Resveratrol Promotes in vitro Differentiation of Osteoblastic MC3T3-E1 Cells via Potentiation of the Calcineurin/NFATc1 Signaling Pathway
Y. Huang1#, J. Huo2#, F. Q. Liu3, J. Liu3, X. J. Zhang4, C. H. Guo4, and L. H. Song4,a*
1Department of Biochemistry, Changzhi Medical College, 046000 Changzhi, Shanxi, China2Department of Biology, Changzhi Medical College, 046000 Changzhi, Shanxi, China
3Changzhi Medical College, 046000 Changzhi, Shanxi, China
4Department of Pharmacology, Changzhi Medical College, 046000 Changzhi, Shanxi, China
# These authors contributed equally to this study.
* To whom correspondence should be addressed.
Received March 15, 2019; Revised April 8, 2019; Accepted April 8, 2019
Resveratrol has been shown to stimulate differentiation of osteoblastic MC3T3-E1 cells in vitro; however, the mechanisms underlying the anabolic effect of resveratrol on osteoblasts remain largely unknown. Our study was aimed to investigate the molecular mechanism of resveratrol-induced differentiation of MC3T3-E1 cells. MC3T3-E1 cells were treated for 8 days with different concentrations of resveratrol (10–8-10–6 M) and 10–6 M cyclosporine A (CsA), a specific inhibitor of the calcineurin/NFAT pathway. According to the results of pilot studies of cell proliferation and alkaline phosphatase activity, 10–7 M concentration of resveratrol was used in subsequent experiments. The levels of mRNA expression of the osteosis-related genes CaN, NFATc1, and Runx2 were analyzed by real-time RT-PCR; the levels of the corresponding proteins were estimated by Western blot analysis. Resveratrol upregulated expression of the CaN, NFATc1, and Runx2 genes at both mRNA and protein levels compared to the control group (p < 0.05), while CsA reduced the effects of resveratrol (p < 0.05). Using immunohistochemical staining, we showed that resveratrol induced NFATc1 accumulation in the cell nuclei, and treatment with CsA inhibited resveratrol-mediated induction of NFATc1, suggesting that the calcineurin/NFATc1 signaling pathway plays an important role in the regulatory effect of resveratrol on osteoblasts.
KEY WORDS: resveratrol, cyclosporine A, calcineurin/NFATc1, osteoblast differentiationDOI: 10.1134/S0006297919060117
Abbreviations: ALP, alkaline phosphatase; α-MEM, alpha minimum essential medium; CaN, calcineurin; CsA, cyclosporine A; IHC, immunohistochemistry; MTT, 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide; NFATc1, nuclear factor of activated T cells, cytoplasmic 1; NF-κB, nuclear factor kappa B; pNPP, p-nitrophenyl phosphate; PVDF, polyvinylidene fluoride; RIPA, radioimmunoprecipitation assay; Runx2, Runt-related transcription factor 2.
Phytoestrogens are a group of natural plant compounds with the mammalian
estrogen-like activity. Resveratrol (3,4′,5-trihydroxystilbene)
is an edible polyphenolic stilbene phytoestrogen that can be found in
the skin of most grape cultivars, peanut roots, and a variety of
medicinal herbs [1]. Extensive studies have
established resveratrol as an anti-neoplastic, anti-oxidant,
anti-platelet, and anti-inflammatory agent [2].
Resveratrol was also found to reduce bone loss in ovariectomized rats
[3]. However, the mechanisms underlying the
anabolic effect of resveratrol on osteoblasts remain largely
unknown.
Calcineurin (CaN) is a ubiquitous serine/threonine phosphatase, whose intracellular substrate is the nuclear factor of activated T cells (NFAT). CaN-catalyzed dephosphorylation unmasks nuclear localization signals in the NFAT family factors, which leads to their translocation to the nucleus and gene activation. NFATs are a family of four transcription factors: NFAT1 (p, c2), NFAT2 (c, c1), NFAT3 (c4), and NFAT4 (x, c3). Bone formation is severely impaired in NFATc1–/– embryonic fibroblast cells [4]. At present, most studies are focused on the effects of NFATc1 (NFAT cytoplasmic 1) activation on osteoclastogenesis [5, 6]. However, the specific role of NFATc1 in osteoblastic differentiation is not well understood, and, in some instances, the reports on this subject are contradictory. Runt-related transcription factor 2 (Runx2) is the major regulator of osteoblastic differentiation [7, 8]. No osteoblast differentiation is observed in the absence of Runx2; Runx2 deficiency results in the lack of bone formation and lesser transcription of Runx2 target genes [9-11].
