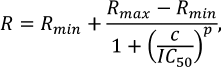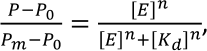The Effect of Antitumor Antibiotic Olivomycin A and Its New Semi-synthetic Derivative Olivamide on the Activity of Murine DNA Methyltransferase Dnmt3a
A. V. Sergeev1,a*, A. N. Tevyashova2,3, A. P. Vorobyov1, and E. S. Gromova1
1Lomonosov Moscow State University, Faculty of Chemistry, 119991 Moscow, Russia2Gause Institute of New Antibiotics, 119021 Moscow, Russia
3D. Mendeleev University of Chemical Technology of Russia, 125047 Moscow, Russia
* To whom correspondence should be addressed.
Received July 5, 2018; Revised September 28, 2018; Accepted September 28, 2018
Olivomycin A is a highly active antitumor drug that belongs to the family of aureolic acid antibiotics. The antitumor effect of olivomycin A is related to its ability to bind to the DNA minor groove in GC-rich regions as Mg2+-coordinated complexes. Characterization of cellular targets of olivomycin A and its mechanism of action is crucial for the successful application of this antibiotic in clinical practice and development of semi-synthetic derivatives with improved pharmacological properties. Previously, we have shown that minor groove ligands are able to disrupt the key epigenetic process of DNA methylation. In this paper, we have studied the impact of olivomycin A and its improved semi-synthetic analogue N,N-dimethylaminoethylamide of 1′-des-(2,3-dihydroxy-n-butyroyl)-1′-carboxy-olivomycin A (olivamide) on the functioning of de novo DNA methyltransferase Dnmt3a (enzyme that carries out methylation of cytosine residues in the DNA CG-sites in eukaryotic cells) using an in vitro system consisting of the murine Dnmt3a catalytic domain and a 30-mer DNA duplex containing four consecutive GC pairs. We have shown that olivomycin A and olivamide inhibit Dnmt3a with IC50 of 6 ± 1 and 7.1 ± 0.7 μM, respectively. Neither olivomycin A nor olivamide interfered with the formation of the specific enzyme–substrate complex; however, olivomycin A prevented formation of the covalent DNA–Dnmt3a intermediate that is necessary for the methylation reaction to proceed. The inhibitory effects of olivomycin A and olivamide can be explained by the disruption of the enzyme catalytic loop movement through the DNA minor groove (the reaction stage that precedes the covalent bond formation between DNA and the enzyme). The results of this work indicate the epigenetic contribution to the antitumor effect of aureolic acid group antibiotics.
KEY WORDS: DNA methylation, olivomycin A, olivamide, minor groove ligands, DNA methyltransferase Dnmt3a, inhibition of methylationDOI: 10.1134/S0006297919010085
Abbreviations: AA, aureolic acid; AdoHcy, S-adenosyl-L-homocysteine; AdoMet, S-adenosyl-L-methionine; MTase, DNA methyltransferase; P, fluorescence polarization; Z, pyrimidin-2-one.
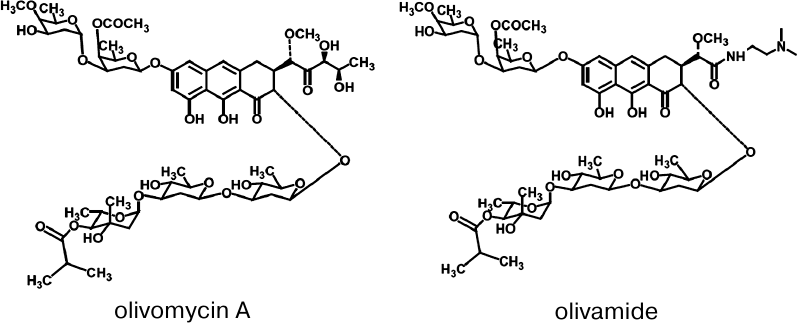
Olivomycin A (Fig. 1) is an antitumor antibiotic
belonging to the aureolic acid (AA) group, that also includes
mithramycin, chromomycins, and duramycin [1].
Clinical indications for the use of mithramycin are testicular cancer
and Paget’s disease [2]. Chromomycin has a
limited use in Japan for the gastric cancer treatment [3]. Olivomycin A is used in the treatment of
testicular cancer, reticulosarcoma, and some other tumor types [4]. However, clinical application of these antibiotics
is limited due to severe side effects. Since olivomycin A has the
highest chemotherapeutic index among the AA group antibiotics, it is
the most promising agent for developing semi-synthetic derivatives that
would feature decreased toxicity and improved pharmacological
characteristics [5, 6]. Among
olivomycin A analogs synthesized at the Gause Institute of New
Antibiotics, N,N-dimethylaminoethylamide
1′-des-(2,3-dihydroxy-n-butyroyl)-1′-carboxy-olivomycin
A (olivamide, LCTA-1599; Fig. 1) demonstrated the
highest activity in different cancer cell lines and exhibited lower
toxicity, as compared to olivomycin A [6]. Thus,
the treatment of human cancer cell lines HCT116 and K562 with olivamide
inhibited the tumor growth. Besides, the antiproliferative effect of
this antibiotic was demonstrated in mice bearing T cell lymphoma P388
or melanoma B16 transplants [6].
Fig. 1. Structures of the antitumor antibiotics olivomycin A and olivamide.
Similarly to other AA group antibiotics, olivomycin A binds to the DNA minor groove in GC-rich regions in a form of Mg2+-coordinated complexes. The cytotoxicity of olivomycin A towards tumor cells is attributed first of all to the impairment of transcription and replication [7, 8]. The AA group antibiotics bind to the GC-rich DNA sequences in the promoter regions of various genes and prevent the binding of transcription factors (e.g., Sp1 and c-Myc), which results in the inhibition of transcription of these genes [9, 10].
