Aim23p Interacts with the Yeast Mitochondrial Ribosomal Small Subunit
I. V. Chicherin1, V. V. Zinina1, S. A. Levitskiy1, M. V. Serebryakova2, and P. A. Kamenski1,a*
1Lomonosov Moscow State University, Faculty of Biology, 119991 Moscow, Russia2Belozersky Institute of Physico-Chemical Biology, Lomonosov Moscow State University, 119991 Moscow, Russia
* To whom correspondence should be addressed.
Received August 23, 2018; Revised September 12, 2018; Accepted September 12, 2018
Protein synthesis in mitochondria is generally organized in a bacterial-like manner but, at the same time, possesses several unique traits. Translation initiation in mitochondria is regulated by two protein factors, mtIF2 and mtIF3. Previously we demonstrated that Saccharomyces cerevisiae Aim23 protein is an ortholog of IF3 in budding yeast. However, the data on the interactions between Aim23p and other proteins are limited. Here, we demonstrated that Aim23p interacts with the yeast mitochondrial ribosomal small subunit both in vivo and in vitro using co-immunoprecipitation and density gradient sedimentation.
KEY WORDS: mitochondria, translation, ribosomes, initiation factor, Aim23pDOI: 10.1134/S000629791901005X
Mitochondria are eukaryotic cell organelles responsible for a variety of functions, including respiration, ATP production, synthesis of fatty acids and iron-sulfur clusters, heme metabolism, and orchestration of programmed cell death (apoptosis). Mitochondria have originated from endosymbiotic α-proteobacteria [1] and preserved their own ancestral genome and protein expression system. Mitochondrial genomes of animals and fungi are relatively small and encode a limited set of transcripts, as most of mitochondrial genes have migrated to the nucleus in the process of evolution. The mitochondrial genome of Saccharomyces cerevisiae budding yeast represents an approximately 85-kb circular DNA bearing eight genes [2], most of which encode membrane-embedded components of the respiratory chain complexes. Proteins encoded by the mitochondrial DNA are COX1, COX2, COX3 (cytochrome c oxidase subunits; complex IV), ATP6, ATP8, ATP9 (ATP synthase subunits; complex V), cytochrome b (complex III), and one mitochondrial ribosomal protein (VAR1).
Mitochondrial translation apparatus is finely tuned for the synthesis of a limited set of proteins, resulting in highly specialized organellar ribosomes [3] and translation factors that considerably differ from the canonical translation factors [4]. Mitochondrial initiation factor 2 (mtIF2) is the only one out of the classical bacterial translation initiation factors (IF1, IF2, and IF3) that is universally conserved in mitochondria. Homologs of bacterial initiation factor IF1 have never been found in S. cerevisiae. It had also been believed that yeast lack IF3 [5]. However, using phylogenetic analysis and in vivo complementation tests, yeast Aim23 protein was identified as a mitochondrial ortholog of bacterial IF3 [6]. Taking into account the critical role of IF3 in the ribosomal functional cycle, it is not surprising that the corresponding gene (infC) is essential in bacteria [7]. However, this is not the case for yeast mitochondria, where deletion of the AIM23 gene failed to arrest mitochondrial translation but rather led to the imbalance in the amounts of proteins synthesized in mitochondria [8]. In particular, the synthesis of Atp9p was upregulated, while the synthesis of complex IV subunits (Cox1p, Cox2p, and Cox3p) was downregulated in the AIM23Δ yeast strain, as compared to the parental wild-type strain. This closely resembles the situation observed for another group of proteins involved in mitochondrial translation in budding yeast, namely translational activators. Translational activators interact with specific mRNAs to facilitate their recruitment to the ribosomes and subsequent translation [9]. They bind to the 5′-UTRs of mRNAs, large and small subunits of mitochondrial ribosomes, and mitochondrial inner membrane. Deletions of genes encoding translational activators result in rapid mRNA degradation [10], translational repression [11], depletion of other translational activators [12], and upregulation of synthesis of other mitochondrial DNA-encoded proteins [13]. Selective effect of AIM23 deletion on the mitochondrial mRNAs encoding subunits of cytochrome c oxidase and ATP synthase suggests that Aim23p may act as a translational activator or be involved in the translational activator network; however, the molecular mechanisms of this phenomenon have never been investigated.
