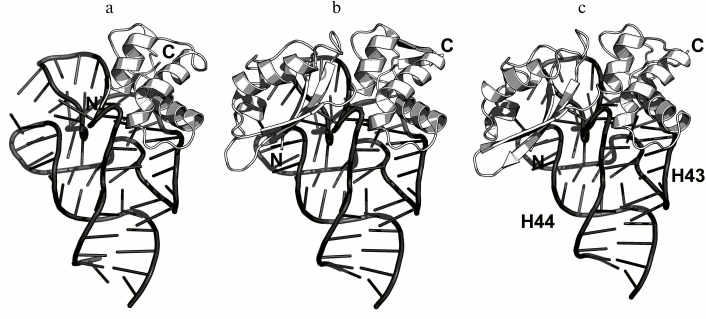REVIEW: Structural Aspects of Ribosomal RNA Recognition by Ribosomal Proteins
A. D. Nikulin
Institute of Protein Research, Russian Academy of Sciences, 142290 Pushchino, Moscow Region, Russia; E-mail: nikulin@vega.protres.ru
Received July 26, 2017; Revision received August 8, 2017
This review is focused on the structural aspects of interaction between ribosomal proteins and ribosomal RNA in bacterial ribosomes and complexes of ribosomal proteins with specific fragments of ribosomal RNA. Special attention is given to the recognition of specific spatial architecture of the double-stranded ribosomal RNA by ribosomal proteins and to the role of unstructured protein regions in stabilization of distant ribosomal RNA segments.
KEY WORDS: ribosome structure, ribosomal proteins, ribosomal RNA, RNA–protein recognition, double-stranded RNA, unstructured proteinsDOI: 10.1134/S0006297918140109
Abbreviations: EM, electron microscopy; LRS, 50S (large) ribosomal subunit; NMR, nuclear magnetic resonance; r-protein, ribosomal protein; rRNA, ribosomal RNA; SRS, 30S (small) ribosomal subunit.
Protein biosynthesis in cells occurs on special cellular organelles
– ribosomes. Ribosomes are complex macromolecular machines
consisting of several ribosomal RNAs (rRNAs) and over 50 ribosomal
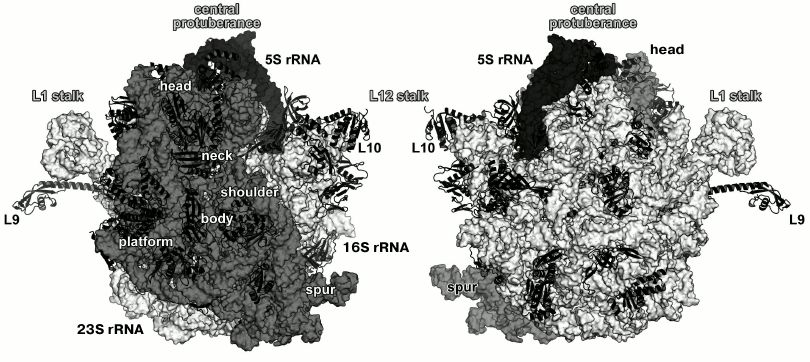
proteins (r-proteins) in bacteria (Fig. 1) and up
to 80 proteins in eukaryotes [1]. Protein
biosynthesis is one of the key cellular processes, and its
understanding requires knowing ribosome organization (structure) and
interactions between its components. The major break in structural
studies of ribosomes happened at the turn of the 21st century, when
development of X-ray crystallography and cryo-electron microscopy
(cryo-EM) methods allowed to solve the structures of entire bacterial
ribosomes and their subunits at a resolution sufficient for detailed
description of all ribosomal components.
The structures of isolated r-proteins and their complexes with specific RNA fragments had been determined even before elucidation of the structure of ribosomes and their subunits. The results of the structural studies of the RNA–protein complexes were the first experimental data describing protein interactions with double-stranded RNA and became an important milestone on the pathway to understanding the principles of RNA–protein recognition. Moreover, structures of isolated r-proteins and RNA–protein complexes have served as a basis for interpretation of first electron density maps of ribosomal subunits and their cryo-EM models. Structures of the rRNA–protein complexes have been very useful for deciphering the architecture of functional sites of the 50S (large) ribosomal subunit (LRS), since these sites are not visible in the electron density maps because of their high flexibility. Further investigations of the structure of ribosomal subunits and ribosomes allowed to classify in detail the structures of r-proteins [2].
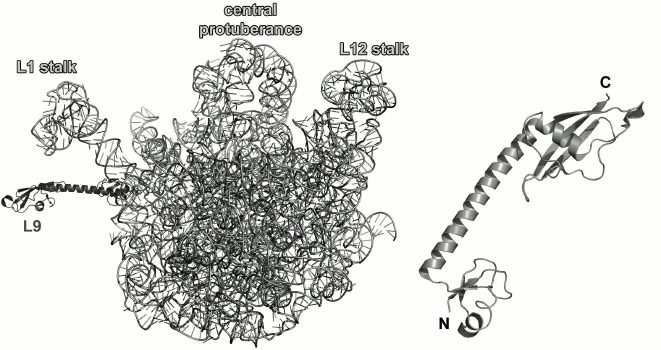
Fig. 1. Structure of Escherichia coli 70S ribosome [12] (created using the PyMol software, ver. 1.8.6.0 Open-Source, Schrödinger LLC, based on PDB 5IT8). Left, view from the 30S (small) ribosomal subunit (SRS); right, view from the 50S (large) ribosomal subunit (LRS). Ribosomal RNAs of the LRS are shown in light grey (23S rRNA) and black (5S rRNA); 16S rRNA of the SRS is shown in grey. The r-proteins are shown as ribbons.
The majority of r-proteins is conserved and could be found in ribosomes of living organisms from all three domains: bacteria, archaea, and eukaryotes [3, 4]. Comparison of amino acid sequences of r-proteins from different organisms showed that they typically have similar size and do not include nonconserved structural elements [5]. In 2014, a common classification of r-proteins was proposed, that since then has received a general acceptance [6]. Many conserved r-proteins are involved in the formation of functional sites in the ribosome and in initial stages of ribosome assembly. In Escherichia coli, primary RNA-binding r-proteins are S4, S7, S8, S15, S17, and S20 of the 30S (small) ribosomal subunit (SRS) [7, 8] and L1, L2, L3, L4, L9, L20, L23, and L24 of the LRS [9, 10]. During in vitro assembly of ribosomal subunits, binding of these proteins to the corresponding rRNAs leads to a significant compaction of ribonucleoprotein particles [11, 12]. According to the new nomenclature, these proteins are referred to as uS4, uS7, uS8, uS15, uS17, bS20 and uL1, uL2, uL3, uL4, bL9, bL20, uL23, uL24, respectively (here, “u” stands for universally conserved r-proteins, whereas “b” denotes proteins specific to bacterial ribosomes [6]).
In this review, we discuss the structural aspects of the interaction between conserved primary binding r-proteins and rRNAs. When possible, structures of bacterial r-proteins in the ribosome are compared to those of the isolated r-proteins or r-proteins complexed with the corresponding rRNA fragments. The “old” nomenclature is used for designation of bacterial r-proteins [1].
RIBOSOMAL PROTEINS OF THE L12 STALK OF THE LRS
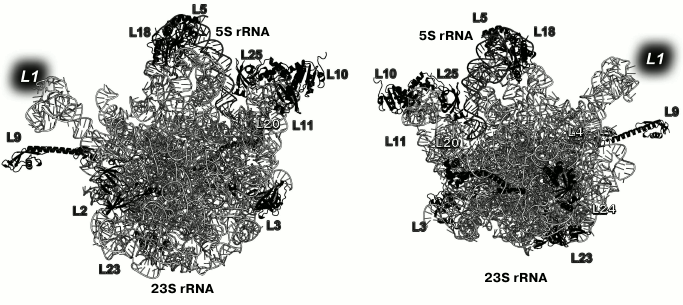
Primary binding proteins of the LRS associate with different 23S rRNA domains and are situated on both the inner (at the intersubunit interface) and outer (solvent side) surfaces of the LRS subunit (Fig. 2) [13]. Primary binding r-protein L1 forms distinct morphological element of the LRS, the so-called L1 protuberance.
Fig. 2. Positions of the primary binding r-proteins in E. coli LRS (based on PDB 5IT8 coordinates). Left, view from the LRS side; right, view from the solvent side. Position of L1 is indicated with a grey background, as the model lacks its coordinates.
On the opposite side of the LRS there is another protuberance – the so-called L12 stalk (P stalk in archaea and eukaryotes) formed by the r-proteins L10 and L11 (uL10 and uL11, according to the new nomenclature, respectively) and several dimers of L12 (bL12). The L12 stalk of the LRS plays an essential role in the ribosome interactions with translation factors and in control of translation fidelity by forming ribosome GTPase site with the 23S rRNA [14, 15]. The L12 stalk r-proteins were among the first, whose structures were determined by X-ray crystallography and NMR analysis in either isolated form or in complexes with rRNA fragments [16, 17].
Ribosomal protein L11. L11 consists of two globular domains connected by a short flexible linker [17]. The C-terminal domain (75 amino acid residues (a.a.)) interacts with the 23S rRNA domain II (region 1051-1108, E. coli numbering) [18]. The N-terminal domain interacts with translation factors on the ribosome and does not affect the specificity of the 23S rRNA binding by the protein [19, 20].
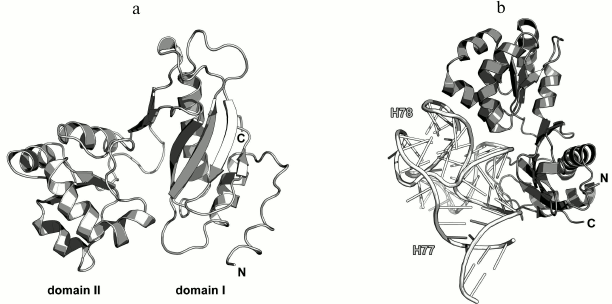
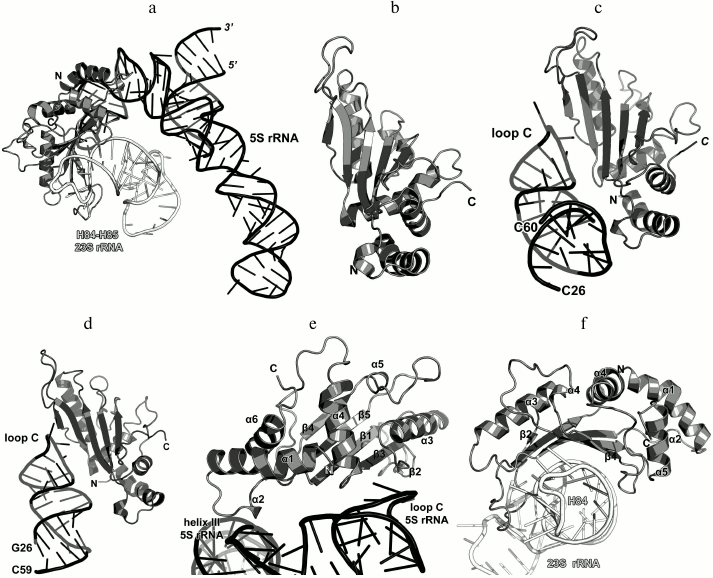
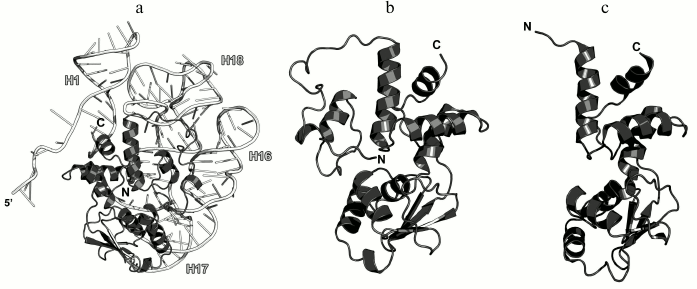
In 1990s, X-ray crystallography was used to solve the structures of the C-terminal domain of L11 from Geobacillus stearothermophilus complexed with an E. coli 23S rRNA fragment including helices H43 and H44 [21] (Fig. 3a) and of the full-size L11 from Thermotoga maritima bound to the same 23S rRNA fragment (Fig. 3b) [20]. Somewhat later, the structure of L11 complexed with the 23S rRNA and antibiotic thiostrepton was determined by NMR analysis [19].