The discovery of new aspects of mechanisms of the AA group antibiotics should significantly broaden their therapeutic potential as antitumor agents [11]. The antitumor activity of olivomycin A and its analogs can also be realized through their effect on the process of DNA methylation by DNA methyltransferases (MTases). The eukaryotic de novo MTase Dnmt3a transfers methyl group from S-adenosyl-L-methionine (AdoMet) to position 5 of the cytosine in CpG sequences during embryogenesis thereby shaping the DNA methylation profile [12]. The C-terminal domains of Dnmt3a and catalytically inactive factor Dnmt3L form a tetrameric complex containing two active sites [13]. X-ray analysis of the Dnmt3a/Dnmt3L complex with a 25-bp DNA substrate has shown that the enzyme active sites are situated at a distance corresponding to one turn of a double-stranded DNA helix from each other [14]. Dnmt3a is able to bind and methylate DNA even in the absence of Dnmt3L. Dnmt3a is also prone to oligomerization on DNA with the formation of long filaments [15, 16]. In mammalian cells, DNA methylation plays a pivotal role in the regulation of gene expression, differentiation, genomic imprinting, and other processes [12, 17]. Alterations of the DNA methylation profile, such as promoter hypermethylation leading to the inactivation of tumor suppressor genes, are typically observed in cancer cells [18, 19]. The effect of AA group antibiotics on DNA methylation has never been studied. Earlier, we have demonstrated that Dnmt3a functioning is affected by other minor groove ligands, dimeric bisbenzimidazoles [20-22]. Неre, we studied the effect of two AA group antibiotics, olivomycin A and its semi-synthetic derivative olivamide, on the functioning of the catalytic domain of murine Dnmt3a (Dnmt3a-CD) in vitro by using a 30-bp DNA duplex with four consecutive GC-pairs as a substrate.
MATERIALS AND METHODS
Reagents. All oligonucleotides (Table 1) were purchased from Syntol, Russia. Some of the oligonucleotides were modified with the fluorescent label 6-carboxyfluorescein (FAM, f) covalently attached to the 5′-end phosphate through the amino linker -NH-(CH2)6-. Oligonucleotides containing pirimidin-2-one (Z) were synthesized by S. N. Mikhailov (Institute of Molecular Biology, Russian Academy of Sciences). The concentration of oligonucleotides was determined spectrophotometrically as described earlier [23]. S-adenosyl-L-methionine (AdoMet) and S-adenosyl-L-homocysteine (AdoHcy) were from Sigma (Germany). Buffer A contained 20 mM HEPES-NaOH (pH 7.5), 100 mM KCl, 1 mM EDTA, and 1 mM 1,4-dithiothreitol (DTT).
Table 1. DNA duplexes used in the study
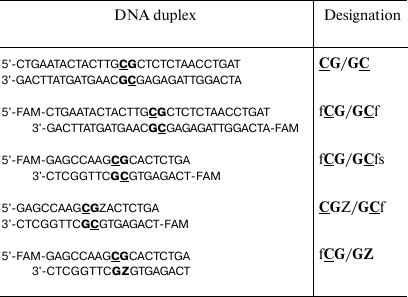
Note: FAM (f), 6-carboxyfluorescein; Z (pyrimidin-2-one) and CG
dinucleotides are written in bold. Target cytosine residues are
underlined.
Enzymes. Restriction endonuclease Hin6I was from Sibenzyme, Russia. Dnmt3a-CD was expressed in E. coli BL21(DE3) cells (Stratagene, USA) carrying the pET28a plasmid with the cloned gene for Dnmt3a-CD containing the N-terminal His6 tag. The enzyme was purified by metal-chelating chromatography on Co2+-charged Talon® resin (GE Healthcare, USA) according to the previously described technique [24]. The purity of the obtained protein preparations was >95%, as assessed by 12.5% Laemmli polyacrylamide gel electrophoresis [25]. The concentration of Dnmt3a-CD was determined per monomer using the Bradford procedure [26]. Protein preparations were stored at –80°C.
Olivomycin A binding to DNA in the presence of Mg2+ ions. Formation of olivomycin A–DNA complex was monitored by the increase in the fluorescence intensity of the antibiotic upon its binding to the DNA substrate [27]. CG/GC (300 nM) was incubated with olivomycin A (5 μM) in buffer A in the presence of 0-50 mM Mg2+ for 1 h at 25°C. The fluorescence spectra of the obtained complexes were registered in the wavelength range from 430 to 650 nm with a Cary Eclipse fluorescence spectrophotometer (Varian, USA) at λex 420 nm using a 100-μl microcuvette (1-cm light path length). The concentration of Mg2+ ions sufficient for complete DNA duplex binding was determined from the dependence of olivomycin A fluorescence intensity at 550 nm on the Mg2+ concentration.
Effect of olivomycin A and olivamide on DNA methylation by Dnmt3a-CD. FAM-labeled fCG/GCf (300 nM) was incubated in buffer A containing 3 mM Mg2+ for 1 h at room temperature either in the presence or absence of olivomycin A or olivamide (0-19 μM) and then methylated with 2 μM Dnmt3a-CD in the presence of 25 μM AdoMet for 1 h at 37°C [28]. After methylation, the DNA duplex was digested with Hin6I for 1 h at 37°C in the same buffer (DNA duplex cleaved without prior methylation was used as a control). The reaction mixtures were evaporated, resuspended in 10 μl of 80% formamide with dyes, and analyzed by electrophoresis in 20% polyacrylamide gel in the presence of 7 M urea. The fluorescence intensities of the original DNA and reaction products were determined by scanning the gels with a Typhoon FLA 9500 scanner (GE Healthcare Life Sciences, Great Britain). The extent of DNA cleavage (w) was calculated in each lane as a ratio of fluorescence intensity of the cleaved DNA to the total fluorescence intensity of the intact and cleaved DNA.