Interaction between Aim23p and yeast mitochondrial ribosome has been demonstrated in two independent studies. Kehrein at al. detected Aim23p in complexes with GFP-tagged mitochondrial ribosomes immunoprecipitated with anti-GFP antibodies [14]. Interestingly, Aim23p, as well as several classical translational activators, was found to interact with the ribosomes if the latter were isolated under the low-salt conditions. When mitochondria were isolated in the presence of high salt concentration, no Aim23p was observed in the precipitates. Further evidence came from the study, in which Aim23p was found in the fractions containing mitochondrial ribosomes after fractionation of yeast mitochondrial lysates by centrifugation in a sucrose density gradient [8]. The results of both studies suggest (but do not clearly demonstrate) that Aim23p interacts with yeast mitochondrial ribosome. Thus, Aim23p was not found in high resolution cryo-electron microscopy structures of yeast mitochondrial ribosomes [15] or their large subunits [16]. To better understand Aim23p interactions with yeast mitochondrial ribosomes, we have exploited two independent experimental setups that allowed us to corroborate the observation that Aim23p interacts (most likely, directly) with the mitochondrial ribosomal small subunit both in vivo and in vitro.
MATERIALS AND METHODS
Yeast strains. Two Saccharomyces cerevisiae strains further referred to as wt [D273-10B DUL2, MATa mal (lys2, ura3)] (kind courtesy of Dr. T. Fox, Cornell University, USA) and Aim23Δ [D273-10B DUL2, MATa mal (lys2, ura3) AIM23::KanMX4] were used in this work.
Yeast media and growth conditions. Yeast were grown at 30°C with shaking at 200 rpm in YPGal medium (1% yeast extract, 2% peptone, 2% galactose) (Amresco, USA) supplemented with antibiotic G418 (Sigma, USA) when required.
Isolation of mitochondria. Yeast were grown in 1 liter of YPGal medium up to OD600 = 2.0-3.0. The cells were collected by centrifugation (3000g, 5 min), washed with milliQ water, weighed, resuspended in mitochondria isolation buffer (20 mM HEPES-KOH, pH 7.4, 2 mM EGTA, 0.6 M sorbitol, 0.2% Blue Dextran, 1× proteinase inhibitor cocktail) at 1 g of wet yeast biomass per 1 ml, and mixed with glass beads (1 g of beads per gram of wet yeast biomass). The cells were disrupted by shaking with the beads using an MP Fast Prep Instrument at 4 m/s for 30 s. The resulting suspension was centrifuged at 1000g for 5 min at 4°C to remove the beads. The supernatant was collected in fresh tubes and centrifuged at 3000g for 5 min at 4°C several times to remove cell debris, nuclei, and undisrupted cells. When the pellet was no longer visible, the supernatant was centrifuged at 12,000g for 15 min at 4°C to sediment mitochondria. Mitochondrial pellet was resuspended in a minimal volume of isolation buffer, aliquoted, flash-frozen in liquid nitrogen, and stored at –80°C.
Immunoprecipitation was carried out as described in [17]. Briefly, mitochondria were resuspended in lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM CH3COOK, 2 mM MgCl2, 0.5% Triton X-100, 0.5% NP-40) and incubated for 10 min at 4°C with gentle shaking. The tubes were centrifuged at 20,000g for 10 min at 4°C, and the supernatant was collected. Protein concentration was measured with a Pierce 660 nm protein assay kit (Thermo Scientific, USA). Anti-Aim23p antibody (Almabion LLC, Russia) was added to each sample (6 μg per sample) and the tubes were incubated for 30 min at 4°C with gentle shaking. Then, 50 μl of Protein A-Sepharose slurry (GE Healthcare, Germany) was washed with lysis buffer and mixed with each sample. The samples were incubated for another 30 min at 4°C with gentle shaking. The beads were collected by centrifugation at 300g for 2 min at 4°C, washed twice with 1 ml of lysis buffer, mixed with 100 μl of 2× Laemmli sample buffer, and stored at –20°C.
Methanol–chloroform precipitation of proteins. The volumes of the samples were adjusted to 500 μl with milliQ water. Then, the samples were vigorously mixed with 500 μl of methanol and 125 μl of chloroform, vortexed for 10 s, and centrifuged for 2 min at 16,000g. The upper phase was discarded, and 500 μl of methanol was added to the lower phase and interphase. The tubes were mixed carefully by inverting several times, and the proteins were pelleted at 16,000g for 2 min. The supernatant was discarded, and the pellet was air-dried for 2 min at 80°C and dissolved in Laemmli buffer.
