Potential Markers of Autoimmune Diseases, Alleles rs115662534(T) and rs548231435(C), Disrupt the Binding of Transcription Factors STAT1 and EBF1 to the Regulatory Elements of Human CD40 Gene
L. V. Putlyaeva1,a, D. E. Demin1,2, K. V. Korneev1,3, A. S. Kasyanov4, K. A. Tatosyan1, I. V. Kulakovskiy1,4,5, D. V. Kuprash1,2,3,b, and A. M. Schwartz1,2,c*
1Engelhardt Institute of Molecular Biology, Russian Academy of Sciences, 119991 Moscow, Russia2Moscow Institute of Physics and Technology, Department of Molecular and Biological Physics, 141701 Dolgoprudny, Moscow Region, Russia
3Lomonosov Moscow State University, Faculty of Biology, 119991 Moscow, Russia
4Vavilov Institute of General Genetics, Russian Academy of Sciences, 119333 Moscow, Russia
5Institute of Mathematical Problems of Biology, Keldysh Institute of Applied Mathematics, Russian Academy of Sciences, 142290 Pushchino, Moscow Region, Russia
* To whom correspondence should be addressed.
Received July 13, 2018; Revision received September 4, 2018
CD40 receptor is expressed on B lymphocytes and other professional antigen-presenting cells. The binding of CD40 to its ligand CD154 on the surface of T helper cells plays an important role in the activation of B lymphocytes required for production of antibodies, in particular, against autoantigens. Association of several single nucleotide polymorphisms (SNPs) located in the non-coding areas of human CD40 locus with the elevated risk of autoimmune diseases has been demonstrated. The most studied of these SNPs is rs4810485 located in the first intron of the CD40 gene. Expression of the CD40 gene in B lymphocytes of donors homozygous for the common allelic variant of this polymorphism (G) is higher than in B cells from donors carrying the minor (T) variant. We investigated the enhancer activity of this fragment of the CD40 locus in human B cell lines and showed that it is independent on the rs4810485 alleles. However, the minor allelic variants of the rs4810485-linked SNPs rs548231435 and rs115662534 were associated with a significant decrease in the activity of the CD40 promoter due to the impairments in the binding of EBF1 and STAT1 transcription factors, respectively.
KEY WORDS: autoimmune diseases, polymorphism, CD40, transcription regulationDOI: 10.1134/S0006297918120118
Abbreviations: SNP, single nucleotide polymorphism; TNF, tumor necrosis factor.
The CD40 gene encodes CD40 receptor protein belonging to the
tumor necrosis factor (TNF) superfamily. CD40 is expressed on the
surface of B cells, macrophages, dendritic cells, fibroblasts,
endothelial and smooth muscle cells, etc. . Interactions between CD40
and its cognate ligand CD154 expressed on the T cells play an important
role in the B cell activation, antibody isotype switching, and
formation of memory B cells . CD40 expressed on dendritic cells is
essential for the primary T cell immune response against a broad range
of various immunogens . CD40 expressed on the surface of macrophages
ensures efficient cell response against intracellular bacteria .
Similarly to various regulators of adaptive immunity, CD40 also plays a
role in the development of autoimmune disorders, such as autoimmune
thyroiditis, rheumatoid arthritis, systemic lupus erythematosus,
multiple sclerosis, type 1 diabetes, and psoriasis . Blocking the
CD154–CD40 axis interferes with the development of autoimmune
pathologies in mice and non-human primates . However, clinical trials
with CD40-suppressing antibodies have been terminated mainly due to the
adverse side effects of the tested preparations .
The relationship between single nucleotide polymorphisms (SNPs) in the non-coding region of the CD40 gene and the risk of autoimmune disorders has been demonstrated. Moreover, for some of the SNPs, molecular mechanisms underlying this relationship were proposed. For instance, the rs1883832 polymorphism associated with the risk of Graves’ disease is located within the Kozak sequence, so that the CD40 mRNA carrying the minor T allele is translated 15% less efficiently than the mRNA with the C allele . On the other hand, the polymorphism rs4810485 found in the first intron of the CD40 gene was shown to be associated with a risk of multiple autoimmune diseases . Donors homozygous by the major rs4810485 variant (G) have an elevated level of CD40 mRNA in the peripheral blood cells , as well as an increased number of B cells with upregulated CD40 expression .
Here, we used the B lymphoblastoid MP1 cell line bearing the reporter gene to demonstrate that the activity of the regulatory element within the first intron of the CD40 gene does not depend on the rs4810485 allelic variant. We also searched for the linked polymorphisms that potentially influence the CD40 gene expression using an approach earlier applied to examine the CD25 gene . Two rs4810485-linked candidate SNPs (rs548231435 and rs115662534) in the CD40 gene were identified whose allelic variants significantly affected the activity of the CD40 gene promoter by altering the binding of the EBF1 and STAT1 transcription factors, respectively.