The extent of methylation (R) was calculated using the equation:
where w0 is the extent of DNA cleavage without methylation; wDnmt3a is the extent of DNA cleavage after methylation by Dnmt3a-CD [28].
The IC50 values were calculated by approximation of the dependence of the extent of methylation on the antibiotic concentration using the logistic equation in the OriginPro 2015 software (OriginLab, USA):
where R is the extent of methylation; Rmin and Rmax are the lower and upper asymptotes, respectively; c is an antibiotic concentration (μM); p is the slope factor.
fCG/GCf (300 nM) was incubated in buffer A with 10 µM olivomycin A in the presence of various concentrations of Mg2+ (0-5 mM) for 1 h at room temperature. Prior to the treatment with Hin6I endonuclease, the concentration of Mg2+ in the reaction mixtures containing 0-1 mM Mg2+ was increased to 3 mM. The extent of methylation was calculated using the above-mentioned method.
Dnmt3a-CD–DNA complex formation. Dnmt3a-CD binding to 10 nM DNA in the presence of 0.1 mM AdoHcy and antibiotics was studied using fluorescence polarization (P) [29] of the FAM-labeled fCG/GCf. The reaction mixtures were incubated with olivomycin A or olivamide for 1 h at room temperature in buffer A containing 3 mM Mg2+ and then titrated with Dnmt3a-CD (0-800 nM). FAM fluorescence was excited at 495 nm and registered at 520 nm in a 100-µl microcuvette. Fluorescence polarization (P) was registered 2 min after adding Dnmt3a-CD using a Cary Eclipse fluorescence spectrophotometer. The dissociation constants (Kd) for the complexes were calculated by approximation of the dependences of P on the concentration of Dnmt3a-CD using the Hill equation in the OriginPro 2015 software:
where P0 and Pm are fluorescence polarization values for the free and completely bound FAM-labeled DNA duplexes, respectively; [E] is the Dnmt3a-CD concentration, n is the Hill coefficient.
Formation of covalent intermediates between Dnmt3a and DNA duplexes containing pyrimidin-2-one (Z). FAM-labeled DNA duplexes (300 μM) without (fCG/GCfs) or with (CGZ/GCf and fCG/GZ) the Z residue were incubated with 6 μM Dnmt3a-CD or M.HhaI in buffer A for 1 h at 4°C. The reaction mixtures containing 100 μM AdoHcy, 0 or 3 mM Mg2+, and 0 or 10 μM olivomycin A were analyzed by electrophoresis in 12% polyacrylamide gel with 0.1% SDS. The gels were visualized with a Typhoon FLA 9500 scanner.
RESULTS
We studied the effect of olivomycin A and olivamide on the DNA methylation, as well as on different stages of this reaction. The first step was designing a convenient model system for studying the effects of these antibiotics on the functioning of the Dnmt3a-CD. This domain displays enzymatic activity even in the absence of the enzyme N-terminal regulatory domain [30]. The sequence of murine Dnmt3a-CD is very similar to that of the human enzyme. Thirty- and 18-bp DNA duplexes containing CpG sites were used as the Dnmt3a-CD substrates (Table 1). When designing the DNA duplexes, we considered the ability of the AA group antibiotics to bind to DNA at the regions containing four consecutive GC pairs [31]. DNA duplexes fCG/GCf, fCG/GCfs, CGZ/GCf and fCG/GZ were labeled with FAM at 5′-ends of one or both strands.
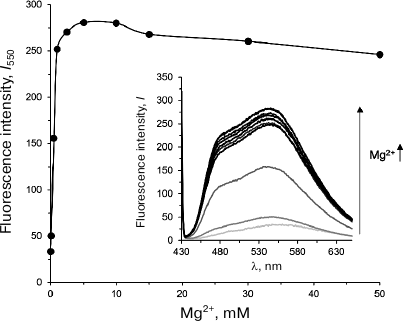
Olivomycin A binding to DNA in the presence of Mg2+ ions. To investigate the formation of complexes between Dnmt3a-CD and DNA in the presence of olivomycin A and olivamide, we first selected conditions for the optimal binding of the studied antibiotics to the DNA duplexes. Because olivomycin A binding to DNA requires the presence of Mg2+ ions [7], we determined the optimal Mg2+ concentrations for olivomycin A binding to DNA, as well as for Dnmt3a-CD to preserve its activity. We used the fact that the intensity of olivomycin A fluorescence increases upon its binding to DNA [6]. The fluorescence spectra of olivomycin A were registered in the presence of CG/GC at different Mg2+ concentrations (Fig. 2). With the increase in Mg2+ concentration from 0 to 5 mM, the fluorescence intensity of olivomycin A at 550 nm increased and reached a plateau at 2.5 mM, which suggests that at this Mg2+ concentration all binding sites on DNA were occupied with the antibiotic. In all further experiments Mg2+ concentration was 3 mM. It was recently demonstrated that the presence of Mg2+ ions is not necessary for olivamide binding to DNA [32], but the stability of olivamide–DNA complexes depends on the solution ionic strength. In this work, the binding of olivomycin A and olivamide was carried out in the presence of Mg2+ ions in order to compare results obtained under the same conditions.
Fig. 2. Dependence of fluorescence intensity of the olivomycin A–CG/GC complex at 550 nm (I550) on Mg2+ concentration. Inset, fluorescence spectra of the olivomycin A–CG/GC complex at 0-50 mM Mg2+ (300 nM CG/GC, 5 μM olivomycin A, buffer A).