Mass-spectrometry. After electrophoresis, gel pieces containing stained protein bands were cut out from the gel, washed twice with 100 μl of 40% acetonitrile in 0.1 M NH4HCO3 for 20 min, and dehydrated with 100 μl of acetonitrile. After acetonitrile was completely removed, 5 μl of 15 μg/ml trypsin (Promega, USA) solution in 0.05 M NH4HCO3 was added to the dried gel piece. Digestion was carried out at 37°C for 4 h. The resulting peptides were eluted with 10 μl of 0.5% TFA in 10% acetonitrile aqueous solution and used for MALDI spectrometry analysis.
The sample (1 µl) was mixed with 0.5 μl of 2,5-dihydroxybenzoic acid (Sigma Aldrich, USA; 30 mg/ml solution in 30% aqueous acetonitrile containing 0.5% TFA) and air-dried on the sample target. Mass-spectra were acquired with an UltrafleXtreme MALDI-TOF/TOF mass spectrometer (Bruker Daltonics, USA) and analyzed using the Flex Analysis 3.3 software (Bruker Daltonics). Proteins were identified with the Mascot software (www.matrixscience.com) by screening the NCBI database. Candidate proteins with a Score >87 were considered to be reliably validated (p < 0.05).
Preparation of mitochondrial ribosomes. Yeast mitochondrial ribosomes were isolated by the protocol described in [16]. Yeast were grown in YPGal media to OD600 ~2, collected by centrifugation at 4000g for 10 min, washed twice with water, and resuspended in dithiothreitol (DTT) buffer (0.1 M Tris-H2SO4, pH 9.0, 10 mM DTT) at 2 ml/g wet biomass ratio. After incubation at 30°C for 30 min, the cells were collected by centrifugation, washed with Zymolyase buffer (20 mM K-phosphate buffer, pH 7.4, 1.2 M sorbitol) at 7 ml/g of wet biomass, and resuspended in the same volume of Zymolyase buffer containing Zymolyase T20 (1 mg/g of yeast biomass). After incubation for 1 h at 30°C in an orbital shaker at 80 rpm, the resulting spheroplast suspension was collected by centrifugation and washed again with Zymolyase buffer at the same ratio. The spheroplasts were then resuspended in homogenization buffer (10 mM Tris-HCl, pH 7.4, 0.6 M sorbitol, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 0.2% BSA) and disrupted in a Dounce homogenizer (10 strokes with tight pestle). The lysate was centrifuged at 3000g for 10 min, and crude mitochondrial fraction was then sedimented from the supernatant at 17,000g for 15 min. The pellet was gently resuspended in a minimal volume of SEM buffer (10 mM MOPS-KOH, pH 7.2, 250 mM sucrose, 1 mM EDTA) and applied on a 15/23/32/60% sucrose step density gradient for ultracentrifugation at 134,000g for 1 h. Pure mitochondrial fraction was collected from the interphase between 60 and 32% sucrose, diluted threefold with SEM buffer, and sedimented at 17,000g for 15 min.
To isolate mitochondrial ribosomes, mitochondria were gently resuspended in three volumes of lysis buffer (25 mM HEPES-KOH, pH 7.5, 100 mM KCl, 20 mM magnesium acetate, 1.7% Triton X-100, 2 mM DTT), incubated for 30 min on ice, and centrifuged at 25,000g for 30 min. The supernatant was applied on 0.5 M sucrose cushion prepared in the same buffer containing 1% Triton X-100. After ultracentrifugation (100,000g, 16 h), sedimented mitochondrial ribosomes were dissolved in a minimal volume of buffer containing 10 mM Tris-HCl, pH 7.0, 60 mM KCl, 60 mM NH4Cl, 10 mM magnesium acetate, 7 mM 2-mercaptoethanol, and 0.25 mM EDTA, applied on a 15-40% sucrose gradient prepared in the same buffer, and ultracentrifuged at 100,000g for 16 h. The fractions (~250 µl each; 40-45 in total) were collected; the presence of mitochondrial ribosomal subunits in the fractions was determined from the absorbance at 260 nm.
