.
28.Thomas-Claudepierre, A. S., Robert, I., Rocha, P.
P., Raviram, R., Schiavo, E., Heyer, V., Bonneau, R., Luo, V. M.,
Reddy, J. K., Borggrefe, T., Skok, J. A., and Reina-San-Martin, B.
(2016) Mediator facilitates transcriptional activation and dynamic
long-range contacts at the IgH locus during class switch recombination,
J. Exp. Med., 213, 303-312.
29.Drogat, J., Migeot, V., Mommaerts, E., Mullier,
C., Dieu, M., Van Bakel, H., and Hermand, D. (2012) Cdk11-cyclinL
controls the assembly of the RNA polymerase II mediator complex,
Cell Rep., 2, 1068-1076.
30.Davis, M. A., Larimore, E. A., Fissel, B. M.,
Swanger, J., Taatjes, D. J., and Clurman, B. E. (2013) The SCF-Fbw7
ubiquitin ligase degrades MED13 and MED13L and regulates CDK8 module
association with Mediator, Genes Dev., 27, 151-156.
31.Pavri, R., Lewis, B., Kim, T. K., Dilworth, F.
J., Erdjument-Bromage, H., Tempst, P., De Murcia, G., Evans, R.,
Chambon, P., and Reinberg, D. (2005) PARP-1 determines specificity in a
retinoid signaling pathway via direct modulation of mediator, Mol.
Cell, 18, 83-96.
32.Alarcon, C., Zaromytidou, A. I., Xi, Q., Gao, S.,
Yu, J., Fujisawa, S., Barlas, A., Miller, A. N., Manova-Todorova, K.,
Macias, M. J., Sapkota, G., Pan, D., and Massague, J. (2009) Nuclear
CDKs drive Smad transcriptional activation and turnover in BMP and
TGF-beta pathways, Cell, 139, 757-769.
33.Fryer, C. J., White, J. B., and Jones, K. A.
(2004) Mastermind recruits CycC:CDK8 to phosphorylate the Notch ICD and
coordinate activation with turnover, Mol. Cell, 16,
509-520.
34.Zhao, X., Feng, D., Wang, Q., Abdulla, A., Xie,
X. J., Zhou, J., Sun, Y., Yang, E. S., Liu, L. P., Vaitheesvaran, B.,
Bridges, L., Kurland, I. J., Strich, R., Ni, J. Q., Wang, C., Ericsson,
J., Pessin, J. E., Ji, J. Y., and Yang, F. (2012) Regulation of
lipogenesis by cyclin-dependent kinase 8-mediated control of SREBP-1,
J. Clin. Invest., 122, 2417-2427.
35.Zhao, J., Ramos, R., and Demma, M. (2013) CDK8
regulates E2F1 transcriptional activity through S375 phosphorylation,
Oncogene, 32, 3520-3530.
36.Bancerek, J., Poss, Z. C., Steinparzer, I.,
Sedlyarov, V., Pfaffenwimmer, T., Mikulic, I., Dolken, L., Strobl, B.,
Muller, M., Taatjes, D. J., and Kovarik, P. (2013) CDK8 kinase
phosphorylates transcription factor STAT1 to selectively regulate the
interferon response, Immunity, 38, 250-262.
37.Knuesel, M. T., Meyer, K. D., Donner, A. J.,
Espinosa, J. M., and Taatjes, D. J. (2009) The human CDK8 subcomplex is
a histone kinase that requires Med12 for activity and can function
independently of mediator, Mol. Cell. Biol., 29,
650-661.
38.Chen, M., Liang, J., Ji, H., Yang, Z., Altilia,
S., Hu, B., Schronce, A., McDermott, M. S. J., Schools, G. P., Lim, C.
U., Oliver, D., Shtutman, M. S., Lu, T., Stark, G. R., Porter, D. C.,
Broude, E. V., and Roninson, I. B. (2017) CDK8/19 Mediator kinases
potentiate induction of transcription by NFkappaB, Proc. Natl. Acad.
Sci. USA, 114, 10208-10213.
39.Poss, Z. C., Ebmeier, C. C., Odell, A. T.,
Tangpeerachaikul, A., Lee, T., Pelish, H. E., Shair, M. D., Dowell, R.
D., Old, W. M., and Taatjes, D. J. (2016) Identification of Mediator
kinase substrates in human cells using cortistatin A and quantitative
phosphoproteomics, Cell Rep., 15, 436-450.
40.Law, M. J., and Finger, M. A. (2017) The
Saccharomyces cerevisiae Cdk8 Mediator represses AQY1
transcription by inhibiting Set1p-dependent histone methylation, G3
(Bethesda), 7, 1001-1010.
41.Donner, A. J., Ebmeier, C. C., Taatjes, D. J.,
and Espinosa, J. M. (2010) CDK8 is a positive regulator of
transcriptional elongation within the serum response network, Nat.