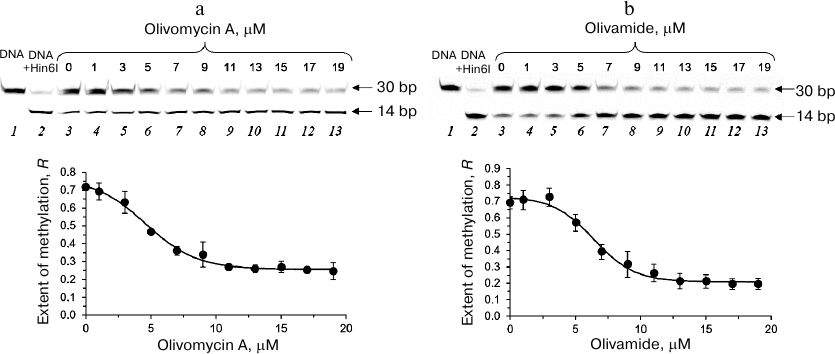
Effect of olivomycin A and olivamide on DNA methylation. To assess the effect of olivomycin A on DNA methylation, we studied the protection of methylated DNA from cleavage with restriction endonucleases [28]. The complexes of fCG/GCf with olivomycin A or olivamide obtained by DNA incubation with the antibiotic for 1 h in the presence of 3 mM Mg2+ were methylated with Dnmt3a-CD. Then, the DNA duplexes were cleaved with Hin6I endonuclease (G↓CGC, the cleavage site is indicated with an arrow) that hydrolyzes only unmethylated DNA. After the cleavage, the reaction mixture was analyzed in 20% denaturing polyacrylamide gel (Fig. 3a). fCG/GCf was cleaved with the formation of a 14-bp fluorescent product. The presence of 3 mM Mg2+ did not interfere with the activity of Dnmt3a-CD (Fig. 3a, lanes 2 and 3). The extent of DNA methylation decreased upon the addition of olivomycin A to the reaction mixture with the IC50 value of 6 ± 1 μM (Fig. 3a, lanes 4-13; Table 2). DNA methylation in the presence of olivamide was examined similarly (Fig. 3b). Olivamide inhibited DNA methylation with IC50 of 7.1 ± 0.7 μM (Table 2). Addition of olivomycin A to the preformed DNA–(Dnmt3a-CD) complex also resulted in the inhibition of DNA methylation (data not shown), which indicates that olivomycin A and Dnmt3a-CD do not compete for the binding sites on DNA.
Fig. 3. Effect of olivomycin A (a) and olivamide (b) on DNA methylation. Upper panel, cleavage of the fCG/GCf with Hin6I endonuclease after methylation by Dnmt3a-CD in the presence of 0-19 μM olivomycin A or olivamide (lanes 3-13) in 20% polyacrylamide gel with 7 M urea. fCG/GCf (lane 1) and products of its hydrolysis with Hin6I (lane 2) are shown as controls. Lower panel, dependence of the extent of DNA methylation on the concentration of olivomycin A and olivamide. Conditions: 300 nM fCG/GCf, 3 mM Mg2+, 25 μM AdoMet, buffer A.
Table 2. Effect of olivomycin A and
olivamide on the fCG/GCf binding to
Dnmt3a-CD and inhibition of DNA methylation
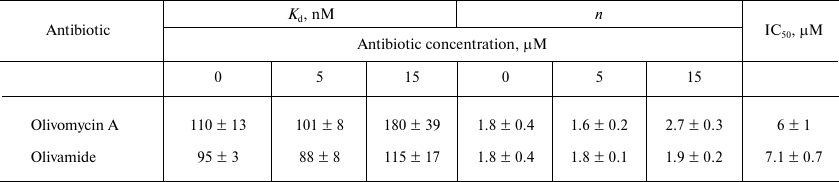
Note: Kd, n, and IC50 values are
given as mean ± standard deviation from three independent
experiments.
Formation of the DNA–(Dnmt3a-CD) complex in the presence of olivomycin A and olivamide. The mechanism of MTases includes several stages: (i) recognition and binding to the DNA substrate; (ii) flipping of the target cytosine from the double helix; (iii) movement of the enzyme catalytic loop through the DNA minor groove; (iv) formation of the enzyme–substrate covalent intermediate; (v) transfer of methyl group to the C5 position of methylated cytosine; (vi) release of the reaction products [12, 33]. To determine which stage of methylation is affected by the studied antibiotics, we investigated the effect of olivomycin A on the formation of enzyme–substrate complex, as well as on the generation of the covalent intermediate.
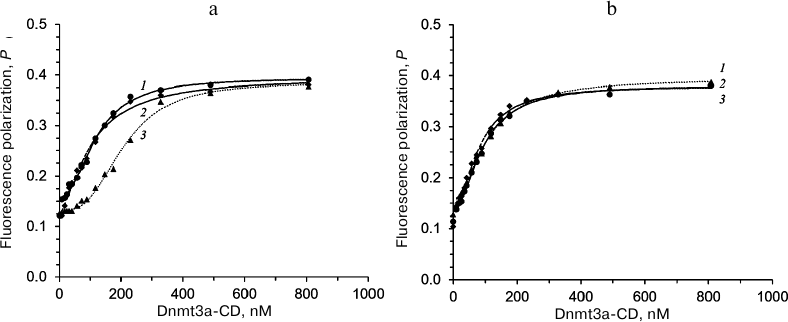
The effect of olivomycin A and olivamide on the binding of Dnmt3a-CD to the substrate DNA was assessed from the fluorescence polarization of the FAM-labeled fCG/GCf in the presence of AdoHcy. Figure 4 shows typical binding curves in the presence of 0, 5, and 15 μM olivomycin A or olivamide. When Dnmt3a-CD was added to the reaction mixture containing fCG/GCf and tested antibiotic, the fluorescence polarization increased and reached a plateau. The obtained binding curves were approximated using the Hill equation, and the Kd values and Hill coefficients (n) were calculated for the DNA–(Dnmt3a-CD) complexes (Table 2). The Kd values did not change significantly upon addition of 5 and 15 μM olivomycin A or olivamide, i.e., neither olivomycin A nor olivamide noticeably impaired enzyme binding to the substrate DNA. The obtained Hill coefficients (n) indicated the existence of positive binding cooperativity [34].