Western blot. Protein samples (10-20 μg) were separated in 12% denaturing polyacrylamide gel according to a standard procedure and transferred on the Hybond ECL nitrocellulose membrane (GE Healthcare, Germany). The membrane was blocked for 1 h in PBS containing 0.1% Tween 20 and 2.5% fat-free dry milk. Primary anti-Aim23p rabbit antibodies (Almabion, Russia) were generated using 6His-tagged recombinant Aim23p as an antigen. Secondary ECL Anti-Rabbit IgG HRP-linked whole antibody from donkey (GE Healthcare) were diluted in PBS containing 0.1% Tween 20.
Northern blot. We used oligonucleotide probes against 25S rRNA (5′-CCATGGCCACCGTCC-3′), 18S rRNA (5′-GTTTCTCAGGCTCCCTC-3′), 21S rRNA (5′-GCGGGACCTCAATTATAGGCC-3′), and 15S rRNA (5′-TAGAACTACACGGGTATCGAATCCG-3′).
The probes were 32P-labelled with T4 polynucleotide kinase (T4 PNK; Thermo Scientific) according to manufacturer’s instructions, purified on Bio-Gel P-6 Micro Bio-Spin columns (Bio-Rad, USA), and diluted in the hybridization buffer. RNA was isolated from the samples using Trizol (Invitrogen, USA) according to manufacturer’s instructions. RNA samples (1-2 μg per lane) were loaded on 1% formaldehyde agarose gel and run in 1× formaldehyde gel-running buffer at 5 V/cm. RNAs were passively transferred on the Hybond N+ membrane (GE Healthcare) overnight in 20× SSC buffer (0.3 M sodium citrate, 3 M NaCl, pH 7.0). The membrane was UV-linked (3 min, 1500 J/cm2) and incubated with the labelled probes. The signals were registered with a STORM Scanner (GE Healthcare) and analyzed with the Image Quant software (GE Healthcare).
RESULTS
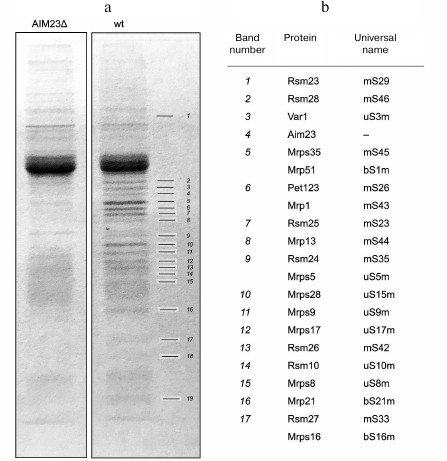
Aim23p co-immunoprecipitates with proteins of the yeast mitochondrial ribosomal small subunit. The protein interactome of Aim23p in yeast mitochondria has never been studied in detail. To identify interacting partners of Aim23p, we isolated mitochondria from two S. cerevisiae strains: wild-type (wt) and Aim23Δ with deleted AIM23 gene (aim23Δ). Isolated mitochondria were lysed and subjected to immunoprecipitation with anti-Aim23p antibodies (as described in “Materials and Methods”). The Aim23Δ strain served as a negative control, since we did not expect to find any Aim23p partners because of the deletion of the corresponding gene. Precipitated proteins from both strains were concentrated and run in 12% denaturing polyacrylamide gel. After the gel was stained with Coomassie Brilliant Blue G-250, we compared the band pattern for the two samples and identified many bands that were present in the wt sample, but not in Aim23Δ sample (Fig. 1a). These bands that presumably represented proteins that bind specifically to Aim23p were excised from the gel and analyzed by mass-spectrometry.
Fig. 1. a) Coomassie stained gel with separated bands of proteins recovered by immunoprecipitation with anti-Aim23p antibodies from the wt and Aim23Δ yeast strains. Differential bands are numbered 1-19. b) Proteins identified by mass-spectrometry.
Proteins identified by mass-spectrometry are shown in Fig. 1b. All proteins specifically enriched in the wt sample belonged to the mitochondrial ribosomal small subunit, except band 4 that corresponded to Aim23p. The proteins listed in Fig. 1b occupy most of the small subunit surface, so it was not possible to identify which of them specifically bound Aim23p. Rather the small subunit co-immunoprecipitates with Aim23p as a whole unit. These results clearly indicate that Aim23p physically interacts with the yeast mitochondrial ribosomal small subunit in vivo.