The main objective of this study was to investigate molecular mechanisms of resveratrol-mediated osteoblastic differentiation of MC3T3-E1 cells by analyzing the levels of mRNA and protein expression for the osteosis-related genes CaN, NFATc1, and Runx2. To confirm the effect of resveratrol on osteoblastic differentiation, 10–6 M cyclosporine A (CsA), a specific inhibitor of CaN, was used based on the preliminary experimental results of this study, as well as published data [12].
MATERIALS AND METHODS
Cell culture. MC3T3-E1 cells were obtained from the Shanghai Institute of Biochemistry and Cell Biology, Shanghai, China. Cell culture media and 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide (MTT) from BD Company (USA), fetal calf serum from Invitrogen (USA), and CsA from ALEXIS Laboratories (USA) were used.
MC3T3-E1 cells were maintained in the growth medium (Phenol red-free alpha minimum essential medium, α-MEM) containing 10% (v/v) fetal bovine serum, 5·10—3 M β-glycerophosphate, and 25 g/liter ascorbic acid at 37°C in 5% CO2. Resveratrol and CsA were added every other day, and the medium was replaced every 4 days thereafter.
MTT assays. Briefly, the cells were incubated with different concentrations of resveratrol (10—8-10—6 M) for 8 days. The cells were then seeded in a 96-well plate at a density of 3·103 cells/well. A 20-µl sample of MTT stock solution (5 mg/ml) in phosphate buffered saline was added into each well to a final MTT concentration of 0.5 mg/liter, and the cells were incubated at 37°C for another 4 h. The medium with MTT was removed and the formed purple formazan crystals were dissolved in 150 μl of dimethyl sulfoxide. The absorbance [abs] in the wells was read at 570 nm using a microplate reader. The control group with 100% viability was used as a negative control. Cell proliferation (%) of the treated group was calculated as follows:
[abs]treated group/[abs]control group × 100.
Alkaline phosphatase assays. MC3T3-E1 cells were cultured in α-MEM medium at an initial density of 2·104 cells/well in a 24-well culture plate and collected after 8 days of culturing. After different treatments, the cells were assayed for alkaline phosphatase (ALP) activity using p-nitrophenyl phosphate (pNPP) liquid substrate (Sigma, USA) following the manufacturer’s instructions. First, the cells were lysed in a lysis buffer, incubated for 30 min at 37°C, and centrifuged at 12,000g at 4°C for 10 min. Clear cell lysate was transferred into a new 1.5-ml centrifuge tube for the following ALP assays: 10 μl of clear cell lysate was added into a 96-well transparent plate; subsequently, 200 μl of pNPP solution was added to each well. The plate was incubated in the dark for approximately 30 min at room temperature and absorbance in the wells was read at 405 nm with a multiwell plate reader (San Jose, CA, USA).
Real-time RT-PCR. Total RNA for the real-time reverse transcription-polymerase chain reactions (RT-qPCR) was extracted using the MiniBEST Universal RNA Extraction kit (TaKaRa, Japan). cDNA was synthesized using the PrimeScript RT Reagent kit with gDNA Eraser (Perfect Real Time) (TaKaRa), and qPCR was performed using the SYBR Green Mix kit (TaKaRa). All samples were processed in triplicates in the eight-strip optical tubes (Axygen, USA). Primer sequences are shown in Table 1. The qPCR regime was as follows: initial heating at 95°C for 30 s followed by 40 cycles of 95°C (denaturation) for 5 s and 60°C (annealing/elongation) for 30 s, and then a dissociation. Relative expression of the target genes was quantified using the 2 −ΔΔCt method [13].