Fig. 4. fCG/GCf binding to Dnmt3a-CD in the presence of 0-15 μM olivomycin A (a) or olivamide (b) assessed by the fluorescence polarization: 1) 0 μM; 2) 5 μM; 3) 15 μM olivomycin A or olivamide. Conditions: 10 nM fCG/GCf, 3 mM Mg2+, 100 μM AdoHcy, buffer A.
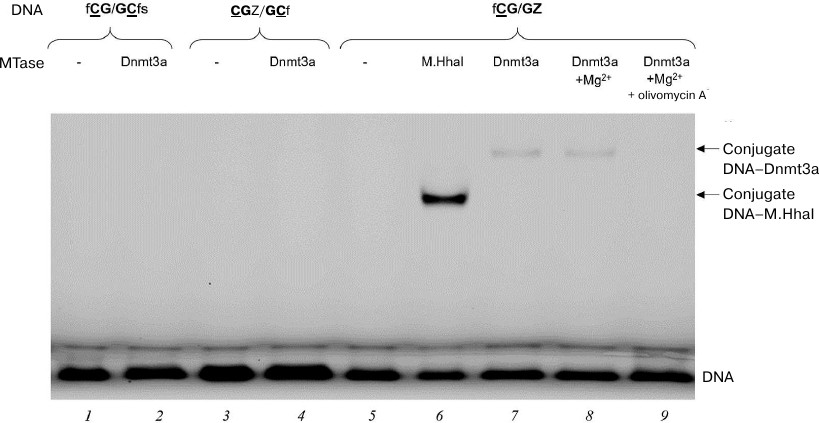
Formation of DNA–(Dnmt3a-CD) covalent intermediates in the presence of olivomycin A. We studied the effect of olivomycin A on one of the key stages of DNA methylation – formation of covalent intermediate during nucleophilic attack at the C6 position of the target cytosine by a highly conserved cysteine residue of the C5 MTase active site [12, 35]. The intermediate is unstable and rapidly decomposes with the release of the substrate DNA carrying 5-methylcytosine residue. It has been shown earlier that FAM-labeled DNA substrates containing pyrimidin-2-one (Z) instead of target cytosine form significantly more stable covalent intermediates with M.SssI and Dnmt3a MTases, that can be detected by the fluorescence of FAM in a polyacrylamide gel [36]. Recently, a covalent intermediate formed by a 25-bp Z-containing DNA duplex and human DNMT3A–DNMT3L tetramer was analyzed by X-ray crystallography [14]. We used 18-bp DNA duplexes (Table 1) labeled with FAM at 5′-ends and containing the Z residue either located near the CG site (CGZ/GCf) or substituting one of the target cytosines (fCG/GZ). The duplexes were incubated with Dnmt3a-CD for 1 h at 4°C, and the reaction mixtures were then analyzed in 12% polyacrylamide gel with 0.1% SDS (Fig. 5). Prokaryotic MTase M.HhaI was used as a positive control for the covalent intermediate formation (Fig. 5, lane 6). The duplexes that did not contain Z residues (fCG/GCfs) or contained Z close to the CG site (CGZ/GCf) did not form stable covalent intermediates (Fig. 5, lanes 2 and 4). fCG/GZ formed covalent intermediate with Dnmt3a-CD, even in the presence of 3 mM Mg2+ required for olivomycin A binding to DNA (Fig. 5, lane 8). When olivomycin A was introduced into the reaction mixture prior to adding Dnmt3a-CD, no intermediate formation was observed (Fig. 5, lanes 7 and 9). Therefore, olivomycin A prevented formation of the covalent intermediate in the reaction mixture.
Fig. 5. Effect of olivomycin A on the formation of covalent intermediates (conjugates) between Dnmt3a-CD and DNA duplexes containing or lacking the Z residue. Reaction mixtures contained 300 nM DNA duplex fCG/GCfs (lanes 1, 2), CG{{anchor|GoBack}} Z/GCf (lanes 3, 4), or fCG/GZ (lanes 5-9), 6 μM Dnmt3a-CD (lanes 2, 4, 7, 8, 9) or M.HhaI (lane 6), 100 μM AdoHcy (lanes 1-9), 3 mM Mg2+ (lanes 8, 9), and 10 μM olivomycin A (lane 9). The mixtures were separated in 12% polyacrylamide gel with 0.1% SDS.
The possibility of olivomycin A binding to Dnmt3a-CD. Could the inhibition of methylation be explained by antibiotic binding to Dnmt3a-CD rather than to the DNA duplex? First, as it was shown above, the Kd values in the presence of 0, 5, or 15 μM olivomycin A were virtually the same (Table 2). If the DNA duplex and olivomycin A competed for the binding to the enzyme active site, an increase in the antibiotic concentration should have led to a significant increase in the Kd value.