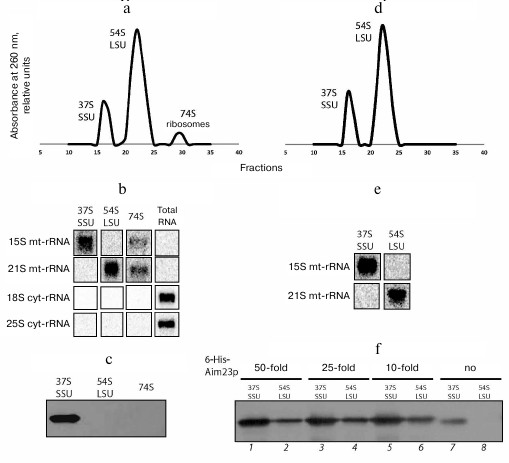
Aim23p is co-purified with the yeast mitochondrial ribosomal small subunit in a sucrose density gradient. To provide additional evidence that Aim23p interacts with the yeast mitochondrial ribosomal small subunit, we isolated ribosomes from the purified mitochondria of the wt strain, separated them into large and small subunits by centrifugation in a sucrose density gradient, and probed for the presence of Aim23p by Western blotting (Fig. 2, a-c).
Fig. 2. a) UV absorbance profiles of fractions collected after separation of yeast mitochondrial ribosomes in a sucrose density gradient. Positions of the small subunit (37S SSU), large subunit (54S LSU), and monosomes (74S ribosome) are indicated. b) Northern blot analysis of peak fractions from panel (a). Oligonucleotide probes against small (15S) and large (21S) subunits of mitochondrial rRNAs, as well as against small (18S) and large (25S) subunits of cytosolic rRNAs, were used. c) Western blot analysis of peak fractions from panel (a) with anti-Aim23p antibodies. d) UV absorbance profile of fractions collected after separation of yeast mitochondrial ribosomes preincubated with 6His-tagged Aim23p in a sucrose density gradient. Positions of small (37S SSU) and large subunits (54S LSU) are indicated. e) Northern blot analysis of peak fractions from panel (d). Oligonucleotide probes were the same as in panel (b). f) Western blot analysis of fractions from panel (d) with anti-Aim23p antibodies.
Fractionation of mitochondrial ribosomes in a sucrose density gradient produced clear peaks corresponding to small (37S) and large (54S) ribosomal subunits and monosomes (74S) (Fig. 2a). Northern blot analysis with oligonucleotide probes complementary to cytosolic and mitochondrial ribosomal RNAs showed that the obtained fractions were free from cytosolic ribosomal contamination (25S and 18S rRNA) and contained mitochondrial ribosomal RNAs (15S and 21S mt-rRNA; Fig. 2b). Western blot analysis of proteins in these fractions showed that Aim23p was present exclusively in the fractions of small subunits (Fig. 2c). These data confirm our conclusion that Aim23p physically interacts with the yeast mitochondrial ribosomal small subunit in vivo.
Recombinant Aim23p interacts with the yeast mitochondrial ribosomal small and large subunits of in vitro. After confirming Aim23p interactions with the yeast mitochondrial ribosomal small subunit in vivo, we checked if the recombinant yeast 6His-tagged Aim23p interacts with either large or small subunits in vitro. For this, we incubated purified mitochondrial ribosomes (the same preparation as in the previous experiment) with 50-, 25- and 10-fold molar excesses of 6His-tagged Aim23p at 30°C for 30 min and then separated the resulting complexes in a sucrose density gradient. The presence of Aim23p in the fractions was monitored by Western blotting.
The A260 profiles were almost identical for all samples obtained using different quantities of recombinant Aim23p and contained two peaks corresponding to the large and small subunits of mitochondrial ribosomes (Fig. 2d demonstrates the profile for the ribosome complex with 25-fold molar excess of Aim23p; all other profiles look identical). The peak of monosomes was absent, probably due to the IF3-like subunit dissociating activity of Aim23p. Northern blotting confirmed that the peaks contained mitochondrial rRNAs (Fig. 2e). The fractions were not analyzed for cytosolic contamination this time, as it its absence has been already demonstrated for the same preparation of mitochondrial ribosomes (Fig. 2b). Aim23p was identified with both types of mitochondrial ribosome subunits; however, large subunits clearly contained substantially less Amp23p (Fig. 2f). We also analyzed small and large subunits (Fig. 2f, lanes 7 and 8) from the fractions collected in the previous experiment that contained no recombinant Aim23p (Fig. 2, a-c) and found that native Aim23p was associated only with small subunits (Fig. 2f, lane 7). This experiment demonstrated that recombinant 6His-tagged Aim23p is capable of interacting with both subunits of mitochondrial ribosomes in vitro.