Fig. 3. Structures of L11 proteins associated with specific 23S rRNA fragments. a) G. stearothermophilus L11 C-terminal domain in complex with an E. coli 23S rRNA fragment; 2.8 Å resolution (PDB 1QA6 [20]). b) T. maritima L11 in complex with the same 23S rRNA fragment; 2.6 Å resolution (PDB 1MMS [19]). c) L11 and a fragment of the 23S rRNA within the structure of E. coli ribosome (PDB 5IT8). Here and in the following figures, 23S rRNA helices are indicated.
The N-terminal domain of L11 consists of a three-stranded β-sheet with two α-helices located on one of its sides. According to the Structural Classification of Proteins (SCOP) [22], this α+β domain structure is unique. The C-terminal domain of L11 consists of three α-helices with a short β-hairpin and two long loops, which are disordered in the absence of rRNA [20]. According to the SCOP, this domain has the DNA/RNA-binding three-helical bundle fold and belongs to the all-alpha protein class.
In all the determined structures, L11 protein binds the 23S rRNA via the C-terminal helix α5, that is positioned alongside the minor groove of the rRNA and forms the majority of contacts with it (Fig. 3). The area of contact is shielded by residues of the helix α3 and loops α3–β4 and α4–α5, which are situated on both sides of the helix [20]. The majority of L11–rRNA contacts are hydrogen bonds between atoms of the protein main chain and the rRNA sugar-phosphate backbone, although there are also a number of contacts between the protein and the rRNA nucleotide bases [21]. Similar mode of recognition via α-helix is shared by many DNA-interacting transcription factors (e.g., Cro repressor) [23, 24]. In their cases, protein α-helix is located in the major groove of B-DNA, where atoms of amino acid residues interact with atoms of DNA bases, thereby providing specific recognition of the DNA sequence by the protein [25]. Narrow major groove in the A-form double-stranded RNA does not allow such recognition; therefore, it was assumed that L11 recognizes a spatial fold of the rRNA rather than its nucleotide sequence [20, 21].
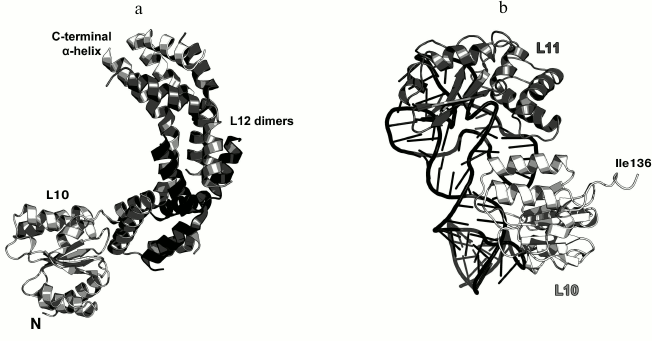
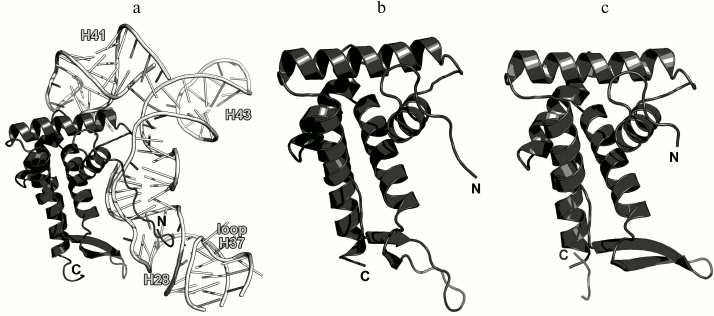
Ribosomal protein L10. L10 is an important element of the L12 stalk. It acts as a bridge between L12 and the 23S rRNA [15, 26]. L10 consists of two domains. The N-terminal α/β domain has a compact three-layer structure and interacts with a region of the 23S rRNA helices H42-H44 close to L11 [27]. The C-terminal domain is a long α-helix that binds the L12 dimers [15] (Fig. 4). The two domains of L10 are connected by a flexible linker; for this reason, its C-terminal domain with bound L12 dimers is extremely mobile and cannot be seen in the ribosome crystal structures. The structural features of the ribosomal L12 stalk have been described based on the cryo-EM reconstruction of ribosomes. In 2005, the structure of the L10 protein bound to three L12 dimers from the bacterium T. maritima was used to create the most interesting structural model of this LRS functional domain in the archaeon Haloarcula marismortui [15].
Fig. 4. a) Structure of L10 from T. maritima in a complex with three dimers of the N-terminal domain of L12; 2.1 Å resolution (PDB 1ZAX). b) Fragment of the E. coli 70S ribosome structure (PDB 5IT8), including nucleotides 1038-1117 of the 23S rRNA, N-terminal domain of L10, and L11. Position of Ile136 in L10 is indicated.
Recently obtained high-resolution structures of bacterial ribosomes from E. coli [28, 29] and Thermus thermophilus [30, 31] allow to analyze the contacts between the N-terminal domain of L10 and the 23S rRNA in detail. Interaction of L10 with the 23S rRNA is realized through amino acid residues of protein helices α1 and α3, which are located at the rRNA minor groove in the region of nucleotides 1038-1117 in E. coli 23S rRNA (Fig. 4). Amino acid residues of L10 in all the available structures do not form hydrogen bonds with atoms of RNA nucleotide bases, but only with the RNA sugar-phosphate backbone. Therefore, L10 recognizes specific spatial rRNA arrangements.
L1 STALK OF THE LRS
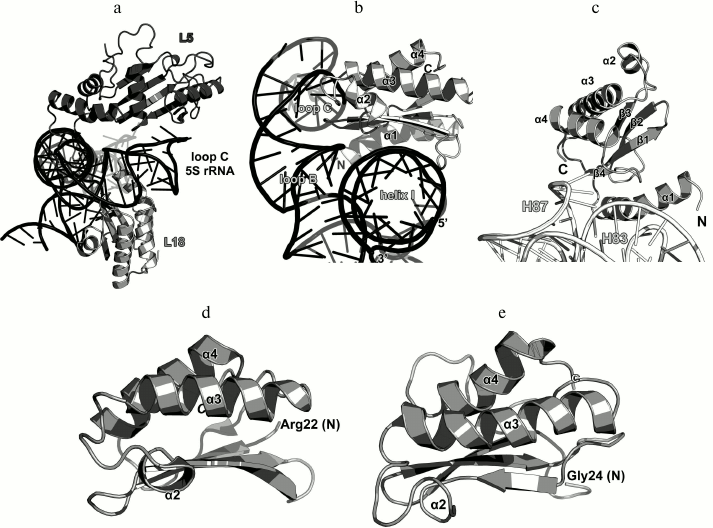
Ribosomal protein L1. The L1 stalk of the LRS is formed by the r-protein L1 and helices H76-H78 of the 23S rRNA. It is located on the opposite side of the LRS from the L12 stalk (Fig. 2). L1 could not be seen on the X-ray structures of ribosomes due to the high mobility of the L1 stalk, which can move a considerable distance and rotate around the axis of the 23S rRNA helix H76 depending on the presence of tRNA in the ribosomal E site [13, 32]. It is believed that the mobility of this region is required for its participation in the tRNA translocation on the ribosome, particularly, in the relocation of deacylated tRNA to the E site with subsequent removal of tRNA from the ribosome [33]. Due to the high mobility of the L1 stalk, the pioneer works on the crystallography of ribosomes had been able to describe only a part of the L1 structure that was a fragment of the 23S rRNA without the protein itself [34, 35]. This gap in the understanding of the ribosome structure was filled by determining the structure of L1 in a complex with the corresponding 23S rRNA fragment.
In 1996, the structure of L1 from the thermophilic bacterium T. thermophilus was determined for the first time in our laboratory [36]. It was shown that the protein consists of two domains. The first domain includes both the N-terminal and C-terminal parts of the protein, whereas the second domain consists of the middle component of the polypeptide chain (Fig. 5a). The first domain has the two-layer split β-α-β structure; the second domain has the three-layer Rossman fold. The domains are connected with a flexible linker and can change mutual orientation in solution [37].
Fig. 5. a) structure of L1 from T. thermophilus [35]; b) structure of L1 from S. solfataricus in a complex with a fragment of the 23S rRNA from T. thermophilus (PDB 1MZP).
In 2003, we solved the structure of L1 from the archaeon Sulfolobus solfataricus in a complex with a fragment of the 23S rRNA from T. thermophilus (Fig. 5b) [38]. Such combination of components from different organisms allowed us to crystallize the complex after almost seven years of searching for proper crystallization conditions after the structure of the isolated L1 had been determined. The obtained structure was used by us and other researchers for describing the L1 stalk in bacterial ribosome models [38].
The structure of the L1–23S rRNA complex has revealed that L1 interacts with RNA through two distant areas located on the surfaces of both domains. The first domain contains four times as many RNA-contacting residues as the second domain, which suggested that it plays the major in the recognition of the 23S rRNA [38]. Similarly to the ribosomal complexes described above, amino acid residues of L1 interact predominantly with atoms of the rRNA sugar-phosphate backbone.
23S rRNA-BINDING RIBOSOMAL PROTEINS OF THE LRS BODY
Ribosomal protein L2. The primary 23S rRNA-binding L2 protein is located close to the base of the L1 stalk in the LRS (Fig. 2) [39]. According to the available data, L2 is required for the association of SRS and LRS and formation of the 70S ribosome, tRNA binding by the ribosome at the A and P sites, and peptide bond formation in the nascent polypeptide [40, 41]. In 70S ribosomes of E. coli, L2 forms several contacts with the 16S rRNA during translation initiation stage with the formation of the bridge B7b, which is then disrupted upon binding of the translation factor EF-G to the ribosome [42]. Bacterial L2 protein can be substituted in the ribosomes by its homologs from the archaeon H. marismortui (HmaL2) or the L8 protein from the human 60S cytoplasmic ribosomal subunit without loss in the ribosome activity [43].
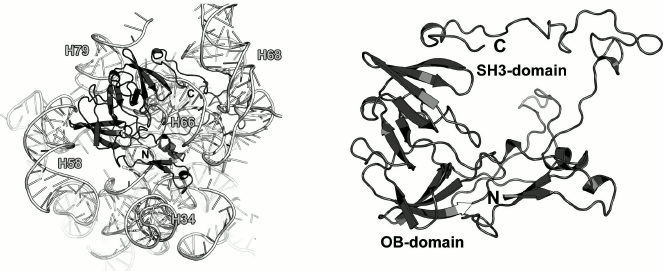
Most part of the 270-a.a. L2 protein does not possess an apparent secondary structure. However, there are two β-structured domains in the central part of the protein molecule that might be classified as the nucleotide-binding OB fold and SH3-like domain (Fig. 6).
Fig. 6. Left, position of L2 on the 23S rRNA in E. coli 70S ribosome (PDB 5IT8). Right, schematic representation of L2 in the ribosome.
As a part of the LRS, L2 interacts with distantly located regions of the 23S rRNA, thereby playing a role of a scaffold protein and connecting 23S rRNA helices (Fig. 6). Analysis of the protein–rRNA contacts shows that only 15% of these interactions are hydrogen bonds between the protein atoms and atoms of the rRNA nucleotide bases, whereas the rest are the contacts with the 23S rRNA sugar-phosphate backbone. An interesting feature of the RNA–protein interactions is that the vast majority of them involve amino acid residues situated in disordered protein regions (loops and turns). Hence, it is difficult to believe that the tertiary structure of L2 provides recognition of the 23S rRNA, rather than secures certain spatial arrangement of the 23S rRNA helices during LRS assembly.