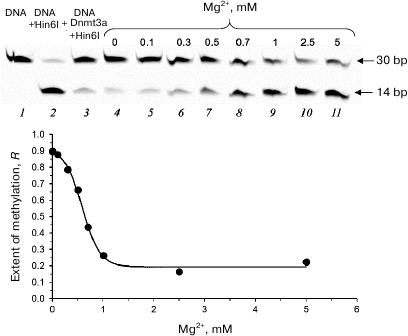
Second, we studied the efficiency of fCG/GCf methylation in the presence of 10 μM olivomycin A and 0-5 mM Mg2+ (Fig. 6). In the absence of Mg2+ ions (required for antibiotic binding to DNA), no inhibition of DNA methylation was observed (Fig. 6, lane 4), while increasing Mg2+ concentration in the reaction mixture resulted in a drastic decrease in the methylation efficiency that reached a plateau at Mg2+ concentrations over 1 mM (Fig. 6, lanes 5-11). Since Mg2+ ions are required for the formation of olivomycin A dimers that bind to GC pairs in DNA [7], no methylation inhibition occurred in the absence of dimer formation.
Fig. 6. Effect of Mg2+ on the functioning of Dnmt3a-CD in the presence of olivomycin A. Cleavage of 300 nM DNA duplex fCG/GCf with endonuclease Hin6I after DNA methylation with Dnmt3a-CD in the presence of 10 μM olivomycin A, 25 μM AdoMet, and 0-5 mM Mg2+. Prior to the cleavage, Mg2+ concentration was adjusted to 3 mM in mixtures containing 0-1 mM Mg2+ (lanes 2-9). Denaturing 20% polyacrylamide gel with 7 M urea and buffer A were used.
The possibility of olivomycin A binding to Dnmt3a-CD was also studied by fluorescent titration of 1 µM olivomycin A with Dnmt3a-CD (0-2 μM) in the presence of 3 mM Mg2+ in buffer A. The fluorescence intensity was measured as described in section “Olivomycin A binding to DNA in the presence of Mg2+ ions” (“Materials and Methods”). We did not observe changes in fluorescence at 550 nm as compared to free antibiotic; the intensity of olivomycin A fluorescence increased only upon the addition of the CG/GC to the reaction mixture (data not shown). Summarizing the obtained results, the effect of olivomycin A on DNA methylation cannot be explained by the antibiotic binding to Dnmt3a-CD.
DISCUSSION
Olivomycin A and its analogs are promising therapeutic agents, whose antitumor activity is determined by the ability to bind to GC-rich regions in the DNA minor groove, affecting DNA-operating enzymes and transcription factors. In this work, we used synthetic DNA substrates to study the effects of olivomycin A and olivamide on the DNA methylation catalyzed by the MTase Dnmt3a-CD that recognizes CpG regions.
When designing the DNA duplexes, we took into account the data on the specificity of binding of olivomycin A to DNA and stability of such complexes (Beniaminov and coworkers, unpublished data). Olivomycin A binds to four consecutive GC pairs in DNA. The strongest complex is formed when the GpC sequence is located in the center of this GC-rich region. In this work, the presence of the CpG site (recognition site for Dnmt3a-CD) was a key criterion for the DNA substrate. GC pairs flanking the central CpG site were selected so that the forming cluster was a recognition site for the Hin6I endonuclease (G↓CGC), which is sensitive to the methylation status of the central cytosine residue. This allowed us to determine the efficiency of methylation of the obtained DNA duplexes by monitoring the extent of DNA protection from cleavage by Hin6I endonuclease after methylation under conditions favoring the binding of olivomycin A or olivamide to the DNA.
The extent of DNA methylation by Dnmt3a-CD in the presence of high concentrations of olivomycin A and olivamide was ~20%, wherein the IC50 values for these antibiotics (6 ± 1 and 7.1 ± 0.7 μM, respectively) did not demonstrate significant differences. This may be explained by highly similar chemical structures of olivomycin A and olivamide (Fig. 1). Besides, the structures of complexes of both antibiotics with DNA are assumed to be similar to that of the complex formed by another AA group antibiotic, chromomycin A3 [37]. Comparison of the IC50 values of olivomycin A and olivamide to those for other minor groove ligands previously studied in our laboratory – dimeric bisbenzimidazoles DB(n) [20], in which bisbenzimidazole fragments are linked by methylene bridges of different length (3-11) – showed that olivomycin A and olivamide inhibited Dnmt3a-CD as efficiently as DB(11), whose IC50 is 5.0 ± 1.5 μM.
Taking into account the putative molecular mechanism of C5 MTases, the effect of minor groove ligands on DNA methylation can be realized through (i) preventing the formation of a complex between Dnmt3a and CpG-containing DNA substrate, (ii) hindering the flipping of the target cytosine from the DNA double helix that occurs through the minor groove (as it was demonstrated for the prokaryotic MTase M.HhaI), and (iii) preventing the movement of the enzyme catalytic loop through the minor groove [12, 38]. Recently, the existence of contacts between Dnmt3a and the DNA minor groove was demonstrated by X-ray crystallography of the catalytically active transient complexes between Z-containing DNA and human DNMT3A–DNMT3L tetramer [14]. Our data on the complex formation between Dnmt3a-CD and the fCG/GCf suggest that olivomycin A and olivamide only slightly affect the enzyme binding to DNA. Therefore, the studied minor groove ligands do not prevent the formation of the enzyme–substrate complex. Analysis of the substrates with different localization of the Z residue demonstrated that Dnmt3a-CD is able to form stable covalent intermediates only with the DNA substrate containing the Z residue instead of the target cytosine in the CpG site (fCG/GZ), whereas no formation of such intermediate was observed in the presence of olivomycin A (Fig. 5). Considering the importance of contacts between Dnmt3a and DNA minor groove [14] for DNA methylation, the absence of the conjugate formation with the Z-containing DNA in the presence of olivomycin A may be explained by the disruption of contacts between the Dnmt3a catalytic loop and a group of atoms exposed in the minor groove. In its turn, it may lead to the inhibition of other stages of methylation, such as flipping of the target cytosine from the DNA double helix.
