Crystal Structure of the PX Domain of SNX27
Y. Li1,a, S. Liao1,b, F. Li1,c, and Z. Zhu1,d*
1Hefei National Laboratory for Physical Sciences at the Microscale and School of Life Sciences, University of Science and Technology of China, 230027 Hefei, China* To whom correspondence should be addressed.
Received July 5, 2018; Revised September 29, 2018; Accepted September 29, 2018
SNX27 is a component of the retromer complex essential for the recycling of transmembrane receptors. SNX27 contains the N-terminal Phox (PX) domain that binds inositol 1,3-diphosphate (Ins(1,3)P2) and is important for the SNX27 localization. Here, we determined the crystal structure of human SNX27 PX domain by X-ray crystallography. We found that the sulfate ion is located in the positively charged lipid-binding pocket of the PX domain, which mimics the phospholipid recognition. In addition, we modelled the SNX27-PX–Ins(1,3)P2 complex to better understand the mechanism of Ins(1,3)P2 recognition by the PX domain of SNX27.
KEY WORDS: SNX27, lipid binding, PX domain, crystal structureDOI: 10.1134/S0006297919020056
Abbreviations: Ins(1,3)P2, inositol 1,3-diphosphate; PX, Phox domain; SNX, sorting nexin (family).
Endocytosis is an energy consuming process that plays an important role
in a variety of cellular trafficking pathways, such as an uptake of
nutrients, molecule recycling, transport of particles, etc. [1, 2]. In particular,
sorting and recycling of transmembrane proteins to the cell surface
through the Golgi apparatus is mediated by endosomes and endocytosis
[3]. Many scaffold molecules, including members of
the Phox (PX) domain-containing sorting nexin (SNX) family, participate
in endocytosis [4-6]. Mammalian
genome encodes over 30 SNXs, all of them possessing the PX domain and
localizing to the cell surface where they mediate signaling pathway
through interactions with receptors [4, 7].
Among the SNXs, SNX27 is of special interest since it includes the PDZ domain at the N-terminus together with the PX domain and three FERM-like domains, implicating its role as a scaffold molecule in cell trafficking [8-11]. It has been reported that SNX27 binds to the Vps26-Vps35-Vps29 complex via its PDZ domain. The PX domain is a lipid-binding domain that tethers SNX27 to early endosome by recognizing inositol 1,3-diphosphate (Ins(1,3)P2) [8, 9]. In addition, SNX27 directly interacts with the WASH complex, thereby inhibiting cargo entry into the lysosomal pathway by recycling endosomes to the cell membrane [9]. In mammals, dysfunctions of SNX27 lead to epileptic encephalopathy, hydrocephalus, growth retardation, pseudosarcomatous fibromatosis, and many other diseases [12, 13].
It has been reported that SNX27 is targeted to early endosome via binding of its PX domain to Ins(1,3)P2 of the endosome [14, 15]. In addition to the PX domain, other modules, such as the PH and FYVE domains [16-18], also possess Ins(1,3)P2-binding ability. Although interaction of the SNX27 PX domain with Ins(1,3)P2 is a key process determining protein localization at the endosome, the molecular mechanism of such interaction remains elusive. Here, we crystallized the PX domain of SNX27 with a sulphate ion mimicking Ins(1,3)P2. Detailed structural analysis revealed that the PX domain recognizes the sulphate ion via a positively charged surface. We also modelled the SNX27–Ins(1,3)P2 complex based on the solved SNX27 structure. The results of our study elucidate the structural mechanism of the PX domain interaction with Ins(1,3)P2, as well as provide insights into Ins(1,3)P2-dependent endosomal localization of SNX27.
MATERIALS AND METHODS
Cloning, protein expression and purification. The coding sequence for the PX domain of SNX27 was cloned into the pET28-MHL vector (Genbank accession number: EF456735), and the resulting constructs were used for the transformation of E. coli BL21(DE3) cells. The cells were grown at 37°C in LB medium containing 50 µg/ml kanamycin to OD600 of 0.8. Expression of the recombinant proteins was induced with 0.2 mM isopropyl β-D-thiogalactopyranoside (IPTG), and the cells were cultivated for another 20 h at 16°C. The cells were then harvested, resuspended in lysis buffer (20 mM Tris-HCl, 0.4 M NaCl, pH 7.5), and disrupted by sonication. The lysate was centrifuged at 14,000g at 4°C for 25 min, and the supernatant containing the N-terminally His6-tagged protein was subjected to chromatography on Ni-NTA-resin (GE Healthcare, USA). The eluted proteins were dialyzed against the cleavage buffer (10 mM Tris-HCl, 0.2 M NaCl, pH 7.5) at 4°C, and the His6-tag was removed by adding tobacco etch virus (TEV) protease. Further purification was performed on a Superdex 75 gel filtration column (HiLoad 16/600; GE Healthcare) and HisTrap SP HP column. The purified protein was concentrated to approximately 15.6 mg/ml.