Ribosomal protein L3. L3 is one of the two initiator proteins (together with L24) in the LRS assembly [44]. It binds to the 3′-end of the 23S rRNA and nucleates rRNA folding [10]. L3 is involved in the formation of the peptidyl transferase site due to the presence of elongated loops that penetrate the LRS [45]. Mutations in the amino acid sequence of L3 might result in the resistance to the antibiotics tiamulin [46, 47] and linezolid [47]. All the amino acid substitutions in this protein were found in the fragment 144-151. The strongest effects were caused by substitutions of the residues 148 and 149 located in the peptidyl transferase site of the ribosome [46].
No structures of the isolated protein and L3 in a complex with a 23S rRNA fragment have been solved. Examination of the L3 structure in the E. coli ribosome shows that L3 is a two-domain protein (Fig. 7). The first domain includes both the N- and C-termini of the protein and represents a β-barrel with a protruding loop of 50 a.a. (residues 115-165). The loop does not form any secondary structure with the exception of a small α-helix and a short β-region. The second domain (residues 30-97) is inserted into the first domain and consists of a β-sheet with few α-helices located on one side of it.
Fig. 7. Left, position of L3 on the 23S rRNA in E. coli 70S ribosome (PDB 5IT8). Right, schematic representation of L3 protein in the ribosome.
Most of the contacts (approximately 80%) between L3 and the 23S rRNA are made by residues 115-165 of the protein loop; some of them are hydrogen bonds with atoms of rRNA nucleotide bases. The β-strands of the second domain interact with the rRNA sugar-phosphate backbone, whereas the first domain binds rRNA through only five amino acid residues. Therefore, the primary rRNA-recognizing unit in L3 is the second domain that can identify the structure of the 23S rRNA during initiation of the LRS assembly. The loop 115-165 makes the majority of contacts with the rRNA, most probably, during subsequent stages of the LRS assembly.
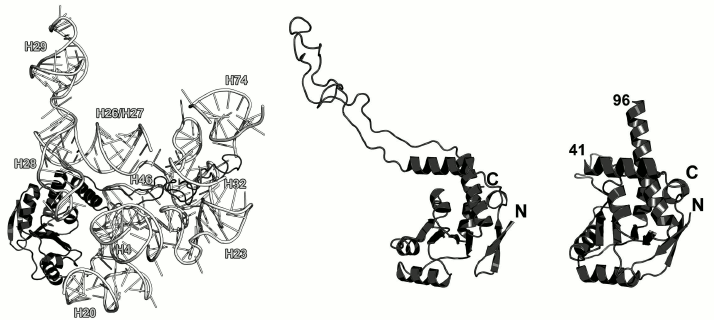
Ribosomal protein L4. L4 is involved in the early stages of the LRS assembly [10]. It binds to the 5′-terminus of the 23S rRNA and forms multiple contacts with different domains of the 23S rRNA in the assembled LRS [48, 49] (Fig. 8). L4 is located at the solvent side of the LRS. However, it possesses an elongated loop that penetrates the entire subunit toward the peptidyl transferase site of the ribosome. This loop has attracted a lot of interest, since it reaches the ribosomal tunnel through which nascent polypeptide passes after its synthesis on the ribosome [50]. Mutations in this loop might make bacteria resistant to macrolide antibiotics [51, 52]. Though the loop of L4 penetrated the entire LRS, it is dispensable for the LRS assembly and might be deleted [53].
Fig. 8. Left, position of L4 on the 23S rRNA in E. coli 70S ribosome (PDB 5IT8). Central panel, structure of L4 in the ribosome. Right, structure of T. maritima L4 (PDB 1DMG). Residues 41 and 96 are the boundaries of the non-described part of the protein.
The structure of the isolated L4 from T. maritima was determined in 2000 [54] except the fragment of 41-96 that corresponds to the long loop, which is disordered if the protein is not bound to rRNA (Fig. 8). Because this region possesses high mobility, it is not fixed in the crystal and has poor electron density, which prevents solving its structure.
According to the SCOP, L4 belongs to the three-layer α/β proteins and contains a four-strand parallel β-sheet. The overall fold of its secondary structure elements is unique and was classified as a separate family of L4 r-proteins (SCOP 52167).
Analysis of contacts between amino acid residues of L4 and the 23S rRNA shows that (similarly to L3), the majority of them are made by residues of the long protein loop and adjacent regions of α-helices (a.a. 40-99). The second protein region involved in the RNA–protein interactions is the helix α2. Since it was shown that the loop in L4 protein is not required for the LRS assembly [53], it is reasonable to assume that the rRNA-recognizing unit of L4 is its structured domain, whose most important element is the helix α2.
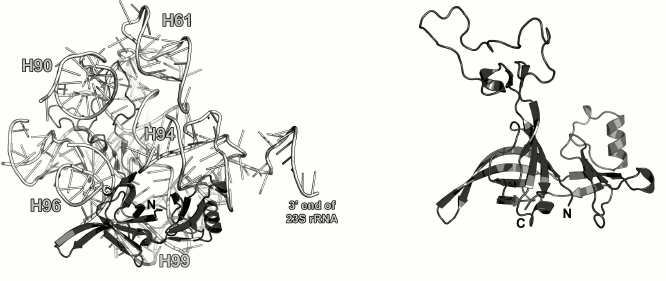
Ribosomal protein L9. L9 is one of the earliest proteins to bind to the 23S rRNA [10]. It interacts with the rRNA 3′-terminus next to the L1-binding site. L9 was found only in bacterial ribosomes [55]. In the LRS, it is located directly under the L1 stalk (Fig. 9).
Fig. 9. Left, position of L9 on the 23S rRNA in E. coli 70S ribosome (PDB 5IT8); view from the intersubunit interface. Right, structure of L9 from G. stearothermophilus (PDB 1DIV).
The structure of L9 was first published in 1994 [55]. L9 consists of two domains: small N-terminal domain (47 a.a.) with the three-layer α/β/α architecture and C-terminal domain (85 a.a.) with the double-layer α+β architecture connected to a long α-helix (37 a.a., 10 turns) that participates in the formation of the domains. The N-terminal domain is structurally similar to the RNA recognition motif (RRM). Both domains are positively charged, so it was assumed that they both interact with the 23S rRNA [55, 56]. However, according to the X-ray structural data, only the N-terminal domain interacts with the 23S rRNA in bacterial ribosomes, whereas the C-terminal domain is distant from the ribosome surface and contacts the SRS of neighboring ribosome in a crystal [57]. Cryo-EM of the ribosomes has demonstrated that the α-helix of the L9 protein can bend so that the C-terminal domain approaches the r-protein S6 [58]. One of the hypotheses that explain L9 location in the ribosome is that it controls the distance between ribosomes in polysomes and suppresses translation frameshifting, thereby increasing the fidelity of mRNA translation by the ribosome [59].
Modeling of interactions between L9 and the 23S rRNA in the E. coli ribosome (PDB 5IT8) showed only a few contacts between them, due probably to the limited resolution of the model and inaccuracy of its refinement at the periphery. However, one may argue that L9 interacts with a region that connects helices H76 and H79 in the 23S rRNA.
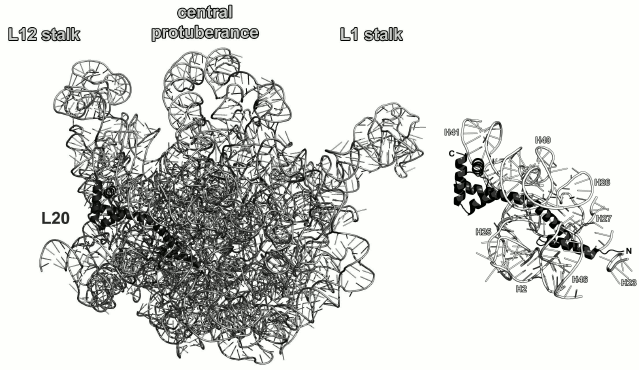
Ribosomal protein L20. L20 binds to the 5′-terminus of the 23S rRNA early in the process of ribosome assembly and is required for the first stage of the LRS assembly in vitro [60, 61]. L20 is not universally conserved and presents in bacterial ribosomes only. However, deletion of the gene encoding L20 protein in E. coli is lethal [61]. It should be noted that L20 can substitute for the L24 protein during initiation of the LRS assembly at low temperatures [62]. It is located on the opposite side of the LRS from the peptidyl transferase site and interacts with several distant regions of the 23S rRNA (Fig. 10).
Fig. 10. Left, position of L20 on the 23S rRNA in E. coli 70S ribosome (PDB 5IT8); view from the solvent side. Right, zoomed view of the L20-binding site.
NMR studies of L20 showed that L20 stays unfolded when not bound to the rRNA [63]. However, it acquires a secondary structure during crystallization [64]. According to SCOP classification, L20 is a single-domain protein containing four orthogonally packed α-helices. Packaging of the helices corresponds to the PAPB domain.
It is believed that L20 becomes structured upon interaction with the 23S rRNA. The C-terminal α-helix of the protein interacts with other r-proteins on the surface of ribosomes, whereas the N-terminal α-helix and long α2-helix interact with rRNA [65]. Indeed, analysis of interactions between L20 and the 23S rRNA in ribosomes shows a large area of contact between these helices and the sugar-phosphate backbone of the 23S rRNA helices H40-H41 on one side of the protein and helices H25-H27 on the other side (Fig. 10). However, amino acid residues of helices α3 and α4 also form a number of contacts with the sugar-phosphate backbone of the 23S rRNA helix H41; therefore, the interactions between the L20 C-terminal domain and the 23S RNA cannot be ignored. Analysis of the L20 interaction with rRNA reveals the role of this protein in the LRS assembly: L20 neutralizes negative charges of phosphates groups in the 23S rRNA and brings its distant domains close together [61, 65].
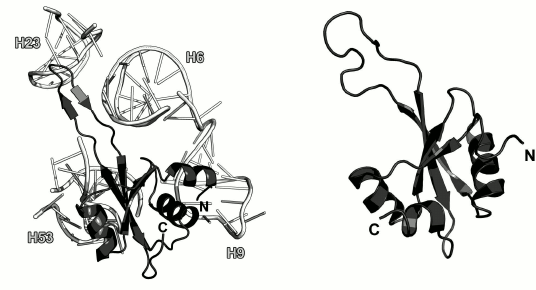
Ribosomal protein L23. L23 binds to the third domain of the 23S rRNA at the early stages of the LRS assembly [10, 49]. It is located on the LRS close to the exit of the nascent polypeptide from the outer side of the ribosome (Fig. 2). Some experimental data indicate that L23 interacts with the trigger factor [66] and signal recognition particle [67, 68] on the ribosomes and might be involved in the folding of the growing polypeptide chain [66].
The spatial structure of isolated L23 was determined by the NMR analysis (Fig. 11) [69]. It was found that L23 belongs to the three-layer α+β protein family (SCOP 54189). Its structure is conserved and does not significantly changes upon protein binding to the 23S rRNA.
Fig. 11. Left, position of L23 on the 23S rRNA in E. coli 70S ribosome (PDB 5IT8). Right, structure of L23 from T. thermophilus as determined by NMR analysis (PDB 1N88).
In the ribosomal subunit, L23 interacts with the sugar-phosphate backbone of the 23S rRNA helices H6, H9, H23 and H53 (Fig. 11) predominantly through a long loop and adjacent termini of the β-strands and through residues of the α2-helix that form contacts with the sugar-phosphate backbone of H53.
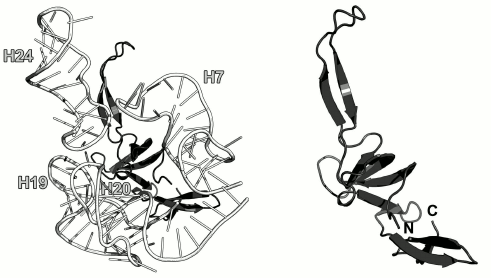
Ribosomal protein L24. L24, along with the L3 protein, is the initiator of the LRS assembly in bacteria [44, 70]. It interacts with the 23S rRNA 5′-terminus and nucleates the LRS folding [10]. In the E. coli LRS, L24 is located close to L23 in the vicinity of the nascent polypeptide exit from the ribosome and interacts with the 23S rRNA helices H7, H18-H20 and H24 (Fig. 12) [70, 71].
Fig. 12. Left, position of L24 on the 23S rRNA in E. coli 70S ribosome (PDB 5IT8). Right, schematic representation of L24 protein in the ribosome.