Collectively, our results suggest an epigenetic contribution, e.g., inhibition of the Dnmt3a activity, to the mechanism of antitumor activity of the AA group antibiotics.
Funding
This work was supported by the Russian Foundation for Basic Research (projects nos. 18-34-00364 and 16-04-01087).
Acknowledgements
Authors express gratitude to A. Jeltsch for kindly providing a plasmid for expression of Dnmt3a-CD, S. Klimasauskas for kindly providing an M.HhaI preparation, and S. N. Mikhailov for synthesis of oligonucleotides containing pyrimidin-2-one.
Conflict of Interest
Authors declare no conflict of interest in financial or any other sphere.
REFERENCES
1.Tevyashova, A. N. (2016) Olivomycin A – an
antitumor antibiotic of the aureolic acid group, Pharm. Chem.
J., 50, 425-430.
2.DeVita, V. T. J., Hellman, S., and Rosenberg, S. A.
(2005) in Cancer: Principles and Practice of Oncology, 7th Edn.,
Lippincott Williams and Wilkins, Philadelphia.
3.Ogawa, M. (1978) A recent overview of chemotherapy
for advanced stomach cancer in Japan, Antibiot. Chemother.,
24, 149-159.
4.Gause, G. F. (1975) Chromomycin, olivomycin,
mithramycin, in Antineoplastic and Immunosuppressive Agents.
Handbook of Experimental Pharmacology (Sartorelli, A. C., et al.,
eds.) Springer-Verlag, Berlin-Heidelberg, Vol. 38, pp. 615-622.
5.Kumar, V., Remers, W. A., and Bradner, W. T. (1980)
Preparation and antitumor activity of olivomycin A analogs, J. Med.
Chem., 23, 376-379.
6.Tevyashova, A. N., Shtil, A. A., Olsufyeva, E. N.,
Luzikov, Y. N., Reznikova, M. I., Dezhenkova, L. G., and Kuzmin, V. A.
(2011) Modification of olivomycin A at the side chain of the aglycon
yields the derivative with perspective antitumor characteristics,
Bioorg. Med. Chem., 19, 7387-7393.
7.Lombo, F., Menendez, N., Salas, J. A., and Mendez,
C. (2006) The aureolic acid family of antitumor compounds: structure,
mode of action, biosynthesis, and novel derivatives, Appl.
Microbiol. Biot., 73, 1-14.
8.Cheglakov, I. B., Tevyashova, A. N., Kurbatov, L.
K., Tatarsky, V. V., Samusenko, A. V., Preobrazhenskaya, M. N., and
Shtil, A. A. (2010) Altered transcription and replication are the
mechanisms of cytotoxicity of antitumor antibiotic olivomycin A,
Dokl. Biochem. Biophys., 435, 320-322.
9.Durandin, N., Vinogradov, A., Shtil, A., and
Kuzmin, V. (2013) Inhibition of c-Myc transcription by olivomycin A
involves preferential drug binding to NFAT/Sp1 promoter site, FEBS
J., 280, 86-87.
10.Fernandez-Guizan, A., Mansilla, S., Barcelo, F.,
Vizcaino, C., Nunez, L. E., Moris, F., Gonzalez, S., and Portugal, J.
(2014) The activity of a novel mithramycin analog is related to its
binding to DNA, cellular accumulation, and inhibition of Sp1-driven
gene transcription, Chem.-Biol. Interact., 219,
123-132.
11.Gonzalez-Sabin, J., and Moris, F. (2013)
Exploring novel opportunities for aureolic acids as anticancer drugs,
Biochem. Pharmacol., 2, 1-3.
12.Jurkowska, R. Z., Jurkowski, T. P., and Jeltsch,
A. (2011) Structure and function of mammalian DNA methyltransferases,
ChemBioChem, 12, 206-222.
13.Jia, D., Jurkowska, R. Z., Zhang, X., Jeltsch,
A., and Cheng, X. (2007) Structure of Dnmt3a bound to Dnmt3L suggests a
model for de novo DNA methylation, Nature, 449,
248-251.
14.Zhang, Z. M., Lu, R., Wang, P., Yu, Y., Chen, D.,
Gao, L., Liu, S., Ji, D., Rothbart, S. B., Wang, Y., and Wang, G. G.
(2018) Structural basis for DNMT3A-mediated de novo DNA
methylation, Nature, 554, 387-391.
15.Jurkowska, R. Z., Rajavelu, A., Anspach, N.,
Urbanke, C., Jankevicius, G., Ragozin, S., Nellen, W., and Jeltsch, A.
(2011) Oligomerization and binding of the Dnmt3a DNA methyltransferase
to parallel DNA molecules heterochromatic localization and role of
Dnmt3L, J. Biol. Chem., 286, 24200-24207.
16.Jeltsch, A., and Jurkowska, R. Z. (2013)
Multimerization of the Dnmt3a DNA methyltransferase and its functional
implications, Prog. Mol. Biol. Transl., 117, 445-464.
17.Bird, A. (2002) DNA methylation patterns and
epigenetic memory, Gene Dev., 16, 6-21.
18.Eden, A., Gaudet, F., Waghmare, A., and Jaenisch,
R. (2003) Chromosomal instability and tumors promoted by DNA
hypomethylation, Science, 300, 455.
19.Jones, P. A., and Baylin, S. B. (2007) The
epigenomics of cancer, Cell, 128, 683-692.
20.Cherepanova, N. A., Ivanov, A. A., Maltseva, D.
V., Minero, A. S., Gromyko, A. V., Streltsov, S. A., and Gromova, E. S.
(2011) Dimeric bisbenzimidazoles inhibit the DNA methylation catalyzed
by the murine Dnmt3a catalytic domain, J. Enzyme Inhib. Med.