Struct. Mol. Biol., 17, 194-201.
42.Galbraith, M. D., Allen, M. A., Bensard, C. L.,
Wang, X., Schwinn, M. K., Qin, B., Long, H. W., Daniels, D. L., Hahn,
W. C., Dowell, R. D., and Espinosa, J. M. (2013) HIF1A employs
CDK8-mediator to stimulate RNAPII elongation in response to hypoxia,
Cell, 153, 1327-1339.
43.Van de Peppel, J., Kettelarij, N., Van Bakel, H.,
Kockelkorn, T. T., Van Leenen, D., and Holstege, F. C. (2005) Mediator
expression profiling epistasis reveals a signal transduction pathway
with antagonistic submodules and highly specific downstream targets,
Mol. Cell, 19, 511-522.
44.Adler, A. S., McCleland, M. L., Truong, T., Lau,
S., Modrusan, Z., Soukup, T. M., Roose-Girma, M., Blackwood, E. M., and
Firestein, R. (2012) CDK8 maintains tumor dedifferentiation and
embryonic stem cell pluripotency, Cancer Res., 72,
2129-2139.
45.Vogl, M. R., Reiprich, S., Kuspert, M., Kosian,
T., Schrewe, H., Nave, K. A., and Wegner, M. (2013) Sox10 cooperates
with the mediator subunit 12 during terminal differentiation of
myelinating glia, J. Neurosci., 33, 6679-6690.
46.Zhou, H., Spaeth, J. M., Kim, N. H., Xu, X.,
Friez, M. J., Schwartz, C. E., and Boyer, T. G. (2012) MED12 mutations
link intellectual disability syndromes with dysregulated GLI3-dependent
Sonic Hedgehog signaling, Proc. Natl. Acad. Sci. USA,
109, 19763-19768.
47.Kuuluvainen, E., Hakala, H., Havula, E., Sahal
Estime, M., Ramet, M., Hietakangas, V., and Makela, T. P. (2014)
Cyclin-dependent kinase 8 module expression profiling reveals
requirement of mediator subunits 12 and 13 for transcription of
Serpent-dependent innate immunity genes in Drosophila, J.
Biol. Chem., 289, 16252-16261.
48.Kagey, M. H., Newman, J. J., Bilodeau, S., Zhan,
Y., Orlando, D. A., Van Berkum, N. L., Ebmeier, C. C., Goossens, J.,
Rahl, P. B., Levine, S. S., Taatjes, D. J., Dekker, J., and Young, R.
A. (2010) Mediator and cohesin connect gene expression and chromatin
architecture, Nature, 467, 430-435.
49.Whyte, W. A., Orlando, D. A., Hnisz, D., Abraham,
B. J., Lin, C. Y., Kagey, M. H., Rahl, P. B., Lee, T. I., and Young, R.
A. (2013) Master transcription factors and mediator establish
super-enhancers at key cell identity genes, Cell, 153,
307-319.
50.Grunberg, S., Henikoff, S., Hahn, S., and
Zentner, G. E. (2016) Mediator binding to UASs is broadly uncoupled
from transcription and cooperative with TFIID recruitment to promoters,
EMBO J., 35, 2435-2446.
51.Poss, Z. C., Ebmeier, C. C., and Taatjes, D. J.
(2013) The Mediator complex and transcription regulation, Crit. Rev.
Biochem. Mol. Biol., 48, 575-608.
52.Yin, J. W., and Wang, G. (2014) The Mediator
complex: a master coordinator of transcription and cell lineage
development, Development, 141, 977-987.
53.Jeronimo, C., Langelier, M. F., Bataille, A. R.,
Pascal, J. M., Pugh, B. F., and Robert, F. (2016) Tail and kinase
modules differently regulate core Mediator recruitment and function
in vivo, Mol. Cell, 64, 455-466.
54.Petrenko, N., Jin, Y., Wong, K. H., and Struhl,
K. (2016) Mediator undergoes a compositional change during
transcriptional activation, Mol. Cell, 64, 443-454.
55.Grants, J. M., Ying, L. T., Yoda, A., You, C. C.,
Okano, H., Sawa, H., and Taubert, S. (2016) The Mediator kinase module
restrains epidermal growth factor receptor signaling and represses
vulval cell fate specification in Caenorhabditis elegans,
Genetics, 202, 583-599.
56.McDermott, M. S., Chumanevich, A. A., Lim, C. U.,
Liang, J., Chen, M., Altilia, S., Oliver, D., Rae, J. M., Shtutman, M.,
Kiaris, H., Gyorffy, B., Roninson, I. B., and Broude, E. V. (2017)
Inhibition of CDK8 mediator kinase suppresses estrogen dependent
transcription and the growth of estrogen receptor positive breast
cancer, Oncotarget, 8, 12558-12575.