MATERIALS AND METHODS
Cell line, transfection, EBF1 and STAT1 gene knockdown, luciferase reporter assay. MP1 B lymphoblastoid cells (kindly provided by E. A. Clark, University of Washington, USA) were grown in RPMI1640 medium (PanEko, Russia) supplemented with 10% fetal bovine serum (Biosera, France), 2 mM L-glutamine, penicillin/streptomycin, and 1 mM sodium pyruvate (Thermo Fisher Scientific, USA). EBF1 and STAT1 gene knockdowns were performed as published in using short interfering RNAs (siRNAs) described in and control siRNAs designed using the Invivogen siRNA Wizard v3.1 tool (Table 1). Electroporation and luciferase reporter assay were performed as described in .
Table 1. RNA oligonucleotides bearing two
3-deoxyribonucleotides for generating siRNAs
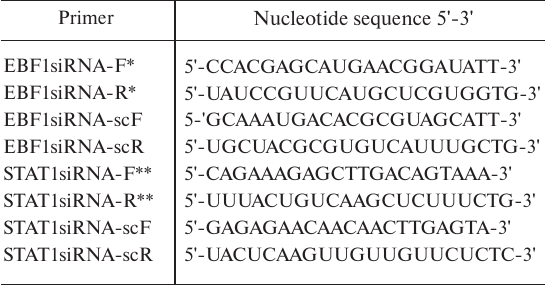
* See [22]
** See [21]
Table 2. Primers for cloning DNA regulatory
elements and for introduction of minor polymorphic variants

Plasmid constructs bearing the reporter gene. Plasmids containing the promoter (886 bp) and putative enhancer (1353 bp) sequences from the CD40 gene and the control element from the first intron of the STAT3 gene (~1100 bp) lacking the enhancer activity were amplified with the primers shown in Table 2 using MP1 cell genomic DNA as a template. Genomic DNA was isolated with a Genomic DNA Purification Kit (Thermo Fisher Scientific). The promoter sequence was cloned into the pGL3-basic luciferase reporter vector (Promega, USA) by the HindIII and PagI sites. The putative enhancer and the control element were cloned by the BamHI and SalI sites into the pGL3-basic vector bearing the CD40 gene promoter. Single nucleotide substitutions were introduced into the promoter and putative enhancer sequences by the two-stage PCR with polymorphism-specific internal overlapping primers and common external primers (CD40pr-H3F/Pag1R and CD40enh1-BH1F/SalR) (Table 2). The regimes for the single- and two-stage PCRs were described in . All mutant PCR products were used to replace the original sequences in the corresponding luciferase reporter vectors. The plasmids were isolated using NucleoBond Xtra Midi Kit (Macherey-Nagel, Germany) and verified by Sanger sequencing (Genome Common Use Center, Moscow).
Table 3. Primers for generation and
quantitative analysis of amplicons used in immunoprecipitation
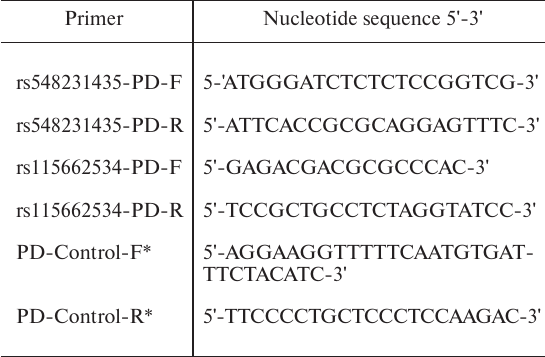
* See [18].
In vitro immunoprecipitation of DNA–protein complexes (pull-down assay). To evaluate the effect of various allelic polymorphisms on the binding of transcription factors, we studied immunoprecipitation of the obtained PCR products as described in . For this, we amplified DNA sequences containing both variants of the rs548231435 and rs115662534 polymorphisms with the rs548231435-PD-F/R and rs115662534-PD-F/R primer pairs (Table 3) using the plasmids carrying these polymorphic variants as templates. A fragment of the IL2RA gene lacking the binding sites for EBF1 and STAT1 was amplified with the PD-Control-F/R primers ([18], Table 3) and MP1 cell genomic DNA as a template to be used as a control. All amplified fragments had similar length: amplicon with the rs548231435 polymorphism – 196 bp, amplicon with the rs115662534 polymorphism – 197 bp, control amplicon – 189 bp. DNA–protein complexes were immunoprecipitated from the MP1 cell nuclear extracts as described in using the following antibodies: rabbit polyclonal IgGs against EBF1 (Antibodies-online Inc., USA) and ESR2 (Thermo Fisher Scientific), anti-STAT1 and anti-ESR1 mouse IgGs (Abcam, UK).
Table 4. Primers for analysis of
transcription factor expression

RNA isolation, cDNA synthesis, and quantitative real-time PCR. Total RNA from MP1 cells was isolated using the ExtractRNA reagent (Evrogen, Russia). Reverse transcription was performed by using 2 µg total RNA and an MMLV RT Kit (Evrogen). Quantitative PCR was performed as published before using the primers shown in Table 4.