Chem., 26, 295-300.
21.Darii, M. V., Rakhimova, A. R., Tashlitsky, V.
N., Kostyuk, S. V., Veiko, N. N., Ivanov, A. A., Zhuze, A. L, and
Gromova, E. S. (2013) Dimeric bisbenzimidazoles: cytotoxicity and
effects on DNA methylation in normal and cancer human cells, Mol.
Biol., 47, 259-266.
22.Kostyuk, S. V., Kvasha, M. A., Khrabrova, D. A.,
Kirsanova, O. V., Ershova, E. S., Malinovskaya, E. M., Veiko, N. N.,
Ivanov, A. A., Koval, V. S., Zhuze, A. L., Tashlitsky, V. H.,
Umriukhin, P. E., Kutsev, S. I., and Gromova, E. S. (2018) Symmetric
dimeric bisbenzimidazoles DBP(n) reduce methylation of RARB and PTEN
while significantly increase methylation of rRNA genes in MCF-7 cancer
cells, PloS One, 13, e0189826.
23.Baskunov, V. B., Subach, F. V., Kolbanovskiy, A.,
Kolbanovskiy, M., Eremin, S. A., Johnson, F., Bonala, R., Geacintov, N.
E., and Gromova, E. S. (2005) Effects of benzo[a]pyrene-deoxyguanosine
lesions on DNA methylation catalyzed by EcoRII DNA methyltransferase
and on DNA cleavage effected by EcoRII restriction endonuclease,
Biochemistry, 44, 1054-1066.
24.Lukashevich, O. V., Baskunov, V. B., Darii, M.
V., Kolbanovskiy, A., Baykov, A. A., and Gromova, E. S. (2011)
Dnmt3a-CD is less susceptible to bulky benzo[a]pyrene diol
epoxide-derived DNA lesions than prokaryotic DNA methyltransferases,
Biochemistry, 50, 875-881.
25.Laemmli, U. K. (1970) Cleavage of structural
proteins during the assembly of the head of bacteriophage T4,
Nature, 227, 680-685.
26.Bradford, M. M. (1976) A rapid and sensitive
method for the quantitation of microgram quantities of protein
utilizing the principle of protein–dye binding, Anal.
Biochem., 72, 248-254.
27.Tevyashova, A. N., Durandin, N. A., Vinogradov,
A. M., Zbarsky, V. B., Reznikova, M. I., Dezhenkova, L. G., Bykov, E.
E., Olsufyeva, E. N., Kuzmin, V. A., Shtil, A. A., and
Preobrazhenskaya, M. N. (2013) Role of the acyl groups in carbohydrate
chains in cytotoxic properties of olivomycin A, J. Antibiot.,
66, 523-530.
28.Sergeev, A. V., Kirsanova, O. V., Loiko, A. G.,
Nomerotskaya, E. I., and Gromova, E. S. (2018) Detection of DNA
methylation by Dnmt3a methyltransferase using methyl-dependent
restriction endonucleases, Mol. Biol., 52, 272-278.
29.Minero, A. S., Lukashevich, O. V., Cherepanova,
N. A., Kolbanovskiy, A., Geacintov, N. E., and Gromova, E. S. (2012)
Probing murine methyltransferase Dnmt3a interactions with
benzo[a]pyrene modified DNA by fluorescence methods, FEBS
J., 279, 3965-3980.
30.Gowher, H., and Jeltsch, A. (2002) Molecular
enzymology of the catalytic domains of the Dnmt3a and Dnmt3b DNA
methyltransferases, J. Biol. Chem., 277, 20409-20414.
31.Carpenter, M. L., Marks, J. N., and Fox, K. R.
(1993) DNA‐sequence binding preference of the
GC‐selective ligand mithramycin, FEBS J., 215,
561-566.
32.Beniaminov, A. D., Dezhenkova, L. G., Mamaeva, O.
K., Shchyolkina, A. K., Tevyashova, A. N., Kaluzhny, D. N., and Shtil,
A. A. (2018) Divalent cations are dispensable for binding to DNA of a
novel positively charged olivomycin A derivative, PloS One,
13, e0191923.
33.Vilkaitis, G., Serva, S., Weinhold, E., and
Klimasauskas, S. (2001) The mechanism of DNA cytosine-5 methylation.
Kinetic and mutational dissection of HhaI methyltransferase, J.
Biol. Chem., 276, 20924-20934.
34.Maltseva, D., Baykov, A., Jeltsch, A., and
Gromova, E. (2009) Impact of 7,8-dihydro-8-oxoguanine on methylation of
the CpG Site by Dnmt3a, Biochemistry, 48, 1361-1368.
35.Kirsanova, O. V., Cherepanova, N. A., and
Gromova, E. S. (2009) Inhibition of C5-cytosine-DNA-methyltransferases,
Biochemistry (Moscow), 74, 1175-1186.
36.Cherepanova, N. A., Zhuze, A. L., and Gromova, E.
S. (2010) Inhibition of murine DNA methyltransferase Dnmt3a by DNA
duplexes containing pyrimidine-2(1H)-one, Biochemistry (Moscow),
75, 1115-1125.
37.Hou, M. H., Robinson, H., Gao, Y. G., and Wang,
A. H. J. (2004) Crystal structure of the
[Mg2+‐(chromomycin A3)2]–d(TTGGCCAA)2 complex
reveals GGCC binding specificity of the drug dimer chelated by a metal
ion, Nucleic Acids Res., 32, 2214-2222.
38.Gerasimaite, R., Merkiene, E., and Klimasauskas,
S. (2011) Direct observation of cytosine flipping and covalent
catalysis in a DNA methyltransferase, Nucleic Acids Res.,
39, 3771-3780.