57.Yamamoto, S., Hagihara, T., Horiuchi, Y., Okui,
A., Wani, S., Yoshida, T., Inoue, T., Tanaka, A., Ito, T., Hirose, Y.,
and Ohkuma, Y. (2017) Mediator cyclin-dependent kinases upregulate
transcription of inflammatory genes in cooperation with NF-kappaB and
C/EBPbeta on stimulation of Toll-like receptor 9, Genes Cells,
22, 265-276.
58.Xie, X. J., Hsu, F. N., Gao, X., Xu, W., Ni, J.
Q., Xing, Y., Huang, L., Hsiao, H. C., Zheng, H., Wang, C., Zheng, Y.,
Xiaoli, A. M., Yang, F., Bondos, S. E., and Ji, J. Y. (2015)
CDK8-cyclin C mediates nutritional regulation of developmental
transitions through the ecdysone receptor in Drosophila, PLoS
Biol., 13, e1002207.
59.Malik, S., and Roeder, R. G. (2016) Mediator: a
drawbridge across the enhancer–promoter divide, Mol. Cell,
64, 433-434.
60.Wong, K. H., Jin, Y., and Struhl, K. (2014) TFIIH
phosphorylation of the Pol II CTD stimulates mediator dissociation from
the preinitiation complex and promoter escape, Mol. Cell,
54, 601-612.
61.Lacombe, T., Poh, S. L., Barbey, R., and Kuras,
L. (2013) Mediator is an intrinsic component of the basal RNA
polymerase II machinery in vivo, Nucleic Acids Res.,
41, 9651-9662.
62.Harlen, K. M., and Churchman, L. S. (2017) The
code and beyond: transcription regulation by the RNA polymerase II
carboxy-terminal domain, Nature Rev. Mol. Cell Biol., 18,
263-273.
63.Eychenne, T., Novikova, E., Barrault, M. B.,
Alibert, O., Boschiero, C., Peixeiro, N., Cornu, D., Redeker, V.,
Kuras, L., Nicolas, P., Werner, M., and Soutourina, J. (2016)
Functional interplay between Mediator and TFIIB in preinitiation
complex assembly in relation to promoter architecture, Genes
Dev., 30, 2119-2132.
64.Jishage, M., Malik, S., Wagner, U., Uberheide,
B., Ishihama, Y., Hu, X., Chait, B. T., Gnatt, A., Ren, B., and Roeder,
R. G. (2012) Transcriptional regulation by Pol II(G) involving mediator
and competitive interactions of Gdown1 and TFIIF with Pol II, Mol.
Cell, 45, 51-63.
65.Johnson, K. M., and Carey, M. (2003) Assembly of
a mediator/TFIID/TFIIA complex bypasses the need for an activator,
Curr. Biol., 13, 772-777.
66.Esnault, C., Ghavi-Helm, Y., Brun, S.,
Soutourina, J., Van Berkum, N., Boschiero, C., Holstege, F., and
Werner, M. (2008) Mediator-dependent recruitment of TFIIH modules in
preinitiation complex, Mol. Cell, 31, 337-346.
67.Seizl, M., Lariviere, L., Pfaffeneder, T.,
Wenzeck, L., and Cramer, P. (2011) Mediator head subcomplex Med11/22
contains a common helix bundle building block with a specific function
in transcription initiation complex stabilization, Nucleic Acids
Res., 39, 6291-6304.
68.Eyboulet, F., Wydau-Dematteis, S., Eychenne, T.,
Alibert, O., Neil, H., Boschiero, C., Nevers, M. C., Volland, H.,
Cornu, D., Redeker, V., Werner, M., and Soutourina, J. (2015) Mediator
independently orchestrates multiple steps of preinitiation complex
assembly in vivo, Nucleic Acids Res., 43,
9214-9231.
69.Malik, S., Molina, H., and Xue, Z. (2017) PIC
activation through functional interplay between Mediator and TFIIH,
J. Mol. Biol., 429, 48-63.
70.Petrenko, N., Jin, Y., Wong, K. H., and Struhl,
K. (2017) Evidence that Mediator is essential for Pol II transcription,
but is not a required component of the preinitiation complex in
vivo, eLife, 6.
71.Boeing, S., Rigault, C., Heidemann, M., Eick, D.,
and Meisterernst, M. (2010) RNA polymerase II C-terminal heptarepeat
domain Ser-7 phosphorylation is established in a mediator-dependent
fashion, J. Biol. Chem., 285, 188-196.
72.Jeronimo, C., and Robert, F. (2014) Kin28
regulates the transient association of Mediator with core promoters,
Nat. Struct. Mol. Biol., 21, 449-455.
73.Tantale, K., Mueller, F., Kozulic-Pirher, A.,
Lesne, A., Victor, J. M., Robert, M. C., Capozi, S., Chouaib, R.,
Backer, V., Mateos-Langerak, J., Darzacq, X., Zimmer, C., Basyuk, E.,
and Bertrand, E. (2016) A single-molecule view of transcription reveals
convoys of RNA polymerases and multi-scale bursting, Nat.