Table 1. Primers used for qPCR

Western blotting. The cells were washed twice with ice-cold phosphate buffered saline and lysed with radioimmunoprecipitation assay (RIPA) lysis buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, and 0.1% SDS) to assess the amounts of CaN, NFATc1, and Runx2. Total protein content in the cell lysates was quantitated using the bicinchoninic acid protein assay reagent (Beyond, China). Samples containing equal amounts of total protein were separated by 10% SDS-PAGE and transferred onto polyvinylidene fluoride (PVDF) membranes. The membranes were then probed with antibodies against CaN (1 : 2000 dilution; ab137335; Abcam, UK), NFATc1 (H-10; 1 : 500 dilution; Santa Cruz Biotechnology, USA), and Runx2 (1 : 3000 dilution; ab23981; Abcam) at 4°C overnight, washed thrice with Tris-buffered saline containing 0.1% Tween 20, and incubated with goat anti-rabbit horseradish peroxidase-conjugated secondary antibody (1 : 2000 dilution; A0208; Beyotime Institute of Biotechnology, China) at room temperature for 2 h. β-Actin stained with mouse polyclonal antibody (1 : 5000 dilution; ab20272; Abcam) was used as a loading control. The proteins were detected using enhanced chemiluminescence system (Amersham Biosciences, Germany) with an ImageQuant LAS 4000 imager (GE Healthcare Life Sciences, UK); protein bands were quantified with the Image J 1.37v software (National Institutes of Health, Bethesda, MD, USA).
Immunohistochemistry (IHC). MC3T3-E1 cells were seeded on the coverslips in a 6-well plate (1.0·105 cells/well) and cultured in the presence of 10—7 M resveratrol and 10—6 M CsA for 8 days. Then, the cells were fixed with 4% paraformaldehyde for 15 min, permeabilized with 0.5% Triton X-100 for 10 min, and incubated with blocking buffer and then with anti-NFATc1 mouse monoclonal antibody (1 : 200) at 37°C for 2 h. Mouse two-step kit (PV-6002) was used according to the manufacturer’s protocol (ZSGB-BIO Co., Ltd., China). All images were obtained under a light microscope (Olympus BX51; Olympus Corporation, Japan) at a 400× magnification. According to the staining intensity, the levels of NFATc1 staining were graded as – (no staining), 1 (+), 2 (++), and 3 (+++). Based on the proportion of NFATc1-positive cells, the scores were assigned as follows: – (0-1%; score 0); + (1-24%; score 1); ++ (25-49%; score 2); and +++ (50-100%; score 3). The final NFATc1 staining scores were calculated based on the sum of the intensity and percentage scores, which were defined as follows: –, no expression (total score 0); +, weak expression (total score 1-2); 2+, moderate expression (total score 3-4); and 3+, strong expression (total score 5-6). The average values were obtained based on the values from 10 randomly selected fields.
Data analysis and statistics. The data were represented as mean ± standard deviation. According to the experimental design, a one-way analysis of variance was used for comparison among groups, followed by the least significant difference t-test. The difference was considered as statistically significant at p < 0.05.
RESULTS
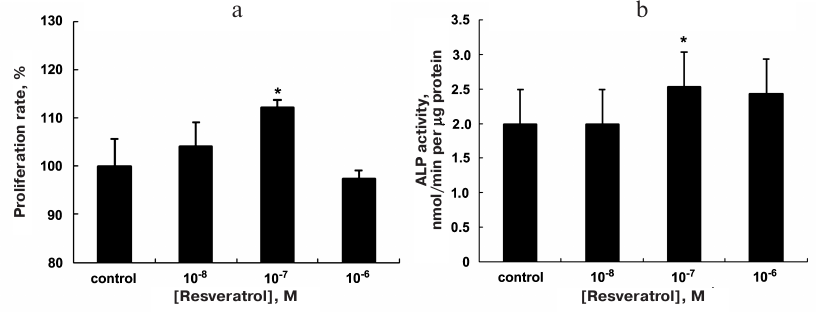
Effect of resveratrol on cell proliferation. Based on the MTT assay results, the optimal treatment concentration of resveratrol was determined to be 10–7 M and used in subsequent experiments (Fig. 1a).
Fig. 1. Proliferation of MC3T3-E1 cells after treatment with resveratrol (n = 6) according to the MTT assay results; the 10—7 M treatment group exhibited the highest proliferation compared to the control group (a). Cells differentiation was assessed based on the ALP activity (b); * p < 0.05 vs. the control group.