Crystallization, data collection, and structure determination. The crystals were grown using the sitting drop vapor diffusion method at 18°C. The protein solution was mixed with an equal volume of crystallization buffer (0.1 M LiSO4, 0.1 M Tris-HCl, pH 8.5, 25% PEG3350), and crystals appeared after 20 h. Before the data collection, the crystals were soaked in cryo-protectant containing 90% reservoir solution and 10% glycerol and stored in liquid nitrogen. Diffraction data were collected on the BL18U1 beamline at the Shanghai Synchrotron Facility (SSRF). The data sets were collected at 0.9778 Å and processed using the HKL2000 program [19]. The initial structure of the SNX27 PX domain was obtained by PHASER [20] using the PX domain of SNX17 as a molecular replacement model (PDB: 3LUI, 21). The resulting structure was manually built with Coot [21] and refined by PHENIX [22]. The statistics for the data collection and structural refinement are summarized in the table.
Ins(1,3)P2-binding model. Docking was performed using the PYRX software [23]. Ins(1,3)P2 was docked into the sulfate anion-binding pocket of the PX domain. The most reasonable model was presented.
RESULTS AND DISCUSSION
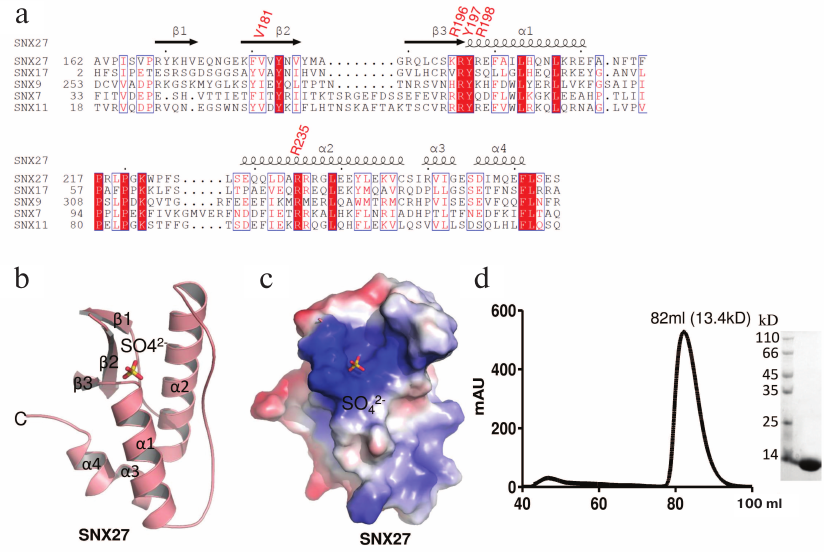
The PX domain of SNX27 was expressed, purified, and crystallized, and its structure was solved at a 1.78 Å resolution (table). The coordinates and structural factors of the SNX27 PX domain were deposited in the Protein Databank (accession number 5ZN9). There are two molecules (RMSD = 0.469) in the asymmetric unit (ASU). Although the PX domain of SNX27 contains a pair of cysteines, gel filtration showed that its molecule is a monomer (Fig. 1d). In addition to the residues at both termini, the electron density of chain A residues 183 to 187 was also missing, which may be due to the molecule internal flexibility. The SNX27 PX domain adopts a canonical PX fold [24] consisting of three anti-parallel β-strands (β1-β3) and four α-helices packed against one side of the β-strands (Figs. 1a and 1b).
Crystallographic data and refinement statistics
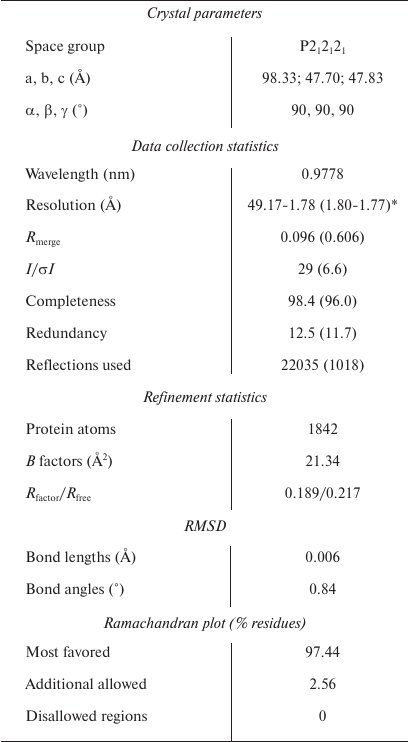
Note: RMSD, root mean square deviation between predicted and
experimental data.