The structure of the isolated L24 has not been determined. In ribosomes, it is a β-protein, whose central part is folded into the SH3-like barrel (SCOP 50105) (Fig. 12). Interestingly, both the N- and C-termini of the L24 protein are located in the same domain; the rest of the polypeptide chain forms distinct domains and elongated loops. Similar architecture is also observed in the L1, L3, L4, and L23 protein reviewed above. It is possible that such spatial arrangement of the secondary structure elements facilitates the formation of a stable protein core, while “extensions” allow these proteins to reach the regions of the 23S rRNA that are distant from the area of the main interaction, thereby promoting correct folding of rRNA during ribosome assembly.
Analysis of contacts between amino acid residues of L24 and the 23S rRNA shows that the protein mainly interacts with atoms of the sugar-phosphate backbone of the double-stranded rRNA regions.
5S rRNA-BINDING RIBOSOMAL PROTEINS OF THE LRS
Along with the large 23S rRNA (2904 nucleotides in E. coli), bacterial LRS also contains small 5S rRNA (120 nucleotides) [72]. There are three r-proteins that bind the 5S rRNA: L5, L18 and L25 [73] (uL5, uL18, and bL25, respectively, according to the new nomenclature [6]). Together they form the central protuberance of the LRS that plays the key role in the ribosome functioning (Fig. 2) [74, 75].
Ribosomal protein L5. L5 binds to the loop C that covers the 5S rRNA helix III [76]. Its rRNA-binding properties are significantly promoted in the presence of L18 protein on the 5S rRNA [77, 78]. L5 interacts with the 23S rRNA helices H84-H85, thereby providing attachment of the 5S rRNA–protein complex to the LRS (Fig. 13a) [9, 79].
Fig. 13. a) Interaction of L5 with the 5S rRNA and 23S rRNA in E. coli 70S ribosome (PDB 5IT8). b) Crystal structure of L5 from G. stearothermophilus (PDB ID 1IQ4). c) The interaction site of E. coli L5 with the 5S rRNA (nucleotides 26-60) in the ribosome (PDB 5IT8). d) Crystal structure of L5 from T. thermophilus in a complex with the 5S rRNA fragment from E. coli (PDB ID 1MJI). Orientation of L5 in the panels (b)-(d) is the same. e) Zoomed view of the region of interaction of L5 and 5S rRNA. f) Zoomed view of the region of interaction of L5 and 23S rRNA.
As a part of the 70S ribosome, L5 protein interacts with the S13 protein of the SRS at the translation initiation stage. According to the 70S ribosome dynamics modeling, this interaction is affected by the binding of the translation elongation factor EF-G [42]. Comparison of the two structures of E. coli 70S ribosome determined independently by the X-ray analysis at a 3.5 Å resolution confirmed the dynamic nature of the L5 contacts with S13 and S19 [80].
L5 protein interacts with tRNAs in the ribosomal P site and, together with the 5S rRNA and other 5S rRNA-associated proteins, participates in the tRNA positioning in this ribosomal site [78, 80, 81].
The structure of the isolated L5 from G. stearothermophilus was determined in 2001 [82]. In 2003, the structure of the complex of L5 from T. thermophilus with a 5S rRNA fragment from E. coli was published (Fig. 13) [83]. Comparison of the structures of L5 alone, in a complex with the 5S rRNA fragment, and in ribosome demonstrated that, with an exception of short mobile protein fragments (e.g., helix α1, whose residues forms additional contacts with the 5S rRNA in the ribosome), the protein overall structure remains invariable [83].
L5 is an α+β protein with the antiparallel β-sheet and belongs to the RL5-like protein superfamily (SCOP 55282). Packaging of the secondary structure elements in L5 is similar to that in the RNP (ribonucleoprotein) domain [83] that binds RNA through the β-sheet surface. In the ribosome, L5 protein makes contacts with the 5S rRNA via amino acid residues of the loop α1-α2, strand β2, and terminus of α4 helix (Fig. 13e). Therefore, the 5S rRNA adjoins the edge of the β-sheet. This interaction involves multiple contacts between the side chains of the L5 amino acid residues and atoms of the 5S rRNA nucleotide bases. This could be explained by the fact that the site of L5 binding includes not only the double-stranded 5S rRNA region but also the loop C, which is a single-stranded rRNA region exposed to the intermolecular contacts.
L5 protein interacts with the 23S rRNA via the β-sheet surface that is typical for the RNP domains (Fig. 13f) [83]. It should be noted that the number of L5 contacts with the 23S rRNA is somewhat larger than with the 5S rRNA. These two facts correspond well to the data on the importance of L5 for binding of the 5S rRNA–protein complex to the LRS [78, 82].
Ribosomal protein L18. As it was mentioned above, L18 significantly increases the affinity of the L5 protein to the 5S rRNA [77, 78]. It is believed that L5 and L18 bind to the 5S rRNA in a cooperative manner [84]. Both proteins interact with a region of helix III and loop C of the 5S rRNA (Fig. 14a), so the cooperativity of L5 and L18 binding to the 5S rRNA is not surprising.
Fig. 14. a) Interaction of L5 and L18 with the 5S rRNA in E. coli 70S ribosome (PDB 5IT8); b) zoomed view of the region of interaction between L18 and 5S rRNA; c) zoomed view of the region of interaction between L18 and 23S rRNA; d) the NMR structure of L18 from T. thermophilus (PDB 1ILY); the N-terminal residue of the molecule portion with the determined structure (Arg22) is indicated; e) the NMR structure of L18 from G. stearothermophilus (PDB 1OVY); the N-terminal residue of the molecule portion with the determined structure (Gly24) is indicated.
The structure of the isolated L18 protein from T. thermophilus was determined by NMR in 2002 [85]. Two years later, the structure of L18 from G. stearothermophilus was also solved [86]. L18 belongs to the three-layer α/β proteins of the RNase H-like type (SCOP 53138). Twenty N-terminal amino acid residues that do not form any secondary structure in a solution were missing from the both structures (Fig. 14). In ribosomes, the N-terminal part of the protein acquires the α-helical structure and interacts with the 23S rRNA domain V, including the H83-H85 helix junction and helix H87 (Fig. 14c). This rRNA region also contacts with the edge of the β4-strand and the adjacent loop. The area of contact between L18 and the 5S rRNA is much broader and includes the β-sheet surface, which interacts with the 5S rRNA helix I, and the terminal parts of all α-helices that interact with the loops B and C of the 5S rRNA helix III (Fig. 14b). By comparing the interactions of proteins L5 and L18 with the 5S rRNA and 23S rRNA, one may see that they are “mirror-like”: L18 forms more contacts with the 5S rRNA and interacts with it via the β-sheet, whereas L5 makes more contacts with the 23S rRNA and also interacts with it via the β-sheet. At the same time, L18 binds to two spatially distant 5S rRNA helices – I and III, which should stabilize their mutual arrangement and the three-dimensional structure of 5S rRNA in the ribosome [87]. These structural features of the L5 and L18 interactions with rRNA correspond well to the previously defined roles of these proteins in the ribosome assembly [75, 78, 84, 85, 88].
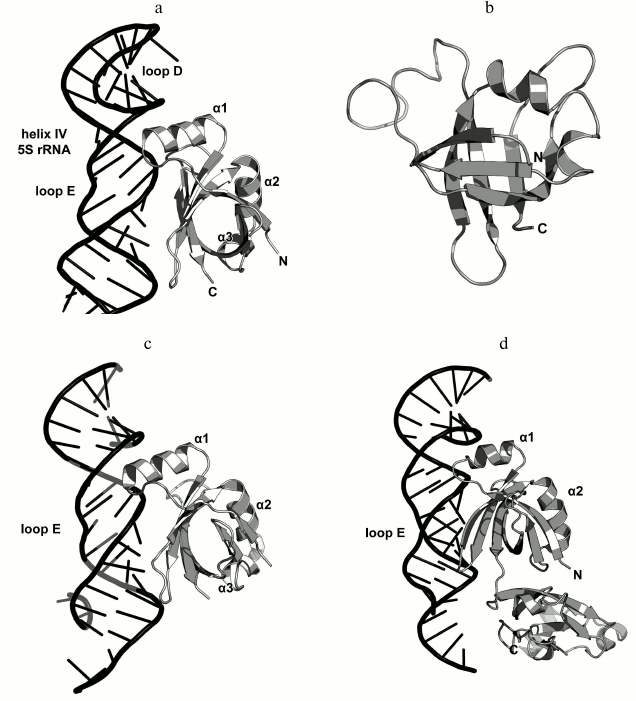
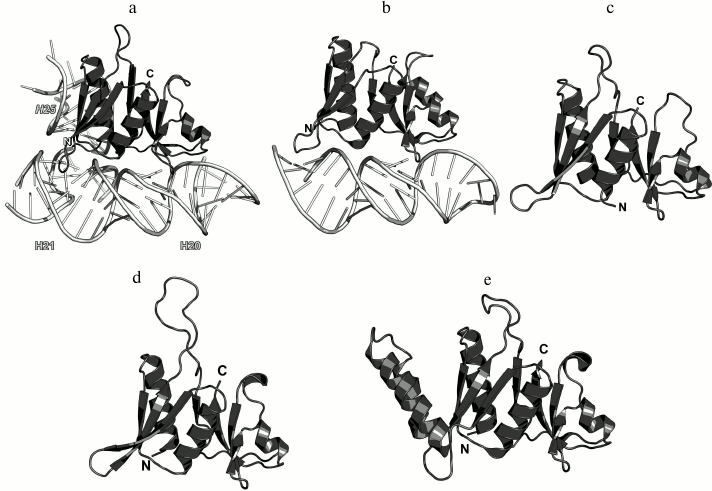
Ribosomal protein L25/TL5. The third 5S rRNA-binding protein is L25 (according to E. coli nomenclature). It binds to the 5S rRNA independently of L5 and L18 and is not required for attachment of the 5S rRNA–protein complex to the 23S rRNA [77, 89]. L25 interacts with the 5S rRNA loop E region situated between helices IV and V, which is the most distant from the site of L5 and L18 binding (Fig. 2) [73, 90]. In contrast to L5 and L18, L25 is not required for the survival of bacterial cells [73, 79]. Its function in the ribosome is not clear, although it was suggested that L25 contributes to the stabilization and proper operation of such functional centers of bacterial LRS as the A and P sites [73, 91].
The structure of the isolated L25 from E. coli was determined by the NMR (Fig. 15) [92]; the structure of L25 bound to the 5S rRNA fragment was solved using X-ray crystallography [93]. According to the SCOP classification, L25 protein belongs to a family of β-proteins with the β-barrel topology (SCOP 50714), although it also contains three α-helices. In the L25–5S rRNA complex, the protein interacts with the sugar-phosphate backbone of the loop E and recognizes its specific spatial structure [93] via amino acid residues of the β-sheet surface and the α1 helix (Fig. 15).
Fig. 15. a) Interaction of L25 with the 5S rRNA in E. coli 70S ribosome (PDB 5IT8); b) NMR structure of E. coli L25 (PDB 1B75); c) crystal structure of E. coli L25 in a complex with the 5S rRNA fragment (PDB 1DFU); d) crystal structure of T. thermophilus TL5 in a complex with the 5S rRNA fragment (PDB 1FEU).
One year later, the structure of the complex of a 5S rRNA fragment with the TL5 protein from T. thermophilus (Fig. 15d) was determined [94]. The N-terminal domain of TL5 is homologous to the E. coli L25 protein. It was demonstrated that TL5 can functionally substitute L25 in the reconstituted E. coli ribosomes [95]. In the complex, only the N-terminal domain, but not the C-terminal domain, interacts with the 5S rRNA fragment. An interesting feature of the TL5 N-terminal domain is that the helix α1 has less turns as compared to E. coli L25 and almost does not form contacts with the 5S rRNA. Therefore, the recognition of the RNA fragment by the TL5 protein is provided exclusively by the amino acid residues of the β-sheet surface of the N-terminal domain.