Effect of resveratrol on the ALP activity. The ALP activity was examined to evaluate the effect of resveratrol on the osteogenic differentiation of cells. The ALP activity per unit protein in the MC3T3-E1 cells was examined after the 8-day treatment with different concentrations of resveratrol. The maximal ALP activity was observed in the 10—7 M group, which showed a significant increase in the ALP activity compared to the control group (Fig. 1b).
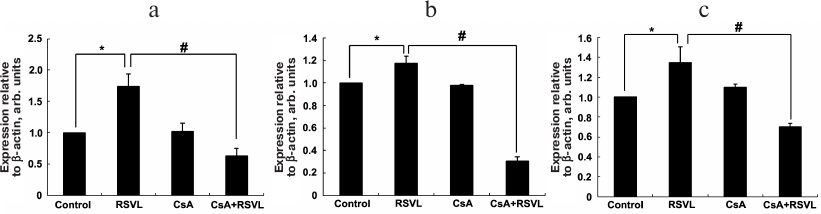
Effect of resveratrol on the mRNA levels of osteosis-related genes CaN, NFATc1, and Runx2. The effect of resveratrol on the expression of CaN, NFATc1, and Runx2 mRNAs was analyzed using RT-qPCR (Fig. 2).
Fig. 2. Expression of mRNAs of the osteosis-related genes. MC3T3-E1 cells were subjected to different treatments for 8 days. Expression of CaN (a), NFATc1 (b), and Runx2 (c) mRNAs was quantified using RT-qPCR. The data are presented as mean ± standard deviation (n = 3); * p < 0.05 vs. the control group; # p < 0.05 between the resveratrol (RSVL) and cyclosporine A/resveratrol (CsA + RSVL) groups.
A significant increase in the expression levels of CaN mRNA was observed in the resveratrol-treated group compared with the control group (p < 0.05). However, the (resveratrol + CsA)-treated group showed a significant decrease in the CaN mRNA expression compared to the resveratrol group (p < 0.05). Similar results were observed for the expression of NFATc1 and Runx2 mRNAs (p < 0.05).
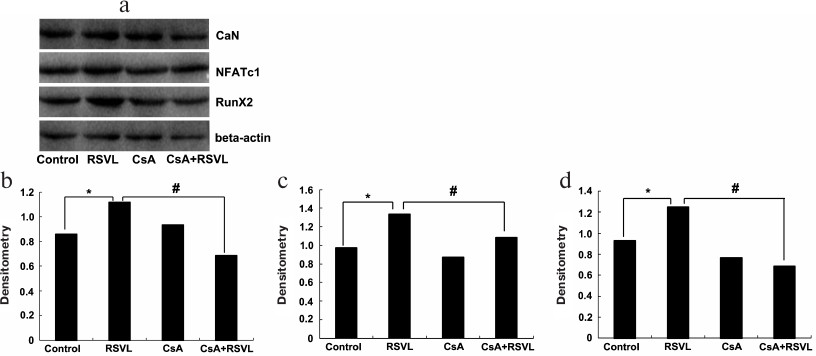
Resveratrol increases the levels of CaN, NFATc1, and Runx2 proteins in vitro. The levels of CaN, NFATc1, and Runx2 proteins in MC3T3-E1 cells were determined to evaluate the effect of resveratrol on the cells. As shown in Fig. 3, resveratrol significantly increased the levels of CaN, NFATc1, and Runx2 proteins (p < 0.05); at the same time, the contents of these proteins in the (resveratrol + CsA)-treated group were significantly decreased compared with the resveratrol group.
Fig. 3. Levels of osteoporosis-related proteins after different treatments. The levels of CaN, NFATc1, and Runx2 proteins (a) and densitometric analysis of CaN (b), NFATc1 (c), and Runx2 (d) content in the cells. β-Actin was used as a loading control; * p < 0.05 vs. the control group; # p < 0.05 between the resveratrol (RSVL) and cyclosporine A/resveratrol (CsA + RSVL) groups.