Commun., 7, 12248.
74.Kremer, S. B., Kim, S., Jeon, J. O., Moustafa, Y.
W., Chen, A., Zhao, J., and Gross, D. S. (2012) Role of Mediator in
regulating Pol II elongation and nucleosome displacement in
Saccharomyces cerevisiae, Genetics, 191,
95-106.
75.Scheidegger, A., and Nechaev, S. (2016) RNA
polymerase II pausing as a context-dependent reader of the genome,
Biochem. Cell Biol., 94, 82-92.
76.Malik, S., Barrero, M. J., and Jones, T. (2007)
Identification of a regulator of transcription elongation as an
accessory factor for the human Mediator coactivator, Proc. Natl.
Acad. Sci. USA, 104, 6182-6187.
77.Guglielmi, B., Soutourina, J., Esnault, C., and
Werner, M. (2007) TFIIS elongation factor and Mediator act in
conjunction during transcription initiation in vivo, Proc.
Natl. Acad. Sci. USA, 104, 16062-16067.
78.Wery, M., Shematorova, E., Van Driessche, B.,
Vandenhaute, J., Thuriaux, P., and Van Mullem, V. (2004) Members of the
SAGA and Mediator complexes are partners of the transcription
elongation factor TFIIS, EMBO J., 23, 4232-4242.
79.Nock, A., Ascano, J. M., Barrero, M. J., and
Malik, S. (2012) Mediator-regulated transcription through the +1
nucleosome, Mol. Cell, 48, 837-848.
80.Hu, X., Malik, S., Negroiu, C. C., Hubbard, K.,
Velalar, C. N., Hampton, B., Grosu, D., Catalano, J., Roeder, R. G.,
and Gnatt, A. (2006) A Mediator-responsive form of metazoan RNA
polymerase II, Proc. Natl. Acad. Sci. USA, 103,
9506-9511.
81.DeLaney, E., and Luse, D. S. (2016) Gdown1
associates efficiently with RNA polymerase II after promoter clearance
and displaces TFIIF during transcript elongation, PLoS One,
11, e0163649.
82.Wang, W., Yao, X., Huang, Y., Hu, X., Liu, R.,
Hou, D., Chen, R., and Wang, G. (2013) Mediator MED23 regulates basal
transcription in vivo via an interaction with P-TEFb,
Transcription, 4, 39-51.
83.Conaway, R. C., and Conaway, J. W. (2013) The
Mediator complex and transcription elongation, Biochim. Biophys.
Acta, 1829, 69-75.
84.Hertweck, A., Evans, C. M., Eskandarpour, M.,
Lau, J. C., Oleinika, K., Jackson, I., Kelly, A., Ambrose, J., Adamson,
P., Cousins, D. J., Lavender, P., Calder, V. L., Lord, G. M., and
Jenner, R. G. (2016) T-bet activates Th1 genes through Mediator
and the super elongation complex, Cell Rep., 15,
2756-2770.
85.Galli, G. G., Carrara, M., Yuan, W. C.,
Valdes-Quezada, C., Gurung, B., Pepe-Mooney, B., Zhang, T., Geeven, G.,
Gray, N. S., De Laat, W., Calogero, R. A., and Camargo, F. D. (2015)
YAP drives growth by controlling transcriptional pause release from
dynamic enhancers, Mol. Cell, 60, 328-337.
86.Bhagwat, A. S., Roe, J. S., Mok, B. Y. L.,
Hohmann, A. F., Shi, J., and Vakoc, C. R. (2016) BET bromodomain
inhibition releases the Mediator complex from select
cis-regulatory elements, Cell Rep., 15,
519-530.
87.Takahashi, H., Parmely, T. J., Sato, S.,
Tomomori-Sato, C., Banks, C. A., Kong, S. E., Szutorisz, H., Swanson,
S. K., Martin-Brown, S., Washburn, M. P., Florens, L., Seidel, C. W.,
Lin, C., Smith, E. R., Shilatifard, A., Conaway, R. C., and Conaway, J.
W. (2011) Human mediator subunit MED26 functions as a docking site for
transcription elongation factors, Cell, 146, 92-104.
88.Lens, Z., Cantrelle, F. X., Peruzzini, R.,
Hanoulle, X., Dewitte, F., Ferreira, E., Baert, J. L., Monte, D.,
Aumercier, M., Villeret, V., Verger, A., and Landrieu, I. (2017)
Solution structure of the N-terminal domain of Mediator subunit
MED26 and molecular characterization of its interaction with EAF1 and
TAF7, J. Mol. Biol., 429, 3043-3055.