16S rRNA-BINDING RIBOSOMAL PROTEINS OF THE SRS
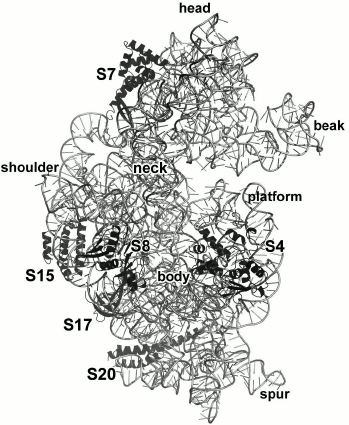
The 16S rRNA is almost twice as short as the 23S rRNA (1542 and 2904 nt in E. coli ribosomes, respectively) [96]. The number of primary 16S rRNA-binding r-proteins is also smaller – S4, S7, S8, S15, S17, and S20 (Fig. 16).
Fig. 16. Position of S4, S7, S8, S15, S17, and S20 in E. coli SRS (PDB 5IT8).
Ribosomal protein S4. S4 is one of the two initiator proteins in the SRS assembly [97]. It binds to the 16S rRNA and nucleates folding of the SRS body [97-99]. The binding site for S4 is situated at a junction of five helices (H3, H4, H16, H17 and H18) at the beginning of the 16S rRNA 5′-domain. The protein interacts with the sugar-phosphate backbones of three of these helices (Fig. 17) and also directly with the 5′-terminus of the 16S rRNA and nucleotides of the helix H21 terminal loop [97, 100]. Such pivotal location of the S4 protein correlates well with the biochemical data on the chemical modifications of nitrogenous bases and protection of the 16S rRNA sugar-phosphate backbone [101].
Fig. 17. a) Interaction of S4 with the 16S rRNA in E. coli SRS (PDB 5IT8); b) S4 structure in E. coli ribosome (PDB 5IT8); c) NMR structure of S4 from G. stearothermophilus (PDB 1C05).
The structure of the isolated S4 from G. stearothermophilus was determined by X-ray crystallography [102] and NMR [103] (Fig. 17). In both cases, the protein lacked 41 N-terminal amino acid residues, as they are disordered in a solution. In the SRS, the N-terminal part of the protein is partially structured and has two small α-helices.
S4 consists of two domains. Again, the second domain is an insert into the first one, which is typical for some r-proteins. The first domain is entirely α-helical; the second one is an α+β domain that consists of a five-strand antiparallel β-sheet with three α-helices located on one of its sides. The spatial arrangement of the secondary structure in the second domain corresponds to the α-L RNA-binding motif (SCOP 55174).
The 16S rRNA interacts with amino acid residues of four α-helices of both domains and two α-helical regions of the unstructured N-terminal part of the protein. The contacts are formed by atoms of the S4 amino acid residues and atoms of the 16S rRNA sugar-phosphate backbone and also some atoms of its nitrogenous bases. Amino acid residues of the β-sheet do not participate in the interaction with the 16S rRNA.
Ribosomal protein S7. S7 is the second initiator protein of SRSs; its binding to the16S rRNA leads to the assembly of SRS head [97]. The protein is situated at the interface side of the SRS close to the SRS decoding center, where it interacts with the mRNA [104]. There are data that S7 contacts tRNA in the P and E sites [105].
The S7-binding site in the 16S rRNA includes the junction of 16S rRNA helices H28, H29, and H43 and nearby helix H41 (Fig. 18a). The N-terminal amino acid residues of the protein form single contacts with the helix H37 loop in E. coli ribosomes (PDB 5IT8). S7 interacts with the 16S rRNA sugar-phosphate backbone predominantly via amino acid residues of α-helices. The long N-terminus of the protein is arranged along the groove of the rRNA helix H43 and forms the surface of contact with the RNA. The small β-hairpin in S7 forms only a couple of contacts with the 16S rRNA. In general, the interaction of S7 with the 16S rRNA is unique: the protein α-helices are not introduced into the grooves of the RNA helices; the β-hairpin is small and does not form a surface for the RNA helix binding. Apparently, the interaction is provided by a complex spatial structure of the 16S rRNA that is specifically recognized by the S7 protein.
Fig. 18. a) Interaction of S7 with the 16S rRNA in E. coli SRS (PDB 5IT8); b) crystal structure of S7 from G. stearothermophilus (PDB 1HUS); c) crystal structure of S7 from T. thermophilus (PDB 1RSS).
The structures of isolated S7 proteins from G. stearothermophilus [106] and T. thermophilus [107] were determined in 1997 (Fig. 18). S7 protein is classified as an all-α protein consisting of five α-helices with an additional α-helix and β-hairpin outside the protein core (SCOP 47974). Comparison of the structures of both S7 proteins in the isolated state and as parts of the ribosome showed that binding to the 16S rRNA does not noticeably change the structure of the proteins.
Ribosomal protein S8. S8 directly interacts with central domain of the 16S rRNA and coordinates assembly of the SRS platform with participation of the proteins S5 and S12 [108, 109]. In the ribosome, it interacts with the conserved region of helix H21 by binding to the sugar-phosphate backbone of double-stranded RNA. It also interacts with the adjacent helix H20 and forms extensive contacts with the 16S rRNA helix H25 (Fig. 19a) [101, 110]. Therefore, S8 protein stabilizes the spatial arrangement of this SRS platform region.
Fig. 19. a) Interaction of S8 with the16S rRNA in E. coli SRS (PDB 5IT8). b) Crystal structure of S8 from the archaeon M. jannaschii in a complex with the 16S rRNA fragment (PDB 1I6U). c) Crystal structure of S8 from G. stearothermophilus (PDB 1SEI). d) Crystal structure of S8 from T. thermophilus (PDB 1AN7). e) Crystal structure of S8 from A. aeolicus (PDB 3RF2).
It was shown that a small fragment of the 16S rRNA helix H21 is sufficient for the S8 binding [111, 112]. In 2001, the structure of archaeal S8 complexed to the corresponding 16S rRNA fragment from Methanococcus jannaschii was published (Fig. 19b) [113] demonstrating that the protein interacts with the RNA minor grove. At the same time, S8 forms a bridge between two turns of the RNA helix by its two domains. It was also established that the majority of contacts are formed between nucleotides and conserved amino acid residues of the C-terminal domain [113].
In 2014, these conclusions were confirmed by the structural studies of the complexes of S8 from Bacillus anthracis with RNA aptamers [114]. It was also shown that nucleotides of free RNA aptamers form numerous non-canonical base pairings, whereas interaction with S8 causes these RNAs to acquire the classical S8-recognized fold and also changes interactions between the aptamer nucleotide bases. It was concluded that S8 is able to change the secondary and tertiary structures of RNA upon binding [114].
The structure of the isolated S8 was determined in 1996 for G. stearothermophilus [115], in 1998 for T. thermophilus [116], and in 2012 for Aquifex aeolicus [117] (Fig. 19). S8 consists of two domains and belongs to the α+β class of proteins. The arrangement of the secondary structure elements corresponds to the four-layer b/a/b/a structure with two subdomains and, according to SCOP, is characteristic of a separate family (SCOP 56048). It should be noted that S8 from A. aeolicus has two additional α-helices in the N-terminal domain (Fig. 19e), which considerably increases its affinity to the region of junction of helices H20, H21, and H22 in the 16S rRNA [117].
Ribosomal protein S15. S15 binds to the 16S rRNA in the vicinity of S8 and also participates in the SRS platform assembly [42, 80]. In E. coli 70S ribosomes, S15 contacts the 23S rRNA of the LRS, thereby forming the intersubunit bridge B4 [42].
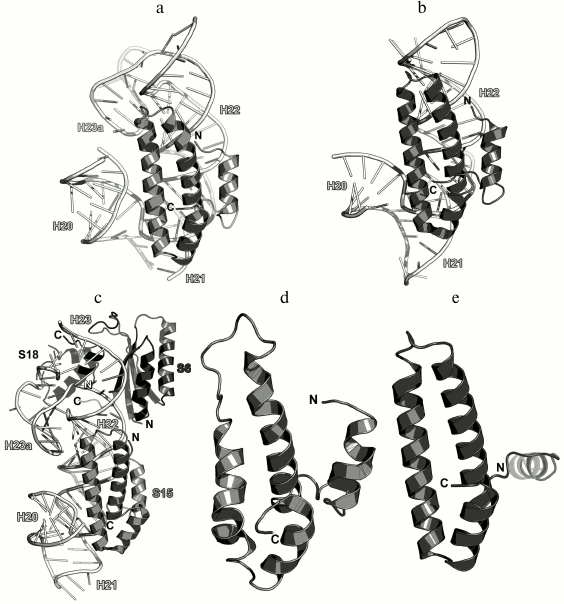
As a part of the SRS, S15 is located along the 16S rRNA helix H22 and forms numerous contacts with its sugar-phosphate backbone (Fig. 20). At the same time, the opposite termini of helices α2 and α3 stabilize two helical junctions formed by helices H20, H21, and H22 and by helices H23a, H22, and H23. As a result, S15 plays a crucial role in the formation of the spatial structure of the 16S rRNA central domain in the region of helix H22 [118, 119].
Fig. 20. a) Interaction of S15 with the 16S rRNA in E. coli SRS (PDB 5IT8); b) crystal structure of S15 from T. thermophilus in a complex with the 16S rRNA (PDB 1DK1); c) crystal structure of the complex of T. thermophilus r-proteins S6, S15, and S18 with the 16S rRNA fragment (PDB 1G1X); d) NMR structure of S15 from T. thermophilus (PDB 1AB3); e) crystal structure of S15 from G. stearothermophilus (PDB 1A32).
Analysis of the structure of the S15 protein from T. thermophilus in a complex with the 16S rRNA fragment demonstrated that the protein stabilizes mutual orientation of three 16S rRNA helices – helix H22 with adjacent fragments of helices H20 add H21 – via interactions with their sugar-phosphate backbones (Fig. 20b) [118]. Concurrently, the structure of the complex of larger 16S rRNA with proteins S6, S15, and S18 was published that uncovered the precise mechanism of the 16S rRNA central domain formation in the SRS (Fig. 20c) [119]. Until now, this complex remains the largest rRNA fragment complexed with several r-proteins.
The structures of isolated S15 proteins from T. thermophilus (Fig. 20d) [120] and G. stearothermophilus were also determined (Fig. 20e) [121]. S15 belongs to the class of all-α proteins. In addition, S15 was assigned to a separate family of S15/NS1 RNA-binding proteins with irregular packing of three α-helices and additional N-terminal α-helix (SCOP 47064). Indeed, comparison of S15 structures suggests the mobility of the N-terminal α-helix relatively to other helices of the protein, which is especially obvious in structure of S15 from G. stearothermophilus.
Ribosomal protein S17. S17 specifically binds to the 5′-terminal domain of the 16S rRNA in the region of helix H11 [108] and stabilizes junction between helices H6, H6a, H7, H11, and H12 [100]. In the SRS, S17 is located close to the primary binding proteins S7 and S15. The loops of this protein reach 16S rRNA fragments that are remote from the helix H11 (Fig. 21). S17 forms single contacts with regions of helices H7, H12, H20, H21, and H44.
Fig. 21. Left, interaction of S17 with helix H11 of the 16S rRNA in E. coli SRS (PDB 5IT8). Right, region of binding of S8, S15 and S17 to the 16S rRNA in E. coli SRS (PDB 5IT8).
S17 plays an important role in the control of the translation fidelity, as amino acid changes in this protein confer resistance to the antibiotic neomycin, which, in its turn, reduces mRNA misreading in vitro [122, 123].
In 1993, the structure of the isolated S17 was determined by the NMR [124], and S17 was classified as an all-β protein. An unusual incident is associated with this PDB entry: the structure of the protein deposited to the data bank turned out to be a mirror image of the real protein structure.
S17 protein is an all-β protein with the widely occurring supersecondary structure of the OB fold type. Its structure represents a β-barrel that belongs to the cold shock DNA-binding domain-like family (SCOP 50282). Despite its resemblance to proteins of this class, virtually all contacts with the16S rRNA are formed by amino acid residues of the loops between the β-strands, whereas residues of the β-sheet form only a small number of contacts with the RNA.