RESULTS
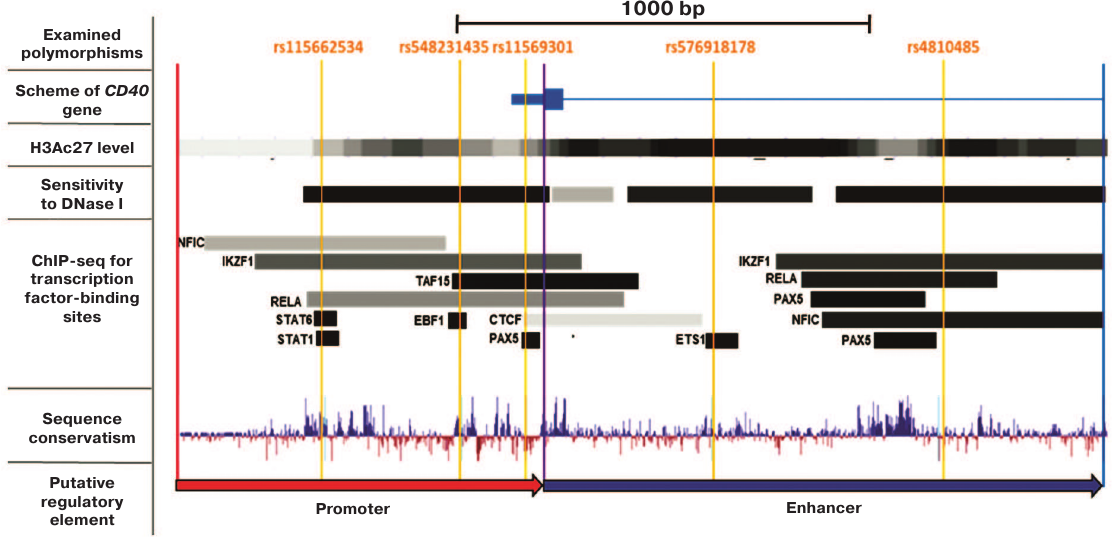
The enhancer activity of the rs4810485-containing region of the CD40 gene first intron does not depend on the SNP allelic variant. The effect of the rs4810485 polymorphism on the CD40 gene expression was examined by cloning the CD40 promoter (–886/0 bp upstream of the translation initiation site) and its putative enhancer (0/1353 bp upstream of the translation initiation site) containing this SNP into the reporter gene plasmid coding for firefly luciferase (FLUC). According to the ENCODE project data , similar regions in the genome of the GM12878 B cell line display features typical for promoter and enhancer elements, e.g., high level of acetylation of lysine 27 in histone H3 (H3K27Ac), hypersensitivity to DNase I, the presence of transcription factor-binding motifs, and sequence conservatism (Fig. 1). Although the region –711/0 bp was described earlier as the CD40 promoter , here, we used the –886/0 bp region containing all transcription factor-binding sites identified by ChIP-Seq assay , 26] (Fig. 1).
Fig. 1. Positions of the examined polymorphisms and putative regulatory elements within the CD40 gene locus (based on data visualized by the UCSC and GTRD genome browser). Red and blue arrows denote the promoter and putative enhancer in the CD40 gene, respectively. Orange lines mark position of the examined polymorphisms. Based on data visualized by UCSC and GTRD genome browser.
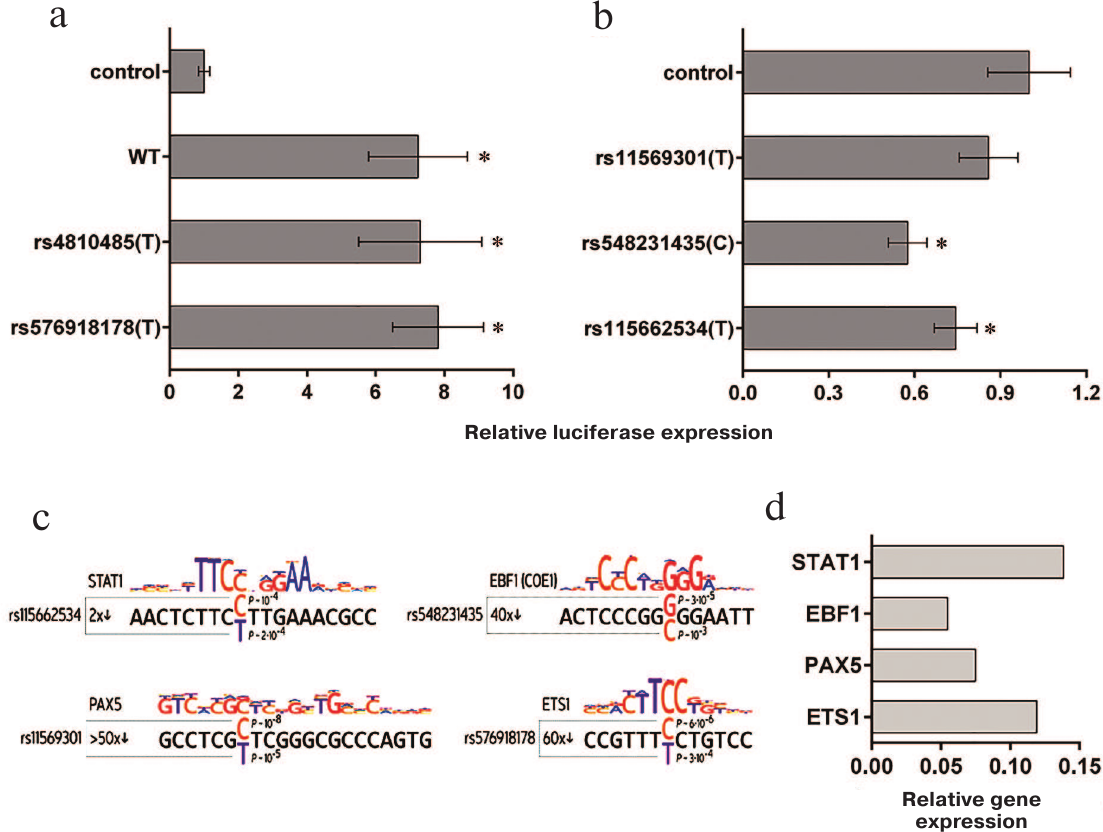
We found that the putative enhancer element was highly efficient in the MP1 cells and upregulated activity of the CD40 gene promoter approximately 7-fold (Fig. 2a). However, no significant difference in the activity of enhancer elements bearing alternative rs4810485 allelic polymorphisms was found (Fig. 2a).