* Statistics for the highest-resolution shell are shown in
parentheses.
Fig. 1. Sequence analysis and structure of the PX domain of SNX27. a) Sequence alignment of PX domains from different human proteins. The residues that form the pocket are R196, Y197, R198, and R235 (UniProt accession numbers: SNX27 (Q96L92); SNX17 (Q15036); SNX9 (Q9Y5X1); SNX7 (Q9UNH6); SNX11 (Q9Y5W9)). b) The overall structure of the SNX27 PX domain. c) The electrostatic potential surface of the SNX27 PX domain. d) Gel filtration of purified SNX27 PX domain and SDS-PAGE indicate that SNX27 PX domain is a monomer.
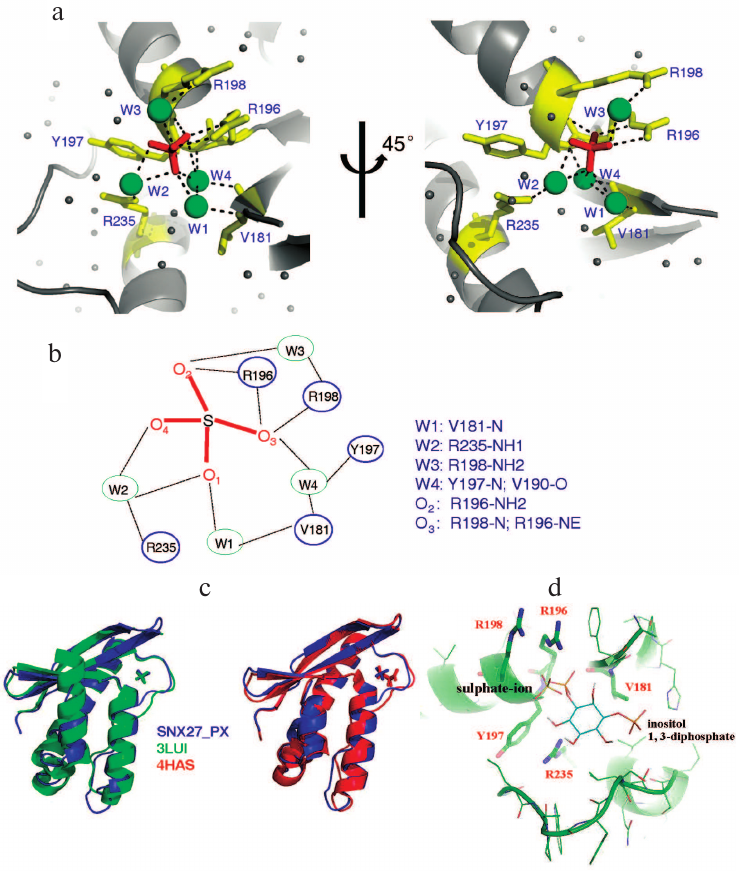
In the ASU, we found three sulfate ions packed with two PX domain molecules; only two of them were located in the lipid-binding pocket. The fact that the third sulfate ion did not localize to the pocket could affect the crystal packing, because crystallization buffer contained sulfate ion. The structure of the PX domain in SNX27 was similar to the structure of the PX domain in SNX17 [25]. The sulphate ion (S1) positioned into the positively charged pocket composed of the β2, β3, α1, and α2 elements (Fig. 1c) forms a hydrogen bond with SNX27 (Figs. 2a and 2c). The backbone and the side chain of Arg205, as well as the backbone of Arg207, are directly connected by hydrogen bonds to S1, while Val190, Tyr206, Arg207, and Arg244 form water-mediated hydrogen bonds with S1 (Figs. 2a and 2c). Sequence alignment of the SNX27 PX domain with those of other SNXs showed that Arg196, Tyr197, and Arg235 of SNX27 are strictly conserved in all aligned sequences (Fig. 1a). Although neither Val181, nor Arg198 of SNX27 are conserved in other SNXs, the interactions are not likely to be strongly affected, since Val181 and Arg198 both form backbone hydrogen bonds with S1 (Figs. 2a and 2b). Recently, the structure of the SNX17 PX domain complex with sulphate ion was solved [25]. Sequence alignment indicated low sequence similarity between different PX domains of human proteins. The most conserved residues localize to the acidic group-binding pocket. Other conserved residues are hydrophobic and located in flexible regions that might be involved in the recognition of fatty acid chains of lipids. Superposition of SNX17 and SNX27 structures showed that both PX domains adopt a very similar architecture and share the same sulphate-binding surface, indicating that the sulphate group-binding pocket is conserved in SNXs (Fig. 2c, left panel). In addition, we compared the structure of the SNX27 PX domain complex with citric acid and demonstrated that the sulfate-binding pocket of the PX domain favors acidic groups (Fig. 2c, right panel).