Ribosomal protein S20. S20 was found only in bacterial ribosomes [6]. It binds to the 16S rRNA 5′-terminal region independently of other bacterial r-proteins [7, 8]. Furthermore, there are two alternative sites of initial protein binding to the rRNA [125]. There are data suggesting that S20 stabilizes the region of helices H6, H7, H8, H9, H11, and H13 [100]. In the ribosome, the protein is located between the above helices and contacts functionally important 16S rRNA helix H44 (Fig. 22). The deficit of S20 significantly reduces the rate of mRNA binding by ribosomes and decreases the SRS affinity to the LRS, which results in the reduced content of 70S ribosomes in vitro [126, 127]. The absence of S20 protein affects the formation of the SRS–fMet-tRNA–mRNA initiation complex [128]. Bacterial strains with deleted S20 protein grow significantly slower than the control bacteria [126]. Therefore, S20 plays an important role in the SRS functioning.
Fig. 22. Interaction of S20 with the 16S rRNA in E. coli SRS (PDB 5IT8). Left, side view at the protein α-helices; right, top view at the protein α-helices.
S20 is a small protein (under 90 a.a.). The structure of the isolated S20 has not been solved yet. S20 is an all-α protein; it contains three α-helices with the spectrin-like fold and, according to the SCOP classification, belongs to a separate protein superfamily (SCOP 46992).
S20 interacts with the 16S rRNA through amino acid residues of its α-helices that are not inserted into the RNA grooves but situated somewhat aside. For example, the second and the third α-helices of the protein contact the 16S rRNA helix H9 by being positioned together in a close vicinity of “ridges” of the sugar-phosphate backbone (Fig. 22).
Analysis of the structural data on the interactions between primary RNA-binding r-proteins and rRNA indicates that these proteins recognize and bind double-stranded RNA regions. The side chains of conserved amino acid residues typically interact with atoms of the RNA sugar-phosphate backbone, i.e., this interaction is a classic example of the recognition of a specific spatial RNA fold. The contacts between amino acid residues and nucleotide bases are rare and typical for the RNA loops, where the bases may be unpaired and accessible for the intermolecular interactions.
The second feature of the interactions between r-proteins and rRNAs is the multiplicity of contacts for the primary binding r-proteins. These contacts stabilize the spatial structure of the 5S rRNA, 16S rRNA, and 23S rRNA in ribosomes and provide formation of functionally important sites, including the areas of the SRS–LRS interaction.
Acknowledgments
The author expresses gratitude to his colleagues for their help in working with the materials of this review, especially to S. V. Tishchenko, N. A. Nevskaya, S. V. Nikonov, and M. B. Garber.
This work was supported by the Molecular and Cell Biology Program of the Presidium of the Russian Academy of Sciences.
REFERENCES
1.Spirin, A. S. (2011) Molecular Biology.
Ribosomes and Protein Biosynthesis [in Russian], Akademiya,
Moscow.
2.Klein, D. J., Moore, P. B., and Steitz, T. A.
(2004) The roles of ribosomal proteins in the structure assembly, and
evolution of the large ribosomal subunit, J. Mol. Biol.,
340, 141-177.
3.Lecompte, O., Ripp, R., Thierry, J.-C., Moras, D.,
and Poch, O. (2002) Comparative analysis of ribosomal proteins in
complete genomes: an example of reductive evolution at the domain
scale, Nucleic Acids Res., 30, 5382-5390.
4.Korobeinikova, A. V., Garber, M. B., and Gongadze,
G. M. (2012) Ribosomal proteins: structure, function and evolution,
Biochemistry (Moscow), 77, 562-574.
5.Hartman, H., Favaretto, P., and Smith, T. F. (2006)
The archaeal origins of the eukaryotic translational system,
Archaea, 2, 1-9.
6Ban, N., Beckmann, R., Cate, J. H., Dinman, J. D., Dragon, F., Ellis,
S. R., Lafontaine, D. L., Lindahl, L., Liljas, A., Lipton, J. M.,
McAlear, M. A., Moore, P. B., Noller, H. F., Ortega, J., Panse, V. G.,
Ramakrishnan, V., Spahn, C. M., Steitz, T. A., Tchorzewski, M.,
Tollervey, D., Warren, A. J., Williamson, J. R., Wilson, D., Yonath,
A., and Yusupov, M. (2014) A new system for naming ribosomal proteins,
Curr. Opin. Struct. Biol., 24, 165-169.
7.Kaczanowska, M., and Ryden-Aulin, M. (2007)
Ribosome biogenesis and the translation process in Escherichia
coli, Microbiol. Mol. Biol. Rev., 71,
477-494.
8.Hamacher, K., Trylska, J., and McCammon, J. A.
(2006) Dependency map of proteins in the small ribosomal subunit,
PLoS Comput. Biol., 2, e10.
9.Rohl, R., and Nierhaus, K. H. (1982) Assembly map
of the large subunit (50S) of Escherichia coli ribosomes,
Proc. Natl. Acad. Sci. USA, 79, 729-733.
10.Herold, M., and Nierhaus, K. H. (1987)
Incorporation of six additional proteins to complete the assembly map
of the 50S subunit from Escherichia coli ribosomes, J. Biol.
Chem., 262, 8826-8833.
11.Spillmann, S., Dohme, F., and Nierhaus, K. H.
(1977) Assembly in vitro of the 50S subunit from Escherichia
coli ribosomes: proteins essential for the first heat-dependent
conformational change, J. Mol. Biol., 115, 513-523.
12.Held, W. A., and Nomura, M. (1973) Rate
determining step in the reconstitution of Escherichia coli 30S
ribosomal subunits, Biochemistry, 12, 3273-3281.
13.Schuwirth, B. S. (2005) Structures of the
bacterial ribosome at 3.5 Å resolution, Science,
310, 827-834.
14.Porse, B. T., and Garrett, R. A. (1999) Ribosomal
mechanics, antibiotics, and GTP hydrolysis, Cell, 97,
423-426.
15.Diaconu, M., Kothe, U., Schlunzen, F., Fischer,
N., Harms, J. M., Tonevitsky, A. G., Stark, H., Rodnina, M. V., and
Wahl, M. C. (2005) Structural basis for the function of the ribosomal
L7/12 stalk in factor binding and GTPase activation, Cell,
121, 991-1004.
16.Leijonmarck, M., and Liljas, A. (1987) Structure
of the C-terminal domain of the ribosomal protein L7/L12 from
Escherichia coli at 1.7 Å, J. Mol.
Biol., 195, 555-579.
17.Mitroshin, I. V., Garber, M. B., and
Gabdulkhakov, A. G. (2016) Investigation of structure of the ribosomal
L12/P stalk, Biochemistry (Moscow), 81, 1589-1601.
18.Xing, Y., and Draper, D. E. (1996) Cooperative
interactions of RNA and thiostrepton antibiotic with two domains of
ribosomal protein L11, Biochemistry, 35, 1581-1588.
19.Jonker, H. R. A., Ilin, S., Grimm, S. K.,
Wohnert, J., and Schwalbe, H. (2006) L11 domain rearrangement upon
binding to RNA and thiostrepton studied by NMR spectroscopy, Nucleic
Acids Res., 35, 441-454.
20.Wimberly, B. T., Guymon, R., McCutcheon, J. P.,
White, S. W., and Ramakrishnan, V. (1999) A detailed view of a
ribosomal active site: the structure of the L11–RNA complex,
Cell, 97, 491-502.
21.Conn, G. L., Draper, D. E., Lattman, E. E., and
Gittis, A. G. (1999) Crystal structure of a conserved ribosomal
protein–RNA complex, Science, 284, 1171-1174.
22.Murzin, A. G., Brenner, S. E., Hubbard, T., and
Chothia, C. (1995) SCOP: a structural classification of proteins
database for the investigation of sequences and structures, J. Mol.
Biol., 247, 536-540.
23.Brennan, R. G., and Matthews, B. W. (1989) The
helix-turn-helix DNA binding motif, J. Biol. Chem., 264,
1903-1906.
24.Matthews, B. W., Ohlendorf, D. H., Anderson, W.
F., and Takeda, Y. (1982) Structure of the DNA-binding region of
lac repressor inferred from its homology with cro
repressor, Proc. Natl. Acad. Sci. USA, 79, 1428-1432.
25.Muller, C. W. (2001) Transcription factors:
global and detailed views, Curr. Opin. Struct. Biol., 11,
26-32.
26.Gudkov, A. T., Tumanova, L. G., Gongadze, G. M.,
and Bushuev, V. N. (1980) Role of different regions of ribosomal
proteins L7 and L10 in their complex formation and in the interaction
with the ribosomal 50S subunit, FEBS Lett., 109,
34-38.
27.Rosendahl, G., and Douthwaite, S. (1993)
Ribosomal proteins L11 and L10-(L12)4 and the antibiotic
thiostrepton interact with overlapping regions of the 23S rRNA backbone
in the ribosomal GTPase center, J. Mol. Biol., 234,
1013-1020.
28.Noeske, J., Wasserman, M. R., Terry, D. S.,
Altman, R. B., Blanchard, S. C., and Cate, J. H. D. (2015)
High-resolution structure of the Escherichia coli ribosome,
Nat. Struct. Mol. Biol., 22, 336-341.
28.Cocozaki, A. I., Altman, R. B., Huang, J.,
Buurman, E. T., Kazmirski, S. L., Doig, P., Prince, D. B., Blanchard,
S. C., Cate, J. H. D., and Ferguson, A. D. (2016) Resistance mutations
generate divergent antibiotic susceptibility profiles against
translation inhibitors, Proc. Natl. Acad. Sci. USA,
113, 8188-8193.
30.Lin, J., Gagnon, M. G., Bulkley, D., and Steitz,
T. A. (2015) Conformational changes of elongation factor G on the
ribosome during tRNA translocation, Cell, 160,
219-227.
31.Gagnon, M. G., Lin, J., and Steitz, T. A. (2016)
Elongation factor 4 remodels the A-site tRNA on the ribosome, Proc.
Natl. Acad. Sci. USA, 113, 4994-4999.
32.Fischer, N., Konevega, A. L., Wintermeyer, W.,
Rodnina, M. V., and Stark, H. (2010) Ribosome dynamics and tRNA
movement by time-resolved electron cryomicroscopy, Nature,
466, 329-333.
33.Trabuco, L. G., Schreiner, E., Eargle, J.,
Cornish, P., Ha, T., Luthey-Schulten, Z., and Schulten, K. (2010) The
role of L1 stalk–tRNA interaction in the ribosome elongation
cycle, J. Mol. Biol., 402, 741-760.
34.Ban, N., Nissen, P., Hansen, J., Moore, P. B.,
and Steitz, T. A. (2000) The complete atomic structure of the large
ribosomal subunit at 2.4 Å resolution, Science,
289, 905-920.
35.Selmer, M. (2006) Structure of the 70S ribosome
complexed with mRNA and tRNA, Science, 313,
1935-1942.
36.Nikonov, S., Nevskaya, N., Eliseikina, I.,
Fomenkova, N., Nikulin, A., Ossina, N., Garber, M., Jonsson, B. H.,
Briand, C., Al-Karadaghi, S., Svensson, A., Aevarsson, A., and Liljas,
A. (1996) Crystal structure of the RNA binding ribosomal protein L1
from Thermus thermophiles, EMBO J., 15,
1350-1359.
37.Tishchenko, S., Nikonova, E., Kostareva, O.,
Gabdulkhakov, A., Piendl, W., Nevskaya, N., Garber, M., and Nikonov, S.
(2011) Structural analysis of interdomain mobility in ribosomal L1
proteins, Acta Crystallogr. D, 67, 1023-1027.
38.Nikulin, A., Eliseikina, I., Tishchenko, S.,
Nevskaya, N., Davydova, N., Platonova, O., Piendl, W., Selmer, M.,
Liljas, A., Drygin, D., Zimmermann, R., Garber, M., and Nikonov, S.
(2003) Structure of the L1 protuberance in the ribosome, Nat.
Struct. Biol., 10, 104-108.
39.Willumeit, R., Forthmann, S., Beckmann, J.,
Diedrich, G., Ratering, R., Stuhrmann, H. B., and Nierhaus, K. H.