Fig. 2. Effect of the examined polymorphisms in the CD40 gene locus on the activity of the CD40 putative enhancer (a) and promoter (b) in MP1 cells. Activity of the reporter plasmids was normalized to the activity of the control plasmid containing the CD40 promoter with major examined polymorphic variants and a fragment of the STAT3 gene locus lacking the enhancer activity (control). The WT plasmid contained the CD40 promoter and putative enhancer bearing all major examined polymorphic variants. The rs4810485(T) and rs576918178(T) vectors carried minor variants of enhancer polymorphisms. Plasmids rs11569301(T), rs548231435(C), and rs115662534(T) differed from the control plasmid by carrying the minor variants of the corresponding polymorphisms. The data from at least seven experiments are presented as mean ± SEM in both plots; *, significant difference between the plasmid with minor polymorphic variants and WT control (p < 0.05, Student’s t-test). c) Examined polymorphic variants and visualization of the sequences involved in the binding of transcription factors (modality of the changes in the affinity and p-values for the alternative alleles) are shown . d) mRNAs levels for transcription factors in MP1 cells normalized to the amount of β-actin mRNA (p < 0.05, Student’s t-test).
Identification and functional analysis of rare rs4810485-linked polymorphic alleles. We suggested that the association between the minor allele of the rs4810485 polymorphism and reduced risk of autoimmune disorders might be related to the effects caused by the minor variants of other linked SNPs that have been overlooked in the current genome-wide studies due to a low population prevalence (less than 5%) and insignificantly elevated morbidity (less than 3-fold) . We have searched for SNPs that were located in the active chromatin region and resided directly within the transcription factor-binding sites, whose interaction with the corresponding transcription factors in B cells have been verified experimentally. The information on the interactions between the examined motifs and various transcription factors was taken from the earlier study , ENCODE project , and new GTRD database containing results from more than 12,000 experiments . Analysis of these data resulted in the identification of another enhancer polymorphism (rs576918178) located in the ETS1-binding site and three polymorphisms (rs11569301, rs548231435, and rs115662534) located in the binding sites for PAX5, EBF1, and STAT1, respectively, in the CD40 promoter (Figs. 1 and 2c).
The effect of alternative alleles of selected polymorphisms on the regulatory activity of the corresponding CD40 gene regions was tested in MP1 B lymphoblastoid cell line. As demonstrated earlier , all genes encoding the above-mentioned transcription factors are expressed in these cells at relatively high levels (Fig. 2d). We found that the activity of the reporter plasmid bearing the CD40 gene promoter and putative enhancer did not depend on the allelic variant of rs576918178 (Fig. 2a), whereas the activity of the vector bearing the CD40 gene promoter only was independent of the rs11569301 allelic variants (Fig. 2b). On the other hand, introduction of the minor allelic variants rs548231435(C) and rs115662534(T) substantially reduced the promoter activity (Fig. 2b).