89.Takahashi, H., Takigawa, I., Watanabe, M., Anwar,
D., Shibata, M., Tomomori-Sato, C., Sato, S., Ranjan, A., Seidel, C.
W., Tsukiyama, T., Mizushima, W., Hayashi, M., Ohkawa, Y., Conaway, J.
W., Conaway, R. C., and Hatakeyama, S. (2015) MED26 regulates the
transcription of snRNA genes through the recruitment of little
elongation complex, Nat. Commun., 6, 5941.
90.Dahlberg, O., Shilkova, O., Tang, M., Holmqvist,
P. H., and Mannervik, M. (2015) P-TEFb, the super elongation complex
and mediator regulate a subset of non-paused genes during early
Drosophila embryo development, PLoS Genet., 11,
e1004971.
91.Uthe, H., Vanselow, J. T., and Schlosser, A.
(2017) Proteomic analysis of the Mediator complex interactome in
Saccharomyces cerevisiae, Sci. Rep., 7, 43584.
92.Huang, Y., Li, W., Yao, X., Lin, Q. J., Yin, J.
W., Liang, Y., Heiner, M., Tian, B., Hui, J., and Wang, G. (2012)
Mediator complex regulates alternative mRNA processing via the MED23
subunit, Mol. Cell, 45, 459-469.
93.Chereji, R. V., Bharatula, V., Elfving, N.,
Blomberg, J., Larsson, M., Morozov, A. V., Broach, J. R., and
Bjorklund, S. (2017) Mediator binds to boundaries of chromosomal
interaction domains and to proteins involved in DNA looping, RNA
metabolism, chromatin remodeling, and actin assembly, Nucleic Acids
Res., 45, 8806-8821.
94.Schneider, M., Hellerschmied, D., Schubert, T.,
Amlacher, S., Vinayachandran, V., Reja, R., Pugh, B. F., Clausen, T.,
and Kohler, A. (2015) The nuclear pore-associated TREX-2 complex
employs Mediator to regulate gene expression, Cell, 162,
1016-1028.
95.Liu, Z., and Myers, L. C. (2012) Med5(Nut1) and
Med17(Srb4) are direct targets of mediator histone H4 tail
interactions, PLoS One, 7, e38416.
96.Zhu, X., Zhang, Y., Bjornsdottir, G., Liu, Z.,
Quan, A., Costanzo, M., Davila Lopez, M., Westholm, J. O., Ronne, H.,
Boone, C., Gustafsson, C. M., and Myers, L. C. (2011) Histone
modifications influence mediator interactions with chromatin,
Nucleic Acids Res., 39, 8342-8354.
97.Lemieux, K., and Gaudreau, L. (2004) Targeting of
Swi/Snf to the yeast GAL1 UAS G requires the Mediator, TAF IIs, and RNA
polymerase II, EMBO J., 23, 4040-4050.
98.Sharma, V. M., Li, B., and Reese, J. C. (2003)
SWI/SNF-dependent chromatin remodeling of RNR3 requires TAF(II)s and
the general transcription machinery, Genes Dev., 17,
502-515.
99.Fukasawa, R., Tsutsui, T., Hirose, Y., Tanaka,
A., and Ohkuma, Y. (2012) Mediator CDK subunits are platforms for
interactions with various chromatin regulatory complexes, J.
Biochem., 152, 241-249.
100.Khorosjutina, O., Wanrooij, P. H., Walfridsson,
J., Szilagyi, Z., Zhu, X., Baraznenok, V., Ekwall, K., and Gustafsson,
C. M. (2010) A chromatin-remodeling protein is a component of fission
yeast mediator, J. Biol. Chem., 285, 29729-29737.
101.Lin, J. J., Lehmann, L. W., Bonora, G.,
Sridharan, R., Vashisht, A. A., Tran, N., Plath, K., Wohlschlegel, J.
A., and Carey, M. (2011) Mediator coordinates PIC assembly with
recruitment of CHD1, Genes Dev., 25, 2198-2209.
102.Bhoite, L. T., Yu, Y., and Stillman, D. J.
(2001) The Swi5 activator recruits the Mediator complex to the HO
promoter without RNA polymerase II, Genes Dev., 15,
2457-2469.
103.Yoon, S., Qiu, H., Swanson, M. J., and
Hinnebusch, A. G. (2003) Recruitment of SWI/SNF by Gcn4p does not
require Snf2p or Gcn5p but depends strongly on SWI/SNF integrity, SRB
mediator, and SAGA, Mol. Cell. Biol., 23, 8829-8845.
104.Ansari, S. A., Paul, E., Sommer, S., Lieleg,
C., He, Q., Daly, A. Z., Rode, K. A., Barber, W. T., Ellis, L. C.,
LaPorta, E., Orzechowski, A. M., Taylor, E., Reeb, T., Wong, J.,
Korber, P., and Morse, R. H. (2014) Mediator, TATA-binding protein, and
RNA polymerase II contribute to low histone occupancy at active gene
promoters in yeast, J. Biol. Chem., 289, 14981-14995.