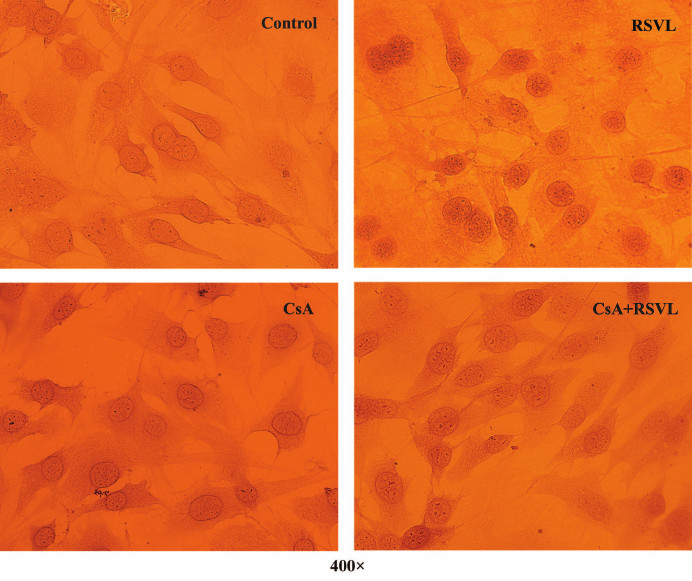
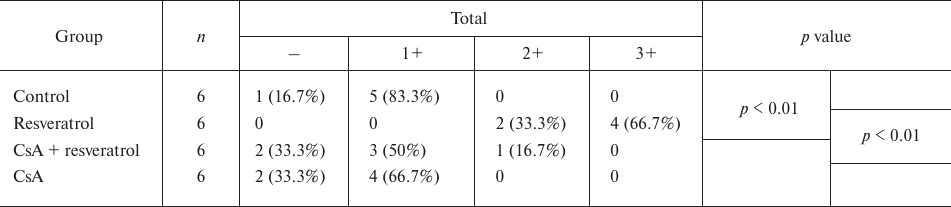
Resveratrol induces NFATc1 accumulation in the nuclei. The NFATc1 levels in MC3T3-E1 cells was evaluated in situ by IHC staining using the anti-NFATc1 antibody (Fig. 4 and Table 2). We found that resveratrol treatment induced NFATc1 accumulation in the cell nuclei, as reflected by increased staining of the nuclei of MC3T3-E1 cells for NFATc1, while CsA inhibited this effect.
Fig. 4. IHC staining of MC3T3-E1 cells for NFATc1.
Table 2. NFATc1 levels in MC3T3-E1 cells as
determined by IHC staining
DISCUSSION
Many drugs have been used to treat osteolytic bone diseases, including bisphosphonates, estrogen, and raloxifene [14]. However, despite the efficacy, most of these drugs have limitations and exhibit side effects, such as thromboembolism and esophageal irritation [15]. The ultimate goal in the development of anti-osteoporotic treatments is finding an agent that can improve bone formation via simultaneous osteoblast stimulation and osteoclast inhibition without undesirable side effects. Natural compounds and phytoestrogens have been examined as candidate anti-osteoporotic agents. Previous studies have shown that resveratrol stimulates proliferation and differentiation of osteoblasts [16-18]. The action mechanisms of resveratrol have also been studied. For instance, it was found that resveratrol prevents apoptosis of osteoblasts by upregulating β-catenin expression [16]. Resveratrol stimulates proliferation and osteoblastic differentiation of human bone marrow-derived mesenchymal stem cells via an ER-dependent mechanism and coupling with ERK1/2 activation [17]. It also prevents bone loss by stimulating the pro-osteogenic factor BMP2 [18]. However, the relationship between resveratrol-induced osteoblast differentiation and the CaN/NFATc1 pathway has not been reported.