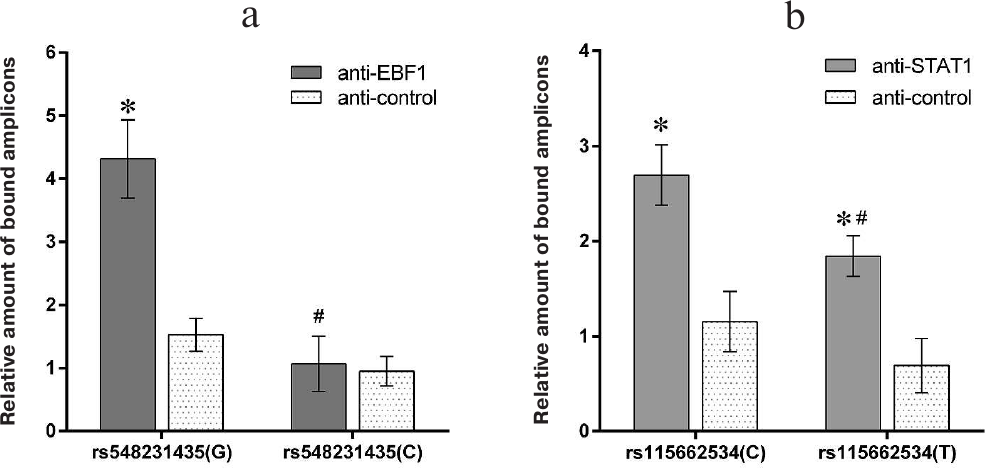
Minor variants rs548231435(C) and rs115662534(T) impair the binding of the EBF1 and STAT transcription factors to the CD40 gene promoter in vitro. Immunoprecipitation of DNA–protein complex (pull-down assay) demonstrated that the amplicon bearing the more common rs548231435(G) allele binds EBF1 three times better compared to the amplicon containing the minor rs548231435(C) allele or the control amplicon fully lacking the EBF1-binding sites (Fig. 3a). Moreover, the binding of STAT1 to the amplicon containing the less common allele rs115662534(T) was lower by 30% that the binding of the more frequent rs115662534(C) polymorphic variant. Both variants, however, bound to STAT1 significantly better than the control amplicon (Fig. 3b), which might be due to the fact that the rs115662534 polymorphism is located outside of the conserved STAT1-binding site (Fig. 2c). The lack of ESR1/2-binding site in all the examined amplicons allowed us to use anti-ESR2 and anti-ESR1 antibodies as isotype controls. All the amplicons were precipitated with control antibodies with a similar efficiency (Fig. 3).
Fig. 3. Effect of the rs548231435 (a) and rs115662534 (b) polymorphisms on the ability of the CD40 gene promoter to bind transcription factors STAT1 and EBF1 in MP1 cells. Amplicons containing various polymorphic variants were precipitated from the nuclear extracts of MP1 cells using anti-STAT1 and anti-EBF1 antibodies. The amount of the amplicons pulled down with the antibodies was estimated by real-time PCR and normalized to the amount of bound amplicon lacking the EBF1- and STAT1-binding sites. Samples treated with the isotype control antibodies were used as a negative control. The data from at least three experiments are presented as mean ± SEM in both plots; *, significant difference between the examined sample and negative control; #, significant difference between vectors bearing minor polymorphic variants and wild-type sequence (p < 0.05, Student’s t-test).
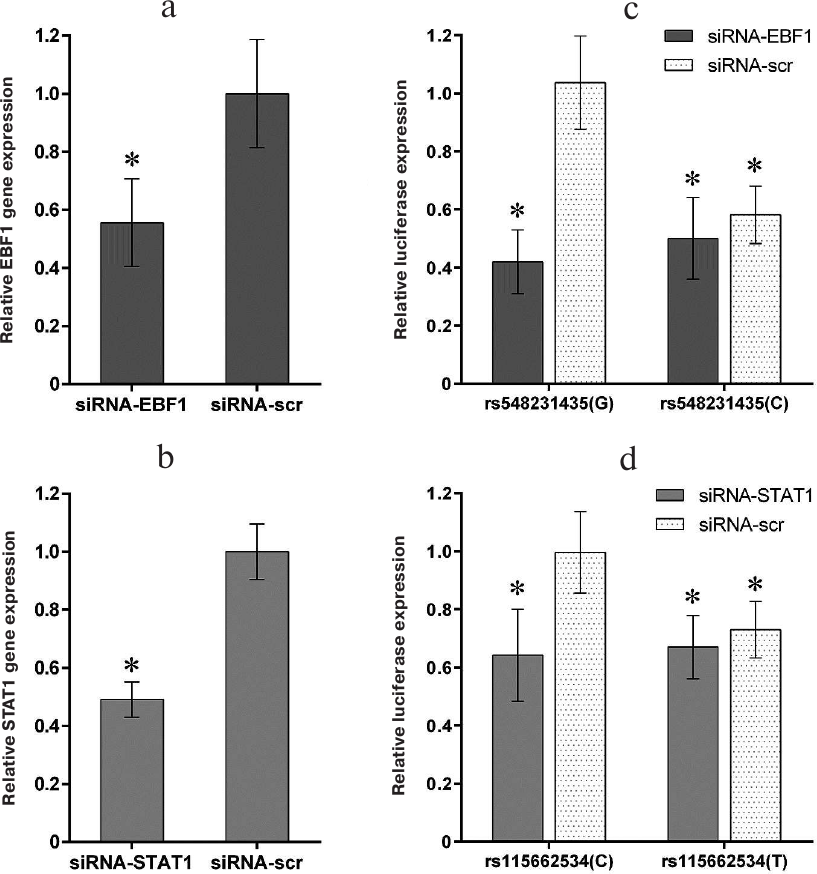
Suppression of the EBF1 and STAT1 expression affects the activity of the CD40 gene promoter depending on the polymorphic allelic variants of rs548231435 and rs115662534. To verify the specificity of EBF1 and STAT1 effects on the transcriptional activity of the examined CD40 gene promoter variants in B cells, expression of the EBF1 and STAT1 genes were downregulated using short interfering RNAs (siRNAs) that reduced the expression of these genes ~2-fold (according to real-time quantitative PCR) (Fig. 4, a and b). Suppression of the EBF1 expression by 2-fold resulted in 50-60% decrease in the promoter activity of the wild-type CD40 gene but had virtually no effect on the activity of the promoter bearing the minor allele rs548231435(C) (Fig. 4c), which correlates well with weak interaction between this rare polymorphic variant of the CD40 gene promoter and EBF1 protein.