(2001) Localization of the protein L2 in the 50S subunit and the 70S
E. coli ribosome, J. Mol. Biol., 305, 167-177.
40.Diedrich, G., Spahn, C. M. T., Stelzl, U.,
Schafer, M. A., Wooten, T., Bochkariov, D. E., Cooperman, B. S., Traut,
R. R., and Nierhaus, K. H. (2000) Ribosomal protein L2 is involved in
the association of the ribosomal subunits, tRNA binding to A and P
sites and peptidyl transfer, EMBO J., 19, 5241-5250.
41.Cooperman, B. S., Wooten, T., Romero, D. P., and
Traut, R. R. (1995) Histidine 229 in protein L2 is apparently essential
for 50S peptidyl transferase activity, Biochem. Cell Biol.,
73, 1087-1094.
42.Gao, H., Sengupta, J., Valle, M., Korostelev, A.,
Eswar, N., Stagg, S. M., Van Roey, P., Agrawal, R. K., Harvey, S. C.,
Sali, A., Chapman, M. S., and Frank, J. (2003) Study of the structural
dynamics of the E. coli 70S ribosome using real-space
refinement, Cell, 113, 789-801.
43.Uhlein, M., Weglohner, W., Urlaub, H., and
Wittmann-Liebold, B. (1998) Functional implications of ribosomal
protein L2 in protein biosynthesis as shown by in vivo
replacement studies, Biochem. J., 331, 423-430.
44.Nowotny, V., and Nierhaus, K. H. (1982) Initiator
proteins for the assembly of the 50S subunit from Escherichia
coli ribosomes, Proc. Natl. Acad. Sci. USA,
79, 7238-7242.
45.Petrov, A., Meskauskas, A., and Dinman, J. D.
(2004) Ribosomal protein L3: influence on ribosome structure and
function, RNA Biol., 1, 59-65.
46.Pringle, M., Poehlsgaard, J., Vester, B., and
Long, K. S. (2004) Mutations in ribosomal protein L3 and 23S ribosomal
RNA at the peptidyl transferase center are associated with reduced
susceptibility to tiamulin in Brachyspira spp. isolates, Mol.
Microbiol., 54, 1295-1306.
47.Klitgaard, R. N., Ntokou, E., Norgaard, K.,
Biltoft, D., Hansen, L. H., Trædholm, N. M., Kongsted, J., and
Vester, B. (2015) Mutations in the bacterial ribosomal protein L3 and
their association with antibiotic resistance, Antimicrob. Agents
Chemother., 59, 3518-3528.
48.Urlaub, H., Kruft, V., Bischof, O., Muller, E.
C., and Wittmann-Liebold, B. (1995) Protein–rRNA binding features
and their structural and functional implications in ribosomes as
determined by cross-linking studies, EMBO J., 14,
4578-4588.
49.Wower, I., Wower, J., Meinke, M., and Brimacombe,
R. (1981) The use of 2-iminothiolane as an RNA–protein
cross-linking agent in Escherichia coli ribosomes, and the
localization on 23S RNA of sites cross-linked to proteins L4, L6, L21,
L23, L27 and L29, Nucleic Acids Res., 9, 4285-4302.
50.Gabashvili, I. S., Gregory, S. T., Valle, M.,
Grassucci, R., Worbs, M., Wahl, M. C., Dahlberg, A. E., and Frank, J.
(2001) The polypeptide tunnel system in the ribosome and its gating in
erythromycin resistance mutants of L4 and L22, Mol. Cell,
8, 181-188.
51.Zaman, S., Fitzpatrick, M., Lindahl, L., and
Zengel, J. (2007) Novel mutations in ribosomal proteins L4 and L22 that
confer erythromycin resistance in Escherichia coli, Mol.
Microbiol., 66, 1039-1050.
52.Chittum, H. S., and Champney, W. S. (1994)
Ribosomal protein gene sequence changes in erythromycin-resistant
mutants of Escherichia coli, J. Bacteriol., 176,
6192-6198.
53.Zengel, J. M. (2003) The extended loops of
ribosomal proteins L4 and L22 are not required for ribosome assembly or
L4-mediated autogenous control, RNA, 9, 1188-1197.
54.Worbs, M., Huber, R., and Wahl, M. C. (2000)
Crystal structure of ribosomal protein L4 shows RNA-binding sites for
ribosome incorporation and feedback control of the S10 operon, EMBO
J., 19, 807-818.
55.Hoffman, D. W., Davies, C., Gerchman, S. E.,
Kycia, J. H., Porter, S. J., White, S. W., and Ramakrishnan, V. (1994)
Crystal structure of prokaryotic ribosomal protein L9: a bi-lobed
RNA-binding protein, EMBO J., 13, 205-212.
56.Hoffman, D. W., Cameron, C. S., Davies, C.,
White, S. W., and Ramakrishnan, V. (1996) Ribosomal protein L9: a
structure determination by the combined use of X-ray crystallography
and NMR spectroscopy, J. Mol. Biol., 264, 1058-1071.
57.Selmer, M., Gao, Y.-G., Weixlbaumer, A., and
Ramakrishnan, V. (2012) Ribosome engineering to promote new crystal
forms, Acta Crystallogr. Sect. D, 68,
578-583.
58.Fischer, N., Neumann, P., Konevega, A. L., Bock,
L. V., Ficner, R., Rodnina, M. V., and Stark, H. (2015) Structure of
the E. coli ribosome–EF-Tu complex at <3 Å
resolution by Cs-corrected cryo-EM, Nature, 520,
567-570.
59.Naganathan, A., Wood, M. P., and Moore, S. D.
(2015) The large ribosomal subunit protein L9 enables the growth of
EF-P deficient cells and enhances small subunit maturation, PLoS
One, 10, e0120060.
60.Raibaud, S., Vachette, P., Guillier, M.,
Allemand, F., Chiaruttini, C., and Dardel, F. (2003) How bacterial
ribosomal protein L20 assembles with 23S ribosomal RNA and its own
messenger RNA, J. Biol. Chem., 278, 36522-36530.
61.Guillier, M., Allemand, F., Graffe, M., Raibaud,
S., Dardel, F., Springer, M., and Chiaruttini, C. (2005) The N-terminal
extension of Escherichia coli ribosomal protein L20 is important
for ribosome assembly, but dispensable for translational feedback
control, RNA, 11, 728-738.
62.Franceschi, F. J., and Nierhaus, K. H. (1988)
Ribosomal protein L20 can replace the assembly-initiator protein L24 at
low temperature, Biochemistry, 27, 7056-7059.
63.Raibaud, S., Lebars, I., Guillier, M.,
Chiaruttini, C., Bontems, F., Rak, A., Garber, M., Allemand, F.,
Springer, M., and Dardel, F. (2002) NMR structure of bacterial
ribosomal protein L20: implications for ribosome assembly and
translational control, J. Mol. Biol., 323, 143-151.
64.Timsit, Y., Allemand, F., Chiaruttini, C., and
Springer, M. (2006) Coexistence of two protein folding states in the
crystal structure of ribosomal protein L20, EMBO Rep., 7,
1013-1018.
65.Timsit, Y., Acosta, Z., Allemand, F.,
Chiaruttini, C., and Springer, M. (2009) The role of disordered
ribosomal protein extensions in the early steps of eubacterial 50S
ribosomal subunit assembly, Int. J. Mol. Sci., 10,
817-834.
66.Kramer, G., Rauch, T., Rist, W., Vorderwulbecke,
S., Patzelt, H., Schulze-Specking, A., Ban, N., Deuerling, E., and
Bukau, B. (2002) L23 protein functions as a chaperone docking site on
the ribosome, Nature, 419, 171-174.
67.Gu, S.-Q., Peske, F., Wieden, H.-J., Rodnina, M.
V., and Wintermeyer, W. (2003) The signal recognition particle binds to
protein L23 at the peptide exit of the Escherichia coli
ribosome, RNA, 9, 566-573.
68.Ullers, R. S., Houben, E. N. G., Raine, A., ten
Hagen-Jongman, C. M., Ehrenberg, M., Brunner, J., Oudega, B., Harms,
N., and Luirink, J. (2003) Interplay of signal recognition particle and
trigger factor at L23 near the nascent chain exit site on the
Escherichia coli ribosome, J. Cell Biol.,
161, 679-684.
69.Ohman, A., Rak, A., Dontsova, M., Garber, M. B.,
and Hard, T. (2003) NMR structure of the ribosomal protein L23 from
Thermus thermophiles, J. Biomol. NMR, 26,
131-137.
70.Spillmann, S., and Nierhaus, K. H. (1978) The
ribosomal protein L24 of Escherichia coli is an assembly
protein, J. Biol. Chem., 253, 7047-7050.
71.Mitra, K., Schaffitzel, C., Shaikh, T., Tama, F.,
Jenni, S., Brooks, C. L., Ban, N., and Frank, J. (2005) Structure of
the E. coli protein-conducting channel bound to a translating
ribosome, Nature, 438, 318-324.
72.Szymanski, M., Barciszewska, M. Z., Erdmann, V.
A., and Barciszewski, J. (2002) 5S ribosomal RNA database, Nucleic
Acids Res., 30, 176-178.
73.Gongadze, G. M., Korepanov, A. P., Korobeinikova,
A. V., and Garber, M. B. (2008) Bacterial 5S rRNA-binding proteins of
the CTC family, Biochemistry (Moscow), 73, 1405-1417.
74.Gongadze, G. M. (2011) 5S rRNA and ribosome,
Biochemistry (Moscow), 76, 1450-1464.
75.Selivanova, O. M., Gongadze, G. M., Gudkov, A.
T., and Vasiliev, V. D. (1986) Structure of protein-deficient 50S
ribosomal subunits. Particles without 5S RNA–protein complex
retain the L7/L12 stalk and associate with 30S subunits, FEBS
Lett., 197, 79-83.
76.Gongadze, G. M., Perederina, A. A.,
Meshcheriakov, V. A., Fedorov, R. V., Moskalenko, S. E., Rak, A. V.,
Serganov, A. A., Shcherbakov, D. V., Nikonov, S. V., and Garber, M. B.
(2001) The Thermus thermophilus 5S rRNA–protein complex:
identifications of specific binding sites for proteins L5 and L18 in 5S
rRNA, Mol. Biol. (Moscow), 35, 610-616.
77.Shpanchenko, O. V., Zvereva, M. I., Dontsova, O.
A., Nierhaus, K. H., and Bogdanov, A. A. (1996) 5S rRNA sugar-phosphate
backbone protection in complexes with specific ribosomal proteins,
FEBS Lett., 394, 71-75.
78.Spierer, P., and Zimmermann, R. A. (1978)
Stoichiometry, cooperativity, and stability of interactions between 5S
RNA and proteins L5, L18, and L25 from the 50S ribosomal subunit of
Escherichia coli, Biochemistry, 17, 2474-2479.
79.Korepanov, A. P., Gongadze, G. M., Garber, M. B.,
Court, D. L., and Bubunenko, M. G. (2007) Importance of the 5S
rRNA-binding ribosomal proteins for cell viability and translation in
Escherichia coli, J. Mol. Biol., 366,
1199-1208.
80.Osswald, M., Doring, T., and Brimacombe, R.
(1995) The ribosomal neighborhood of the central fold of tRNA:
cross-links from position 47 of tRNA located at the A, P or E site,
Nucleic Acids Res., 23, 4635-4641.
81.Korepanov, A. P., Korobeinikova, A. V.,
Shestakov, S. A., Garber, M. B., and Gongadze, G. M. (2012) Protein L5
is crucial for in vivo assembly of the bacterial 50S ribosomal
subunit central protuberance, Nucleic Acids Res.,
40, 9153-9159.
82.Nakashima, T., Yao, M., Kawamura, S., Iwasaki,
K., Kimura, M., and Tanaka, I. (2001) Ribosomal protein L5 has a highly
twisted concave surface and flexible arms responsible for rRNA binding,
RNA, 7, 692-701.
83.Perederina, A., Nevskaya, N., Nikonov, O.,
Nikulin, A., Dumas, P., Yao, M., Tanaka, I., Garber, M., Gongadze, G.,
and Nikonov, S. (2002) Detailed analysis of RNA–protein
interactions within the bacterial ribosomal protein L5/5S rRNA complex,
RNA, 8, 1548-1557.