Fig. 2. Structural analyses of the lipid-binding pocket of the SNX27 PX domain. a) Residues responsible for sulfate ion recognition. The two views of the structure (rotated by 45° around Y-axis). b) Detailed view of interactions between the SNX27 PX domain and sulfate ion. c) Structural comparison of the SNX27 PX domain with 3LUI (SNX17 PX domain with sulfate group) and 4HAS (SNX27 PX domain with citric acid). d) Model of SNX27 PX–Ins(1,3)P2 complex.
It has been reported that the PX domain of SNX27 binds to Ins(1,3)P2 with a Kd of 15 µM [25]. Based on the SNX27–sulphate complex structure, we further modeled Ins(1,3)P2 into the SNX27 complex to replace the sulphate group. In the modeled SNX27–Ins(1,3)P2 complex, Ins(1,3)P2 aromatic ring lies in a positively charged groove composed of α1, α2, β2, and β3 without causing any steric hindrance (Fig. 2d). One phosphate group of Ins(1,3)P2 occupies the site of the sulphate ion, while the other phosphate group interacts with α2 residues (Fig. 2d).
The structure of the complex implies that the interaction of the SNX27 PX domain with Ins(1,3)P2 is specific and depends on both phosphate groups. The interactions between Ins(1,3)P2 and SNX27 suggest that SNX27 could be recruited to early endosome in an Ins(1,3)P2-dependent manner. Therefore, our presented SNX27 structure provides the mechanistic insights into the localizations of SNX27 molecules on early endosome.
Funding
We acknowledge the China National Center for Protein Sciences, Shanghai, for providing the facility support. This work was supported by the National Natural Science Foundation of China (project no. 31500601).
Conflict of Interest
The authors declare no conflict of interest in financial or any other sphere.
REFERENCES
1.Goldstein, J. L., Anderson, R. G., and Brown, M. S.
(1979) Coated pits, coated vesicles, and receptor-mediated endocytosis,
Nature, 279, 679-685.
2.Rejman, J., Oberle, V., Zuhorn, I. S., and
Hoekstra, D. (2004) Size-dependent internalization of particles via the
pathways of clathrin- and caveolae-mediated endocytosis, Biochem.
J., 377, 159-169.
3.Bonifacino, J. S., and Traub, L. M. (2003) Signals
for sorting of transmembrane proteins to endosomes and lysosomes,
Annu. Rev. Biochem., 72, 395-447.
4.Teasdale, R. D., Loci, D., Houghton, F., Karlsson,
L., and Gleeson, P. A. (2001) A large family of endosome-localized
proteins related to sorting nexin 1, Biochem. J., 358,
7-16.
5.Seet, L. F., and Hong, W. (2006) The Phox (PX)
domain proteins and membrane traffic, Biochim. Biophys. Acta,
1761, 878-896.
6.Worby, C. A., and Dixon, J. E. (2002) Sorting out
the cellular functions of sorting nexins, Nat. Rev. Mol. Cell
Biol., 3, 919-931.
7.Sato, T. K., Overduin, M., and Emr, S. D. (2001)
Location, location, location: membrane targeting directed by PX
domains, Science, 294, 1881-1885.
8.Temkin, P., Lauffer, B., Jager, S., Cimermancic,
P., Krogan, N. J., and von Zastrow, M. (2011) SNX27 mediates retromer
tubule entry and endosome-to-plasma membrane trafficking of signalling
receptors, Nat. Cell Biol., 13, 715-721.
9.Steinberg, F., Gallon, M., Winfield, M., Thomas, E.
C., Bell, A. J., Heesom, K. J., Tavare, J. M., and Cullen, P. J. (2013)
A global analysis of SNX27-retromer assembly and cargo specificity
reveals a function in glucose and metal ion transport, Nat. Cell
Biol., 15, 461-471.
10.Lauffer, B. E., Melero, C., Temkin, P., Lei, C.,
Hong, W., Kortemme, T., and von Zastrow, M. (2010) SNX27 mediates
PDZ-directed sorting from endosomes to the plasma membrane, J. Cell
Biol., 190, 565-574.