Fig. 4. Suppression of STAT1 and EBF1 gene expression in MP1 B lymphoblastoid cell line reduces the activity of polymorphic variants of the CD40 promoter bearing rare rs548231435 and rs115662534 alleles: suppression of EBF1 (a) and STAT1 (b) mRNA expression by specific siRNAs in MP1 cells; activity of CD40 gene promoter variants bearing rs548231435 and rs115662534 alleles upon decreased expression of EBF1 (c) and STAT1 (d); siRNA-scr, control siRNA with scrambled nucleotide sequence. The data from at least five experiments are presented as mean ± SEM; * in (a) and (b), significant difference with the control siRNA-scr; * in (c) and (d), significant difference with the wild-type promoter in the absence of specific siRNA (p < 0.05, Student’s t-test).
Downregulation of the STAT1 expression decreased the promoter activity of the wild-type CD40 gene by 30-40%, whereas introduction of the minor rs115662534(T) variant attenuated this effect (Fig. 4d). Because the used siRNAs were strictly specific to the STAT1 gene sequence , it can be suggested that the observed effect was due to the binding of STAT1, and not of other STAT proteins, to the site bearing the rs115662534 polymorphism.
DISCUSSION
It has been demonstrated before that expression of the CD40 protein is elevated during the development of various autoimmune diseases . The upregulation of CD40 expression in lymphocytes and antigen-presenting cells in autoimmune processes may be related to the increased activity of the EBF1 and STAT1 transcription factors . Therefore, it is reasonable to suggest that rare alleles of rs548231435 and rs115662534 polymorphisms could play a protective role in the development of autoimmune diseases via affecting the binding of EBF1 and STAT1 to the CD40 gene promoter. Low frequency of these such protective alleles may result from their rejection in the process of evolution, since low body capacity to efficiently combat infectious diseases poses serious threat, especially in the pre-reproductive period. On the other hand, alleles increasing the risk of autoimmune disorders exhibit activity in adulthood and, therefore, affect less the ability to produce an offspring . Hence, it can be expected that the majority of people possess alleles associated with an increased immune response.
At present, the incidence rate and the threat of numerous infectious diseases have been markedly decreased due to the wide use of vaccines and antibiotics. On the contrary, the morbidity of diseases associated with autoimmune processes (e.g., type I and II diabetes mellitus, rheumatoid arthritis, neurodegenerative diseases, etc.) has substantially increased over the last centuries primarily due to the life expectancy increase . Because of this, identification of rare polymorphisms in genes encoding immune checkpoint regulators associated with a decreased risk of various autoimmune responses is of special importance. With the increasing efficacy and safety of genome editing techniques, introduction of such alleles into the genome of human hematopoietic cells might become an attractive approach for the therapy of autoimmune pathologies.
Funding
This study was performed with financial support from the Russian Science Foundation (project no. 14-14-01140; data shown in Figs. 1-3 and Fig. 4, c and d) and the Program of Fundamental Research of State Academies of Science for 2013-2020 (project no. 01201363817; design of siRNAs specific to the genes encoding transcription factors EBF1 and STAT1 and control RNA; analysis of siRNA effect on the transcription factor expression; Fig. 4, a and b).
Conflicts of Interest
The authors declare no conflicts of interest.
REFERENCES
2.Grewal, I. S., and Flavell, R. A. (1998) CD40 and
CD154 in cell-mediated immunity, Annu. Rev. Immunol., 16,
111-135.
3.Lane, P., Traunecker, A., Hubele, S., Inui, S.,
Lanzavecchia, A., and Gray, D. (1992) Activated human T cells express a
ligand for the human B cell-associated antigen CD40 which participates
in T cell-dependent activation of B lymphocytes, Eur. J.
Immunol., 22, 2573-2578.
4.Cella, M., Scheidegger, D., Palmer-Lehmann, K.,
Lane, P., Lanzavecchia, A., and Alber, G. (1996) Ligation of CD40 on
dendritic cells triggers production of high levels of interleukin-12
and enhances T cell stimulatory capacity: T-T help via APC activation,
J. Exp. Med., 184, 747-752.
5.Summers deLuca, L., and Gommerman, J. L. (2012)
Fine-tuning of dendritic cell biology by the TNF superfamily, Nat.
Rev. Immunol., 12, 339-351.
6.Stout, R. D., Suttles, J., Xu, J., Grewal, I. S.,
and Flavell, R. A. (1996) Impaired T cell-mediated macrophage
activation in CD40 ligand-deficient mice, J. Immunol.,
156, 8-11.
7.Suttles, J., and Stout, R. D. (2009) Macrophage
CD40 signaling: a pivotal regulator of disease protection and
pathogenesis, Semin. Immunol., 21, 257-264.
8.Peters, A. L., Stunz, L. L., and Bishop, G. A.
(2009) CD40 and autoimmunity: the dark side of a great activator,
Semin. Immunol., 21, 293-300.
9.Choi, E. W., Lee, K. W., Park, H., Kim, H., Lee, J.