In our previous work, we have shown that resveratrol increases the level of Runx2 mRNA. Runx2, also known as the core-binding factor alpha 1, is an essential transcription factor for osteoblast differentiation and bone formation. Disruption of the Runx2 expression leads to a complete lack of bone formation in fetal mice [19]. This report suggested an important link between resveratrol and bone-specific transcription function in osteoblasts. Furthermore, it entailed the necessity to search for novel resveratrol-affected intracellular molecules and associated signaling pathways that ultimately direct bone cell differentiation. While searching for the signaling pathway that may link resveratrol to Runx2 activation in MC3T3-E1 cells, we were directed to the CaN/NFATc1 pathway since this pathway is vital for the signal transduction cascade involved in the control of differentiation, apoptosis, and adaptation of a wide range of cell types and tissues [20-22]. Therefore, it seemed reasonable to investigate possible activation mechanism of the CaN/NFATc1 axis under the action of resveratrol. In this study, MC3T3-E1 cells were incubated with 10—7 M resveratrol for 8 days. After resveratrol induction, cell proliferation, expression of mRNAs for the osteosis-related genes (CaN, NFATc1 and Runx2), and the levels of the corresponding proteins, as well the marker of osteogenesis differentiation (ALP activity) were evaluated (Fig. 1b). The results showed that 10—7 M resveratrol increased both the mRNA expression and the protein levels of CaN, NFATc1 and Runx2 (Figs. 2 and 3). IHC staining demonstrated that resveratrol treatment induced NFATc1 accumulation in the cell nuclei (Fig. 4). To further validate the obtained results, we treated MC3T3-E1 cells with CsA, a specific inhibitor of the CaN/NFATc1 pathway. We found that CsA abolished the effect of resveratrol on the mRNA expression and protein levels of CaN, NFATc1 and Runx2 (Figs. 2 and 3), which indicated that the CaN/NFATc1 pathway might participate in the resveratrol-induced osteoblastic differentiation. This observation is in accordance with the results reported by Sun et al. [23], who demonstrated that CaN overexpression enhanced osteoblastogenesis, while CaN deletion or inhibition diminished bone formation and reduced osteoblast differentiation. Moreover, as reported previously, resveratrol prevented the inhibitory effect of CsA on the proliferation and osteoblastic differentiation of bone marrow-derived mesenchymal stem cells [12]. Therefore, this study demonstrated that resveratrol induces in vitro differentiation of osteoblasts through potentiation of the CaN/NFATc1 signaling axis. The effect of resveratrol might be associated with its participation in the regulation of transcription of osteogenic differentiation genes. However, the question whether similar changes can be observed in vivo needs to be studied further.
In conclusion, we analyzed the effects of resveratrol on osteoblasts and showed involvement of the CaN/NFATc1 pathway in the resveratrol-induced osteoblastic stimulation. The results of the present study may shed new light on the molecular mechanism of resveratrol-mediated stimulation of osteoblasts.
Funding. This research was supported by the Changzhi Medical College Science and Technology Innovation Team (CX201413) and Shanxi Province Health and Family Planning Commission research topic (201602024).
Acknowledgements. The authors thank Professor Song for her concentrated guidance during this study.
Conflict of interest. The authors declare no conflict of interest.
REFERENCES
1.Chen, R. S., Wu, P. L., and Chiou, R. Y. (2002)
Peanut roots as a source of resveratrol, J. Agric. Food Chem.,
50, 1665-1667.
2.Fremont, L. (2000) Biological effects of
resveratrol, Life Sci., 66, 663-673.
3.Mizutani, K., Ikeda, K., Kawai, Y., and Yamori, Y.
(2000) Resveratrol attenuates ovariectomy-induced hypertension and bone
loss in stroke-prone spontaneously hypertensive rats, J. Nutr. Sci.
Vitaminol. (Tokyo), 46, 78-83.
4.Koga, T., Matsui, Y., Asagiri, M., Kodama, T.,
Crombrugghe, B., Nakashima, K., and Takayanagi, H. (2005) NFAT and
Osterix cooperatively regulate bone formation, Nat. Med.,
11, 880-885.
5.Penolazzi, L., Zennaro, M., Lambertini, E.,
Tavanti, E., Torreggiani, E., Gambari, R., and Piva, R. (2007)
Induction of estrogen receptor alpha expression with decoy
oligonucleotide targeted to NFATc1 binding sites in osteoblasts,
Mol. Pharmacol., 71, 1457-1462.
6.Zhou, F., Shen, Y., Liu, B., Chen, X., Wan, L., and
Peng, D. (2017) Gastrodin inhibits osteoclastogenesis via
down-regulating the NFATc1 signaling pathway and stimulates
osseointegration in vitro, Biochem. Biophys. Res.
Commun., 484, 820-826.