11.Gallon, M., Clairfeuille, T., Steinberg, F., Mas,
C., Ghai, R., Sessions, R. B., Teasdale, R. D., Collins, B. M., and
Cullen, P. J. (2014) A unique PDZ domain and arrestin-like fold
interaction reveals mechanistic details of endocytic recycling by
SNX27-retromer, Proc. Natl. Acad. Sci. USA, 111,
E3604-3613.
12.Damseh, N., Danson, C. M., Al-Ashhab, M.,
Abu-Libdeh, B., Gallon, M., Sharma, K., Yaacov, B., Coulthard, E.,
Caldwell, M. A., Edvardson, S., Cullen, P. J., and Elpeleg, O. (2015) A
defect in the retromer accessory protein, SNX27, manifests by infantile
myoclonic epilepsy and neurodegeneration, Neurogenetics,
16, 215-221.
13.Small, S. A., and Petsko, G. A. (2015) Retromer
in Alzheimer disease, Parkinson disease and other neurological
disorders, Nat. Rev. Neurosci., 16, 126-132.
14.Cullen, P. J., and Korswagen, H. C. (2011)
Sorting nexins provide diversity for retromer-dependent trafficking
events, Nat. Cell Biol., 14, 29-37.
15.Cai, L., Loo, L. S., Atlashkin, V., Hanson, B.
J., and Hong, W. (2011) Deficiency of sorting nexin 27 (SNX27) leads to
growth retardation and elevated levels of N-methyl-D-aspartate receptor
2C (NR2C), Mol. Cell. Biol., 31, 1734-1747.
16.Ellson, C. D., Gobert-Gosse, S., Anderson, K. E.,
Davidson, K., Erdjument-Bromage, H., Tempst, P., Thuring, J. W.,
Cooper, M. A., Lim, Z. Y., Holmes, A. B., Gaffney, P. R., Coadwell, J.,
Chilvers, E. R., Hawkins, P. T., and Stephens, L. R. (2001) Inositol
1,3-diphosphate regulates the neutrophil oxidase complex by binding to
the PX domain of p40(phox), Nat. Cell Biol., 3,
679-682.
17.Kutateladze, T., and Overduin, M. (2001)
Structural mechanism of endosome docking by the FYVE domain,
Science, 291, 1793-1796.
18.Kutateladze, T. G., Ogburn, K. D., Watson, W. T.,
de Beer, T., Emr, S. D., Burd, C. G., and Overduin, M. (1999)
Phosphatidylinositol 3-phosphate recognition by the FYVE domain,
Mol. Cell, 3, 805-811.
19.Otwinowski, Z., and Minor, W. (1997) Processing
of X-ray diffraction data collected in oscillation mode, Methods
Enzymol., 276, 307-326.
20.McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P.
D., Winn, M. D., Storoni, L. C., and Read, R. J. (2007) Phaser
crystallographic software, J. Appl. Crystallogr.,
40, 658-674.
21.Emsley, P., and Cowtan, K. (2004) Coot:
model-building tools for molecular graphics, Acta Crystallogr. D
Biol. Crystallogr., 60, 2126-2132.
22.Adams, P. D., Grosse-Kunstleve, R. W., Hung, L.
W., Ioerger, T. R., McCoy, A. J., Moriarty, N. W., Read, R. J.,
Sacchettini, J. C., Sauter, N. K., and Terwilliger, T. C. (2002)
PHENIX: building new software for automated crystallographic structure
determination, Acta Crystallogr. D Biol. Crystallogr.,
58, 1948-1954.
23.Dallakyan, S., and Olson, A. J. (2015)
Small-molecule library screening by docking with PyRx, Methods Mol.
Biol., 1263, 243-250.
24.Bravo, J., Karathanassis, D., Pacold, C. M.,
Pacold, M. E., Ellson, C. D., Anderson, K. E., Butler, P. J., Lavenir,
I., Perisic, O., Hawkins, P. T., Stephens, L., and Williams, R. L.
(2001) The crystal structure of the PX domain from p40(phox) bound to
phosphatidylinositol 3-phosphate, Mol. Cell, 8,
829-839.
25.Ghai, R., Mobli, M., Norwood, S. J., Bugarcic,
A., Teasdale, R. D., King, G. F., and Collins, B. M. (2011) Phox
homology band 4.1/ezrin/radixin/moesin-like proteins function as
molecular scaffolds that interact with cargo receptors and Ras GTPases,
Proc. Natl. Acad. Sci. USA, 108, 7763-7768.