H., Song, J. W., Yang, J., Kwon, Y., Kim, T. M., Park, J. B., and Kim,
S. (2018) Therapeutic effects of anti-CD154 antibody in cynomolgus
monkeys with advanced rheumatoid arthritis, Sci. Rep., 8,
2135.
10.'t Hart, B. A., Blezer, E. L., Brok, H. P., Boon,
L., de Boer, M., Bauer, J., and Laman, J. D. (2005) Treatment with
chimeric anti-human CD40 antibody suppresses MRI-detectable
inflammation and enlargement of pre-existing brain lesions in common
marmosets affected by MOG-induced EAE, J. Neuroimmunol.,
163, 31-39.
11.Croft, M., Benedict, C. A., and Ware, C. F.
(2013) Clinical targeting of the TNF and TNFR superfamilies, Nat.
Rev. Drug Discov., 12, 147-168.
12.Jacobson, E. M., Concepcion, E., Oashi, T., and
Tomer, Y. (2005) A Graves' disease-associated Kozak sequence
single-nucleotide polymorphism enhances the efficiency of CD40 gene
translation: a case for translational pathophysiology,
Endocrinology, 146, 2684-2691.
13.Jacobson, E. M., Huber, A. K., Akeno, N., Sivak,
M., Li, C. W., Concepcion, E., Ho, K., and Tomer, Y. (2007) A CD40
Kozak sequence polymorphism and susceptibility to antibody-mediated
autoimmune conditions: the role of CD40 tissue-specific expression,
Genes Immun., 8, 205-214.
14.Lee, Y. H., Bae, S. C., Choi, S. J., Ji, J. D.,
and Song, G. G. (2015) Associations between the functional CD40
rs4810485 G/T polymorphism and susceptibility to rheumatoid arthritis
and systemic lupus erythematosus: a meta-analysis, Lupus,
24, 1177-1183.
15.Raychaudhuri, S., Remmers, E. F., Lee, A. T.,
Hackett, R., Guiducci, C., Burtt, N. P., Gianniny, L., Korman, B. D.,
Padyukov, L., Kurreeman, F. A., Chang, M., Catanese, J. J., Ding, B.,
Wong, S., van der Helm-van Mil, A. H., Neale, B. M., Coblyn, J., Cui,
J., Tak, P. P., Wolbink, G. J., Crusius, J. B., van der Horst-Bruinsma,
I. E., Criswell, L. A., Amos, C. I., Seldin, M. F., Kastner, D. L.,
Ardlie, K. G., Alfredsson, L., Costenbader, K. H., Altshuler, D.,
Huizinga, T. W., Shadick, N. A., Weinblatt, M. E., de Vries, N.,
Worthington, J., Seielstad, M., Toes, R. E., Karlson, E. W., Begovich,
A. B., Klareskog, L., Gregersen, P. K., Daly, M. J., and Plenge, R. M.
(2008) Common variants at CD40 and other loci confer risk of rheumatoid
arthritis, Nat. Genet., 40, 1216-1223.
16.Smets, I., Fiddes, B., Garcia-Perez, J. E., He,
D., Mallants, K., Liao, W., Dooley, J., Wang, G., Humblet-Baron, S.,
Dubois, B., Compston, A., Jones, J., Coles, A., Liston, A., Ban, M.,
Goris, A., and Sawcer, S. (2018) Multiple sclerosis risk variants alter
expression of co-stimulatory genes in B cells, Brain, doi:
10.1093/brain/awx372.
17.Vazgiourakis, V. M., Zervou, M. I., Choulaki, C.,
Bertsias, G., Melissourgaki, M., Yilmaz, N., Sidiropoulos, P., Plant,
D., Trouw, L. A., Toes, R. E., Kardassis, D., Yavuz, S., Boumpas, D.
T., and Goulielmos, G. N. (2011) A common SNP in the CD40 region is
associated with systemic lupus erythematosus and correlates with
altered CD40 expression: implications for the pathogenesis, Ann.
Rheum. Dis., 70, 2184-2190.
18.Schwartz, A. M., Demin, D. E., Vorontsov, I. E.,
Kasyanov, A. S., Putlyaeva, L. V., Tatosyan, K. A., Kulakovskiy, I. V.,
and Kuprash, D. V. (2017) Multiple single nucleotide polymorphisms in
the first intron of the IL2RA gene affect transcription factor binding
and enhancer activity, Gene, 602, 50-56.
19.Pistillo, M. P., Tanigaki, N., Chua, R.,
Mazzoleni, O., and Ferrara, G. B. (1989) Human anti-HLA-DQw2 monoclonal
antibody secreted by an Epstein–Barr-virus-transformed
lymphoblastoid cell line: assessment of the monoclonality,
allospecificity, and target, Hum. Immunol., 24,
253-263.
20.Schwartz, A. M., Putlyaeva, L. V., Covich, M.,
Klepikova, A. V., Akulich, K. A., Vorontsov, I. E., Korneev, K. V.,
Dmitriev, S. E., Polanovsky, O. L., Sidorenko, S. P., Kulakovskiy, I.
V., and Kuprash, D. V. (2016) Early B-cell factor 1 (EBF1) is critical
for transcriptional control of SLAMF1 gene in human B cells,
Biochim. Biophys. Acta, 1859, 1259-1268.