105.Acevedo, M. L., and Kraus, W. L. (2003)
Mediator and p300/CBP-steroid receptor coactivator complexes have
distinct roles, but function synergistically, during estrogen receptor
alpha-dependent transcription with chromatin templates, Mol. Cell.
Biol., 23, 335-348.
106.Aranda-Orgilles, B., Saldana-Meyer, R., Wang,
E., Trompouki, E., Fassl, A., Lau, S., Mullenders, J., Rocha, P. P.,
Raviram, R., Guillamot, M., Sanchez-Diaz, M., Wang, K., Kayembe, C.,
Zhang, N., Amoasii, L., Choudhuri, A., Skok, J. A., Schober, M.,
Reinberg, D., Sicinski, P., Schrewe, H., Tsirigos, A., Zon, L. I., and
Aifantis, I. (2016) MED12 regulates HSC-specific enhancers
independently of mediator kinase activity to control hematopoiesis,
Cell Stem Cell, 19, 784-799.
107.Huang, Z. Q., Li, J., Sachs, L. M., Cole, P.
A., and Wong, J. (2003) A role for cofactor–cofactor and
cofactor–histone interactions in targeting p300, SWI/SNF and
Mediator for transcription, EMBO J., 22, 2146-2155.
108.Ebmeier, C. C., and Taatjes, D. J. (2010)
Activator–Mediator binding regulates Mediator–cofactor
interactions, Proc. Natl. Acad. Sci. USA, 107,
11283-11288.
109.Meyer, K. D., Donner, A. J., Knuesel, M. T.,
York, A. G., Espinosa, J. M., and Taatjes, D. J. (2008) Cooperative
activity of cdk8 and GCN5L within Mediator directs tandem
phosphoacetylation of histone H3, EMBO J., 27,
1447-1457.
110.Bhaumik, S. R., and Green, M. R. (2001) SAGA is
an essential in vivo target of the yeast acidic activator Gal4p,
Genes Dev., 15, 1935-1945.
111.Larschan, E., and Winston, F. (2005) The
Saccharomyces cerevisiae Srb8–Srb11 complex
functions with the SAGA complex during Gal4-activated transcription,
Mol. Cell. Biol., 25, 114-123.
112.Bryant, G. O., and Ptashne, M. (2003)
Independent recruitment in vivo by Gal4 of two complexes
required for transcription, Mol. Cell, 11, 1301-1309.
113.Leroy, C., Cormier, L., and Kuras, L. (2006)
Independent recruitment of mediator and SAGA by the activator Met4,
Mol. Cell. Biol., 26, 3149-3163.
114.Liu, X., Vorontchikhina, M., Wang, Y. L.,
Faiola, F., and Martinez, E. (2008) STAGA recruits Mediator to the MYC
oncoprotein to stimulate transcription and cell proliferation, Mol.
Cell. Biol., 28, 108-121.
115.Malik, S., Wallberg, A. E., Kang, Y. K., and
Roeder, R. G. (2002) TRAP/SMCC/mediator-dependent transcriptional
activation from DNA and chromatin templates by orphan nuclear receptor
hepatocyte nuclear factor 4, Mol. Cell. Biol., 22,
5626-5637.
116.Krebs, A. R., Demmers, J., Karmodiya, K.,
Chang, N. C., Chang, A. C., and Tora, L. (2010) ATAC and Mediator
coactivators form a stable complex and regulate a set of non-coding RNA
genes, EMBO Rep., 11, 541-547.
117.Yao, X., Tang, Z., Fu, X., Yin, J., Liang, Y.,
Li, C., Li, H., Tian, Q., Roeder, R. G., and Wang, G. (2015) The
Mediator subunit MED23 couples H2B mono-ubiquitination to
transcriptional control and cell fate determination, EMBO J.,
34, 2885-2902.
118.Law, M. J., and Ciccaglione, K. (2015)
Fine-tuning of histone H3 Lys4 methylation during pseudohyphal
differentiation by the CDK submodule of RNA polymerase II,
Genetics, 199, 435-453.
119.Ding, N., Zhou, H., Esteve, P. O., Chin, H. G.,
Kim, S., Xu, X., Joseph, S. M., Friez, M. J., Schwartz, C. E., Pradhan,
S., and Boyer, T. G. (2008) Mediator links epigenetic silencing of
neuronal gene expression with x-linked mental retardation, Mol.
Cell, 31, 347-359.