DISCUSSION
In this study we investigated the protein interactome of Aim23p, since the data on its interaction with ribosomal proteins are very scarce. We demonstrated that Aim23p interacts with the mitochondrial ribosomal small subunit, which is in line with its IF3-like role in yeast mitochondria (Aim23p ortholog was found to bind to the ribosomal small subunit in bacteria [18]). However, Aim23p has not been detected in the 74S mitochondrial ribosome in vivo, indicating that it is not a member of the mitochondrial ribosomal proteins or a structural component of the ribosome. Rather, it transiently associates with the small subunit and dissociates later upon progression of the ribosomal cycle. We believe that the co-immunoprecipitation experiment carried out in this work identified only highly represented interaction partners of Aim32p but not the proteins present in yeast mitochondria in small amounts. Therefore, the question of complete interactome of Aim23p remains open. Of particular interest are plausible interactions of Aim23p with low-abundant translational activators in yeast cells [19, 20] that can be proven in further investigations using epitope tagging of translational activators and subsequent co-immunoprecipitation of protein complexes.
The in vitro part of our study carried out with recombinant 6His-tagged Aim23p confirmed that this protein interacts with the yeast mitochondrial ribosomal small subunit. At the same time, it also bound to the large subunit and dissociated the 74S full ribosome. It is not clear if the latter interaction ever occurs in vivo, especially in view of the recent report that demonstrated the presence of an additional binding site for bacterial IF3 on the 50S large ribosomal subunit [21]. Moreover, we have shown recently that Aim23p is able to bind to 30S and 50S subunits of bacterial ribosome in vitro [22]. Extrapolation of these results on the translation initiation in mitochondria is tempting; however, structural studies are required to elucidate if this non-canonical binding of Aim23p to the large subunit takes place.
Funding
This work was supported by the Russian Science Foundation (project no. 17-14-01005).
Acknowledgements
We are grateful to K. Khodosevich (Copenhagen University, Denmark), V. Hauryliuk (Umea University, Sweden, and Tartu University, Estonia), I. Tarassov (Strasbourg University, France), S. Kozlovsky, and A. Kharitonov (Moscow State University, Russia) for sharing materials. We also thank A. Johns (Newcastle University, UK) for critical reading of the manuscript. The technical help of our students A. Mirnaya, A. Kapusta, and M. Chudenkova is greatly appreciated.
Conflict of Interest
Authors declare no conflict of interest in financial or any other sphere.
REFERENCES
1.Andersson, S. G., Zomorodipour, A., Andersson, J.
O., Sicheritz-Ponten, T., Alsmark, U. C., Podowski, R. M., Naslund, A.
K., Eriksson, A. S., Winkler, H. H., and Kurland, C. G. (1998) The
genome sequence of Rickettsia prowazekii and the origin of
mitochondria, Nature, 396, 133-140.
2.Foury, F., Roganti, T., Lecrenier, N., and
Purnelle, B. (1998) The complete sequence of the mitochondrial genome
of Saccharomyces cerevisiae, FEBS Lett., 440,
325-331.
3.Greber, B. J., and Ban, N. (2016) Structure and
function of the mitochondrial ribosome, Annu. Rev. Biochem.,
85, 103-132.
4.Kuzmenko, A., Atkinson, G. C., Levitskii, S.,
Zenkin, N., Tenson, T., Hauryliuk, V., and Kamenski, P. (2014)
Mitochondrial translation initiation machinery: conservation and
diversification, Biochimie, 100, 132-140.
5.Koc, E. C., and Spremulli, L. L. (2002)
Identification of mammalian mitochondrial translational initiation
factor 3 and examination of its role in initiation complex formation
with natural mRNAs, J. Biol. Chem., 277, 35541-35549.
6.Atkinson, G. C., Kuzmenko, A., Kamenski, P.,
Vysokikh, M. Y., Lakunina, V., Tankov, S., Smirnova, E., Soosaar, A.,
Tenson, T., and Hauryliuk, V. (2012) Evolutionary and genetic analyses
of mitochondrial translation initiation factors identify the missing
mitochondrial IF3 in S. cerevisiae, Nucleic Acids Res.,
40, 6122-6134.