84.Spierer, P., Wang, C. C., Marsh, T. L., and
Zimmermann, R. A. (1979) Cooperative interactions among protein and RNA
components of the 50S ribosomal subunit of Escherichia coli,
Nucleic Acids Res., 6, 1669-1682.
85.Woestenenk, E. A., Gongadze, G. M., Shcherbakov,
D. V., Rak, A. V., Garber, M. B., Hard, T., and Berglund, H. (2002) The
solution structure of ribosomal protein L18 from Thermus
thermophilus reveals a conserved RNA-binding fold, Biochem.
J., 363, 553-561.
86.Turner, C. F., and Moore, P. B. (2004) The
solution structure of ribosomal protein L18 from Bacillus
stearothermophilus, J. Mol. Biol., 335, 679-684.
87.Ciesiolka, J., Lorenz, S., and Erdmann, V. A.
(1992) Structural analysis of three prokaryotic 5S rRNA species and
selected 5S rRNA-ribosomal–protein complexes by means of
Pb(II)-induced hydrolysis, Eur. J. Biochem., 204,
575-581.
88.Newberry, V., and Garrett, R. A. (1980) The role
of the basic N-terminal region of protein L18 in 5S RNA–23S RNA
complex formation, Nucleic Acids Res., 8,
4131-4142.
89.Szymanski, M., Barciszewska, M. Z., Erdmann, V.,
and Barciszewski, J. (2003) 5S rRNA: structure and interactions,
Biochem. J., 371, 641-651.
90.Douthwaite, S., Garrett, R. A., Wagner, R., and
Feunteun, J. (1979) A ribonuclease-resistant region of 5S RNA and its
relation to the RNA binding sites of proteins L18 and L25, Nucleic
Acids Res., 6, 2453-2470.
91.Harms, J., Schluenzen, F., Zarivach, R., Bashan,
A., Gat, S., Agmon, I., Bartels, H., Franceschi, F., and Yonath, A.
(2001) High resolution structure of the large ribosomal subunit from a
mesophilic eubacterium, Cell, 107, 679-688.
92.Stoldt, M., Wohnert, J., Gorlach, M., and Brown,
L. R. (1998) The NMR structure of Escherichia coli ribosomal
protein L25 shows homology to general stress proteins and
glutaminyl-tRNA synthetases, EMBO J., 17, 6377-6384.
93.Lu, M., and Steitz, T. A. (2000) Structure of
Escherichia coli ribosomal protein L25 complexed with a 5S rRNA
fragment at 1.8 Å resolution, Proc. Natl. Acad. Sci. USA,
97, 2023-2028.
94.Fedorov, R., Meshcheryakov, V., Gongadze, G.,
Fomenkova, N., Nevskaya, N., Selmer, M., Laurberg, M., Kristensen, O.,
Al-Karadaghi, S., Liljas, A., Garber, M., and Nikonov, S. (2001)
Structure of ribosomal protein TL5 complexed with RNA provides new
insights into the CTC family of stress proteins, Acta Crystallogr.
D, 57, 968-976.
95.Gongadze, G. M., Korepanov, A. P., Korobeinikova,
A. V., and Garber, M. B. (2008) Bacterial 5S rRNA-binding proteins of
the CTC family, Biochemistry (Moscow), 73, 1405-1417.
96.Petrov, A. S., Bernier, C. R., Gulen, B.,
Waterbury, C. C., Hershkovits, E., Hsiao, C., Harvey, S. C., Hud, N.
V., Fox, G. E., Wartell, R. M., and Williams, L. D. (2014) Secondary
structures of rRNAs from all three domains of life, PLoS
One, 9, e88222.
97.Nowotny, V., and Nierhaus, K. H. (1988) Assembly
of the 30S subunit from Escherichia coli ribosomes occurs via
two assembly domains which are initiated by S4 and S7,
Biochemistry, 27, 7051-7055.
98.Stern, S., Powers, T., Changchien, L. M., and
Noller, H. F. (1989) RNA–protein interactions in 30S ribosomal
subunits: folding and function of 16S rRNA, Science, 244,
783-790.
99.Mayerle, M., and Woodson, S. A. (2013) Specific
contacts between protein S4 and ribosomal RNA are required at multiple
stages of ribosome assembly, RNA, 19, 574-585.
100.Ramaswamy, P., and Woodson, S. A. (2009) Global
stabilization of rRNA structure by ribosomal proteins S4, S17, and S20,
J. Mol. Biol., 392, 666-677.
101.Powers, T., and Noller, H. F. (1995) Hydroxyl
radical footprinting of ribosomal proteins on 16S rRNA, RNA,
1, 194-209.
102.Davies, C., Gerstner, R. B., Draper, D. E.,
Ramakrishnan, V., and White, S. W. (1998) The crystal structure of
ribosomal protein S4 reveals a two-domain molecule with an extensive
RNA-binding surface: one domain shows structural homology to the ETS
DNA-binding motif, EMBO J., 17, 4545-4558.
103.Markus, M. A., Gerstner, R. B., Draper, D. E.,
and Torchia, D. A. (1998) The solution structure of ribosomal protein
S4 delta41 reveals two subdomains and a positively charged surface that
may interact with RNA, EMBO J., 17, 4559-4571.
104.Greuer, B., Thiede, B., and Brimacombe, R.
(1999) The cross-link from the upstream region of mRNA to ribosomal
protein S7 is located in the C-terminal peptide: experimental
verification of a prediction from modeling studies, RNA,
5, 1521-1525.
105.Osswald, M., Doring, T., and Brimacombe, R.
(1995) The ribosomal neighborhood of the central fold of tRNA:
cross-links from position 47 of tRNA located at the A, P or E site,
Nucleic Acids Res., 23, 4635-4641.
106.Hosaka, H., Nakagawa, A., Tanaka, I., Harada,
N., Sano, K., Kimura, M., Yao, M., and Wakatsuki, S. (1997) Ribosomal
protein S7: a new RNA-binding motif with structural similarities to a
DNA architectural factor, Structure, 5, 1199-1208.
107.Wimberly, B. T., White, S. W., and
Ramakrishnan, V. (1997) The structure of ribosomal protein S7 at 1.9
Å resolution reveals a beta-hairpin motif that binds
double-stranded nucleic acids, Structure, 5,
1187-1198.
108.Held, W. A., Ballou, B., Mizushima, S., and
Nomura, M. (1974) Assembly mapping of 30S ribosomal proteins from
Escherichia coli, further studies, J. Biol. Chem.,
249, 3103-3111.
109.Ramakrishnan, V., Wimberly, B. T., Brodersen,
D. E., Clemons, W. M., Morgan-Warren, R. J., Carter, A. P., Vonrhein,
C., and Hartsch, T. (2000) Structure of the 30S ribosomal subunit,
Nature, 407, 327-339.
110.Svensson, P., Changchien, L. M., Craven, G. R.,
and Noller, H. F. (1988) Interaction of ribosomal proteins, S6, S8, S15
and S18 with the central domain of 16S ribosomal RNA, J. Mol.
Biol., 200, 301-308.
111.Allmang, C., Mougel, M., Westhof, E.,
Ehresmann, B., and Ehresmann, C. (1994) Role of conserved nucleotides
in building the 16S rRNA binding site of E. coli ribosomal
protein S8, Nucleic Acids Res., 22,
3708-3714.
112.Mougel, M., Allmang, C., Eyermann, F., Cachia,
C., Ehresmann, B., and Ehresmann, C. (1993) Minimal 16S rRNA binding
site and role of conserved nucleotides in Escherichia coli
ribosomal protein S8 recognition, Eur. J. Biochem., 215,
787-792.
113.Tishchenko, S., Nikulin, A., Fomenkova, N.,
Nevskaya, N., Nikonov, O., Dumas, P., Moine, H., Ehresmann, B.,
Ehresmann, C., Piendl, W., Lamzin, V., Garber, M., and Nikonov, S.
(2001) Detailed analysis of RNA–protein interactions within the
ribosomal protein S8–rRNA complex from the archaeon
Methanococcus jannaschii, J. Mol. Biol., 311,
311-324.
114.Davlieva, M., Donarski, J., Wang, J., Shamoo,
Y., and Nikonowicz, E. P. (2014) Structure analysis of free and bound
states of an RNA aptamer against ribosomal protein S8 from
Bacillus anthracis, Nucleic Acids Res.,
42, 10795-10808.
115.Davies, C., Ramakrishnan, V., and White, S. W.
(1996) Structural evidence for specific S8–RNA and
S8–protein interactions within the 30S ribosomal subunit:
ribosomal protein S8 from Bacillus stearothermophilus at 1.9
Å resolution, Structure, 4, 1093-1104.
116.Nevskaya, N., Tishchenko, S., Nikulin, A.,
Al-Karadaghi, S., Liljas, A., Ehresmann, B., Ehresmann, C., Garber, M.,
and Nikonov, S. (1998) Crystal structure of ribosomal protein S8 from
Thermus thermophilus reveals a high degree of structural
conservation of a specific RNA binding site, J. Mol.
Biol., 279, 233-244.
117.Menichelli, E., Edgcomb, S. P., Recht, M. I.,
and Williamson, J. R. (2012) The structure of Aquifex aeolicus
ribosomal protein S8 reveals a unique subdomain that contributes to an
extremely tight association with 16S rRNA, J. Mol. Biol.,
415, 489-502.
118.Nikulin, A., Serganov, A., Ennifar, E.,
Tishchenko, S., Nevskaya, N., Shepard, W., Portier, C., Garber, M.,
Ehresmann, B., Ehresmann, C., Nikonov, S., and Dumas, P. (2000) Crystal
structure of the S15–rRNA complex, Nat. Struct. Mol.
Biol., 7, 273-277.
119.Agalarov, S. C. (2000) Structure of the S15,
S6, S18–rRNA complex: assembly of the 30S ribosome central
domain, Science, 288, 107-112.
120.Berglund, H., Rak, A., Serganov, A., Garber,
M., and Hard, T. (1997) Solution structure of the ribosomal RNA binding
protein S15 from Thermus thermophiles, Nat. Struct.
Biol., 4, 20-23.
121.Clemons, W. M., Davies, C., White, S. W., and
Ramakrishnan, V. (1998) Conformational variability of the N-terminal
helix in the structure of ribosomal protein S15, Structure,
6, 429-438.
122.Yaguchi, M., Wittmann, H. G., Cabezon, T.,
DeWilde, M., Villarroel, R., Herzog, A., and Bollen, A. (1976)
Alteration of ribosomal protein S17 by mutation linked to neamine
resistance in Escherichia coli. II. Localization of the amino
acid replacement in protein S17 from a meaA mutant, J. Mol.
Biol., 104, 617-620.
123.Bollen, A., Cabezon, T., De Wilde, M.,
Villarroel, R., and Herzog, A. (1975) Alteration of ribosomal protein
S17 by mutation linked to neamine resistance in Escherichia
coli. I. General properties of neaA mutants, J. Mol.
Biol., 99, 795-806.
124.Golden, B. L., Hoffman, D. W., Ramakrishnan,
V., and White, S. W. (1993) Ribosomal protein S17: characterization of
the three-dimensional structure by proton and nitrogen-15 NMR,
Biochemistry, 32, 12812-12820.
125.Hedrick, E. G., and Hill, W. E. (2010) Protein
S20 binds two 16S rRNA sites as assembly is initiated, J. Mol.
Biol., 401, 493-502.
126.Tobin, C., Mandava, C. S., Ehrenberg, M.,
Andersson, D. I., and Sanyal, S. (2010) Ribosomes lacking protein S20
are defective in mRNA binding and subunit association, J. Mol.
Biol., 397, 767-776.
127.Ryden-Aulin, M., Shaoping, Z., Kylsten, P., and
Isaksson, L. A. (1993) Ribosome activity and modification of 16S RNA
are influenced by deletion of ribosomal protein S20, Mol.
Microbiol., 7, 983-992.
128.Gotz, F., Dabbs, E. R., and Gualerzi, C. O.
(1990) Escherichia coli 30S mutants lacking protein S20 are
defective in translation initiation, Biochim. Biophys. Acta,
1050, 93-97.