120.Tsutsui, T., Fukasawa, R., Shinmyouzu, K.,
Nakagawa, R., Tobe, K., Tanaka, A., and Ohkuma, Y. (2013) Mediator
complex recruits epigenetic regulators via its two cyclin-dependent
kinase subunits to repress transcription of immune response genes,
J. Biol. Chem., 288, 20955-20965.
121.D’Urso, A., Takahashi, Y. H., Xiong, B.,
Marone, J., Coukos, R., Randise-Hinchliff, C., Wang, J. P.,
Shilatifard, A., and Brickner, J. H. (2016) Set1/COMPASS and Mediator
are repurposed to promote epigenetic transcriptional memory,
eLife, 5.
122.McCleland, M. L., Soukup, T. M., Liu, S. D.,
Esensten, J. H., De Sousa e Melo, F., Yaylaoglu, M., Warming, S.,
Roose-Girma, M., and Firestein, R. (2015) Cdk8 deletion in the Apc(Min)
murine tumour model represses EZH2 activity and accelerates
tumourigenesis, J. Pathol., 237, 508-519.
123.Fukasawa, R., Iida, S., Tsutsui, T., Hirose,
Y., and Ohkuma, Y. (2015) Mediator complex cooperatively regulates
transcription of retinoic acid target genes with Polycomb Repressive
Complex 2 during neuronal differentiation, J. Biochem.,
158, 373-384.
124.Papadopoulou, T., Kaymak, A., Sayols, S., and
Richly, H. (2016) Dual role of Med12 in PRC1-dependent gene repression
and ncRNA-mediated transcriptional activation, Cell Cycle,
15, 1479-1493.
125.Englert, N. A., Luo, G., Goldstein, J. A., and
Surapureddi, S. (2015) Epigenetic modification of histone 3 lysine 27:
mediator subunit MED25 is required for the dissociation of polycomb
repressive complex 2 from the promoter of cytochrome P450 2C9, J.
Biol. Chem., 290, 2264-2278.
126.Zhu, X., Liu, B., Carlsten, J. O., Beve, J.,
Nystrom, T., Myers, L. C., and Gustafsson, C. M. (2011) Mediator
influences telomeric silencing and cellular life span, Mol. Cell.
Biol., 31, 2413-2421.
127.Kim, Y. J., Zheng, B., Yu, Y., Won, S. Y., Mo,
B., and Chen, X. (2011) The role of Mediator in small and long
noncoding RNA production in Arabidopsis thaliana, EMBO
J., 30, 814-822.
128.Oya, E., Kato, H., Chikashige, Y., Tsutsumi,
C., Hiraoka, Y., and Murakami, Y. (2013) Mediator directs
co-transcriptional heterochromatin assembly by RNA
interference-dependent and -independent pathways, PLoS Genet.,
9, e1003677.
129.Thorsen, M., Hansen, H., Venturi, M., Holmberg,
S., and Thon, G. (2012) Mediator regulates non-coding RNA transcription
at fission yeast centromeres, Epigenetics Chromatin, 5,
19.
130.Peng, J., and Zhou, J. Q. (2012) The
tail-module of yeast Mediator complex is required for telomere
heterochromatin maintenance, Nucleic Acids Res., 40,
581-593.
131.Chen, Z., Zhang, C., Wu, D., Chen, H., Rorick,
A., Zhang, X., and Wang, Q. (2011) Phospho-MED1-enhanced UBE2C locus
looping drives castration-resistant prostate cancer growth, EMBO
J., 30, 2405-2419.
132.Park, S. W., Li, G., Lin, Y. P., Barrero, M.
J., Ge, K., Roeder, R. G., and Wei, L. N. (2005) Thyroid
hormone-induced juxtaposition of regulatory elements/factors and
chromatin remodeling of Crabp1 dependent on MED1/TRAP220, Mol.
Cell, 19, 643-653.
133.Saramaki, A., Diermeier, S., Kellner, R.,
Laitinen, H., Vaisanen, S., and Carlberg, C. (2009) Cyclical chromatin
looping and transcription factor association on the regulatory regions
of the p21 (CDKN1A) gene in response to 1alpha,25-dihydroxyvitamin D3,
J. Biol. Chem., 284, 8073-8082.
134.Wang, Q., Carroll, J. S., and Brown, M. (2005)
Spatial and temporal recruitment of androgen receptor and its
coactivators involves chromosomal looping and polymerase tracking,
Mol. Cell, 19, 631-642.
135.Muto, A., Ikeda, S., Lopez-Burks, M. E.,
Kikuchi, Y., Calof, A. L., Lander, A. D., and Schilling, T. F. (2014)
Nipbl and mediator cooperatively regulate gene expression to control
limb development, PLoS Genet., 10, e1004671.
136.Fournier, M., Bourriquen, G., Lamaze, F. C.,
Cote, M. C., Fournier, E., Joly-Beauparlant, C., Caron, V., Gobeil, S.,
Droit, A., and Bilodeau, S. (2016) FOXA and master transcription
factors recruit Mediator and Cohesin to the core transcriptional
regulatory circuitry of cancer cells, Sci. Rep., 6,
34962.