7.Olsson, C. L., Graffe, M., Springer, M., and
Hershey, J. W. (1996) Physiological effects of translation initiation
factor IF3 and ribosomal protein L20 limitation in Escherichia
coli, Mol. Gen. Genet., 250, 705-714.
8.Kuzmenko, A., Derbikova, K., Salvatori, R., Tankov,
S., Atkinson, G. C., Tenson, T., Ott, M., Kamenski, P., and Hauryliuk,
V. (2016) Aim-less translation: loss of Saccharomyces cerevisiae
mitochondrial translation initiation factor mIF3/Aim23 leads to
unbalanced protein synthesis, Sci. Rep., 6, 18749.
9.Herrmann, J. M., Woellhaf, M. W., and Bonnefoy, N.
(2013) Control of protein synthesis in yeast mitochondria: the concept
of translational activators, Biochim. Biophys. Acta,
1833, 286-294.
10.Islas-Osuna, M. A., Ellis, T. P., Marnell, L. L.,
Mittelmeier, T. M., and Dieckmann, C. L. (2002) Cbp1 is required for
translation of the mitochondrial cytochrome b mRNA of
Saccharomyces cerevisiae, J. Biol. Chem., 277,
37987-37990.
11.Manthey, G. M., and McEwen, J. E. (1995) The
product of the nuclear gene PET309 is required for translation of
mature mRNA and stability or production of intron-containing RNAs
derived from the mitochondrial COX1 locus of Saccharomyces
cerevisiae, EMBO J., 14, 4031-4043.
12.De Silva, D., Poliquin, S., Zeng, R.,
Zamudio-Ochoa, A., Marrero, N., Perez-Martinez, X., Fontanesi, F., and
Barrientos, A. (2017) The DEAD-box helicase Mss116 plays distinct roles
in mitochondrial ribogenesis and mRNA-specific translation, Nucleic
Acids Res., 45, 6628-6643.
13.Su, C. H., McStay, G. P., and Tzagoloff, A.
(2014) The Cox3p assembly module of yeast cytochrome oxidase, Mol.
Biol. Cell, 25, 965-976.
14.Kehrein, K., Schilling, R., Moller-Hergt, B. V.,
Wurm, C. A., Jakobs, S., Lamkemeyer, T., Langer, T., and Ott, M. (2015)
Organization of mitochondrial gene expression in two distinct
ribosome-containing assemblies, Cell Rep.,
S2211-1247(15)00025-X.
15.Desai, N., Brown, A., Amunts, A., and
Ramakrishnan, V. (2017) The structure of the yeast mitochondrial
ribosome, Science, 355, 528-531.
16.Amunts, A., Brown, A., Bai, X. C., Llacer, J. L.,
Hussain, T., Emsley, P., Long, F., Murshudov, G., Scheres, S. H., and
Ramakrishnan, V. (2014) Structure of the yeast mitochondrial large
ribosomal subunit, Science, 343, 1485-1489.
17.Gerace, E., and Moazed, D. (2014)
Coimmunoprecipitation of proteins from yeast, Methods Enzymol.,
541, 13-26.
18.Hussain, T., Llacer, J. L., Wimberly, B. T.,
Kieft, J. S., and Ramakrishnan, V. (2016) Large-scale movements of IF3
and tRNA during bacterial translation initiation, Cell,
167, 133-144.e13.
19.Ghaemmaghami, S., Huh, W. K., Bower, K., Howson,
R. W., Belle, A., Dephoure, N., O'Shea, E. K., and Weissman, J. S.
(2003) Global analysis of protein expression in yeast, Nature,
425, 737-741.
20.Derbikova, K. S., Levitsky, S. A., Chicherin, I.
V., Vinogradova, E. N., and Kamenski, P. A. (2018) Activation of yeast
mitochondrial translation: who is in charge? Biochemistry
(Moscow), 83, 87-97.
21.Goyal, A., Belardinelli, R., and Rodnina, M. V.
(2017) Non-canonical binding site for bacterial initiation factor 3 on
the large ribosomal subunit, Cell Rep., 20,
3113-3122.
22.Levitskii, S., Derbikova, K., Baleva, M. V.,
Kuzmenko, A., Golovin, A. V., Chicherin, I., Krasheninnikov, I. A., and
Kamenski, P. (2018) 60S dynamic state of bacterial ribosome is fixed by
yeast mitochondrial initiation factor 3, PeerJ, 6,
e5620.