21.Bachmann, S. B., Frommel, S. C., Camicia, R.,
Winkler, H. C., Santoro, R., and Hassa, P. O. (2014) DTX3L and ARTD9
inhibit IRF1 expression and mediate in cooperation with ARTD8 survival
and proliferation of metastatic prostate cancer cells, Mol.
Cancer, 13, 125.
22.Hasson, S. A., Kane, L. A., Yamano, K., Huang, C.
H., Sliter, D. A., Buehler, E., Wang, C., Heman-Ackah, S. M., Hessa,
T., Guha, R., Martin, S. E., and Youle, R. J. (2013) High-content
genome-wide RNAi screens identify regulators of parkin upstream of
mitophagy, Nature, 504, 291-295.
23.Putlyaeva, L. V., Schwartz, A. M., Korneev, K.
V., Covic, M., Uroshlev, L. A., Makeev, V. Y., Dmitriev, S. E., and
Kuprash, D. V. (2014) Upstream open reading frames regulate translation
of the long isoform of SLAMF1 mRNA that encodes costimulatory receptor
CD150, Biochemistry (Moscow), 79, 1405-1411.
24.Rosenbloom, K. R., Armstrong, J., Barber, G. P.,
Casper, J., Clawson, H., Diekhans, M., Dreszer, T. R., Fujita, P. A.,
Guruvadoo, L., Haeussler, M., Harte, R. A., Heitner, S., Hickey, G.,
Hinrichs, A. S., Hubley, R., Karolchik, D., Learned, K., Lee, B. T.,
Li, C. H., Miga, K. H., Nguyen, N., Paten, B., Raney, B. J., Smit, A.
F., Speir, M. L., Zweig, A. S., Haussler, D., Kuhn, R. M., and Kent, W.
J. (2015) The UCSC Genome Browser database: 2015 update, Nucleic
Acids Res., 43, D670-681.
25.Nguyen, V. T., and Benveniste, E. N. (2000)
Involvement of STAT-1 and Ets family members in interferon-gamma
induction of CD40 transcription in microglia/macrophages, J. Biol.
Chem., 275, 23674-23684.
26.Yevshin, I., Sharipov, R., Valeev, T., Kel, A.,
and Kolpakov, F. (2017) GTRD: a database of transcription factor
binding sites identified by ChIP-seq experiments, Nucleic Acids
Res., 45, D61-D67.
27.Kulakovskiy, I. V., Vorontsov, I. E., Yevshin, I.
S., Sharipov, R. N., Fedorova, A. D., Rumynskiy, E. I., Medvedeva, Y.
A., Magana-Mora, A., Bajic, V. B., Papatsenko, D. A., Kolpakov, F. A.,
and Makeev, V. J. (2018) HOCOMOCO: towards a complete collection of
transcription factor binding models for human and mouse via large-scale
ChIP-Seq analysis, Nucleic Acids Res., 46, D252-D259.
28.McCarthy, M. I., Abecasis, G. R., Cardon, L. R.,
Goldstein, D. B., Little, J., Ioannidis, J. P., and Hirschhorn, J. N.
(2008) Genome-wide association studies for complex traits: consensus,
uncertainty and challenges, Nat. Rev. Genet., 9,
356-369.
29.Toubi, E., and Shoenfeld, Y. (2004) The role of
CD40–CD154 interactions in autoimmunity and the benefit of
disrupting this pathway, Autoimmunity, 37, 457-464.
30.Dong, L., Chen, X., Shao, H., Bai, L., Li, X.,
and Zhang, X. (2018) Mesenchymal stem cells inhibited dendritic cells
via the regulation of STAT1 and STAT6 phosphorylation in experimental
autoimmune uveitis, Curr. Mol. Med., 17, 478-487.
31.Luo, S., Liu, Y., Liang, G., Zhao, M., Wu, H.,
Liang, Y., Qiu, X., Tan, Y., Dai, Y., Yung, S., Chan, T. M., and Lu, Q.
(2015) The role of microRNA-1246 in the regulation of B cell activation
and the pathogenesis of systemic lupus erythematosus, Clin.
Epigenetics, 7, 24.
32.Ramos, P. S., Shedlock, A. M., and Langefeld, C.
D. (2015) Genetics of autoimmune diseases: insights from population
genetics, J. Hum. Genet., 60, 657-664.
33.Itariu, B. K., and Stulnig, T. M. (2014)
Autoimmune aspects of type 2 diabetes mellitus – a mini-review,
Gerontology, 60, 189-196.
34.Mammana, S., Fagone, P., Cavalli, E., Basile, M.
S., Petralia, M. C., Nicoletti, F., Bramanti, P., and Mazzon, E. (2018)
The role of macrophages in neuroinflammatory and neurodegenerative
pathways of Alzheimer’s disease, amyotrophic lateral sclerosis,
and multiple sclerosis: pathogenetic cellular effectors and potential
therapeutic targets, Int. J. Mol. Sci., 19, E831.
35.Nesher, G., and Moore, T. L. (1993) Rheumatoid
arthritis in the aged. Incidence and optimal management, Drugs
Aging, 3, 487-501.