137.Phillips-Cremins, J. E., Sauria, M. E., Sanyal,
A., Gerasimova, T. I., Lajoie, B. R., Bell, J. S., Ong, C. T., Hookway,
T. A., Guo, C., Sun, Y., Bland, M. J., Wagstaff, W., Dalton, S.,
McDevitt, T. C., Sen, R., Dekker, J., Taylor, J., and Corces, V. G.
(2013) Architectural protein subclasses shape 3D organization of
genomes during lineage commitment, Cell, 153,
1281-1295.
138.Krivega, I., and Dean, A. (2017) LDB1-mediated
enhancer looping can be established independent of mediator and
cohesin, Nucleic Acids Res., 45, 8255-8268.
139.Levine, M., Cattoglio, C., and Tjian, R. (2014)
Looping back to leap forward: transcription enters a new era,
Cell, 157, 13-25.
140.Dobi, K. C., and Winston, F. (2007) Analysis of
transcriptional activation at a distance in Saccharomyces
cerevisiae, Mol. Cell. Biol., 27, 5575-5586.
141.Mukundan, B., and Ansari, A. (2013)
Srb5/Med18-mediated termination of transcription is dependent on gene
looping, J. Biol. Chem., 288, 11384-11394.
142.Hsieh, T. H., Weiner, A., Lajoie, B., Dekker,
J., Friedman, N., and Rando, O. J. (2015) Mapping nucleosome resolution
chromosome folding in yeast by micro-C, Cell, 162,
108-119.
143.Lai, F., Orom, U. A., Cesaroni, M., Beringer,
M., Taatjes, D. J., Blobel, G. A., and Shiekhattar, R. (2013)
Activating RNAs associate with Mediator to enhance chromatin
architecture and transcription, Nature, 494, 497-501.
144.Hsieh, C. L., Fei, T., Chen, Y., Li, T., Gao,
Y., Wang, X., Sun, T., Sweeney, C. J., Lee, G. S., Chen, S., Balk, S.
P., Liu, X. S., Brown, M., and Kantoff, P. W. (2014) Enhancer RNAs
participate in androgen receptor-driven looping that selectively
enhances gene activation, Proc. Natl. Acad. Sci. USA,
111, 7319-7324.
145.Step, S. E., Lim, H. W., Marinis, J. M.,
Prokesch, A., Steger, D. J., You, S. H., Won, K. J., and Lazar, M. A.
(2014) Anti-diabetic rosiglitazone remodels the adipocyte transcriptome
by redistributing transcription to PPARgamma-driven enhancers, Genes
Dev., 28, 1018-1028.
146.Siersbaek, R., Madsen, J. G. S., Javierre, B.
M., Nielsen, R., Bagge, E. K., Cairns, J., Wingett, S. W., Traynor, S.,
Spivakov, M., Fraser, P., and Mandrup, S. (2017) Dynamic rewiring of
promoter-anchored chromatin loops during adipocyte differentiation,
Mol. Cell, 66, 420-435.
147.Liu, J., Perumal, N. B., Oldfield, C. J., Su,
E. W., Uversky, V. N., and Dunker, A. K. (2006) Intrinsic disorder in
transcription factors, Biochemistry, 45, 6873-6888.
148.Wright, P. E., and Dyson, H. J. (2015)
Intrinsically disordered proteins in cellular signalling and
regulation, Nat. Rev. Mol. Cell Biol., 16, 18-29.
149.Larson, A. G., Elnatan, D., Keenen, M. M.,
Trnka, M. J., Johnston, J. B., Burlingame, A. L., Agard, D. A.,
Redding, S., and Narlikar, G. J. (2017) Liquid droplet formation by
HP1alpha suggests a role for phase separation in heterochromatin,
Nature, 547, 236-240.
150.Berman, H. M., Westbrook, J., Feng, Z.,
Gilliland, G., Bhat, T. N., Weissig, H., Shindyalov, I. N., and Bourne,
P. E. (2000) The protein data bank, Nucleic Acids Res.,
28, 235-242.
151.Lawson, C. L., Baker, M. L., Best, C., Bi, C.,
Dougherty, M., Feng, P., Van Ginkel, G., Devkota, B., Lagerstedt, I.,
Ludtke, S. J., Newman, R. H., Oldfield, T. J., Rees, I., Sahni, G.,
Sala, R., Velankar, S., Warren, J., Westbrook, J. D., Henrick, K.,
Kleywegt, G. J., Berman, H. M., and Chiu, W. (2011) EMDataBank.org:
unified data resource for CryoEM, Nucleic Acids Res., 39,
456-464.
152.Schneider, E. V., Bottcher, J., Blaesse, M.,
Neumann, L., Huber, R., and Maskos, K. (2011) The structure of
CDK8/CycC implicates specificity in the CDK/cyclin family and reveals
interaction with a deep pocket binder, J. Mol. Biol.,
412, 251-266.