7.Ziros, P. G., Gil, A. P., Georgakopoulos, T.,
Habeos, I., Kletsas, D., Basdra, E. K., and Papavassiliou, A. G. (2002)
The bone-specific transcriptional regulator Cbfa1 is a target of
mechanical signals in osteoblastic cells, J. Biol. Chem.,
277, 23934-23941.
8.Ziros, P. G., Basdra, E. K., and Papavassiliou, A.
G. (2008) Runx2: of bone and stretch, Int. J. Biochem. Cell
Biol., 40, 1659-1663.
9.Vimalraj, S., and Selvamurugan, N. (2013)
MicroRNAs: synthesis, gene regulation and osteoblast differentiation,
Curr. Issues Mol. Biol., 15, 7-18.
10.Kang, J. S., Alliston, T., Delston, R., and
Derynck, R. (2005) Repression of Runx2 function by TGF-beta through
recruitment of class II histone deacetylases by Smad3, EMBO J.,
24, 2543-2555.
11.Jeon, E. J., Lee, K. Y., Choi, N. S., Lee, M. H.,
Kim, H. N., Jin, Y. H., Ryoo, H. M., Choi, J. Y., Yoshida, M., Nishino,
N., Oh, B. C., Lee, K. S., Lee, Y. H., and Bae, S. C. (2006) Bone
morphogenetic protein-2 stimulates Runx2 acetylation, J. Biol.
Chem., 281, 16502-16511.
12.Song, L. H., Pan, W., Yu, Y. H., Quarles, L. D.,
Zhou, H. H., and Xiao, Z. S. (2006) Resveratrol prevents CsA inhibition
of proliferation and osteoblastic differentiation of mouse bone
marrow-derived mesenchymal stem cells through an ER/NO/cGMP pathway,
Toxicol. In Vitro, 20, 915-922.
13.Schmittgen, T. D., and Livak, K. J. (2008)
Analyzing real-time PCR data by the comparative CT method, Nat.
Protoc., 3, 1101-1108.
14.Jakob, F., Genest, F., Baron, G., Stumpf, U.,
Rudert, M., and Seefried, L. (2015) Regulation of bone metabolism in
osteoporosis: novel drugs for osteoporosis in development,
Unfallchirurg, 118, 925-932.
15.Rachner, T. D., Khosla, S., and Hofbauer, L. C.
(2011) Osteoporosis: now and the future, Lancet, 377,
1276-1287.
16.Li, P., Wang, Y., Liu, X., Zhou, Z., Wang, J.,
Zhou, H., Zheng, L., and Yang, L. (2019) Atypical antipsychotics induce
human osteoblasts apoptosis via Wnt/β-catenin signaling, BMC
Pharmacol. Toxicol., 20, 10.
17.Dai, Z., Li, Y., Quarles, L. D., Song, T., Pan,
W., Zhou, H., and Xiao, Z. (2006) Resveratrol enhances proliferation
and osteoblastic differentiation in human mesenchymal stem cells via
ER-dependent ERK1/2 activation, Phytomedicine, 14,
806-814.
18.Zhao, M., Ko, S. Y., Garrett, I. R., Mundy, G.
R., Gutierrez, G. E., and Edwards, J. R. (2018) The polyphenol
resveratrol promotes skeletal growth in mice through a sirtuin
1–bone morphogenic protein 2 longevity axis, Br. J.
Pharmacol., 175, 4183-4192.
19.Maeno, T., Moriishi, T., Yoshida, C. A., Komori,
H., Kanatani, N., Izumi, S., Takaoka, K., and Komori, T. (2011) Early
onset of Runx2 expression caused craniosynostosis, ectopic bone
formation, and limb defects, Bone, 49, 673-682.
20.Rao, A., Luo, C., and Hogan, P. G. (1997)
Transcription factors of the NFAT family: regulation and function,
Annu. Rev. Immunol., 15, 707-747.
21.Flanagan, W. M., Corthesy, B., Bram, R. J., and
Crabtree, G. R. (1991) Nuclear association of a T-cell transcription
factor blocked by FK-506 and cyclosporine A, Nature, 352,
803-807.
22.Zayzafoon, M. (2006) Calcium/calmodulin signaling
controls osteoblast growth and differentiation, J. Cell.
Biochem., 97, 56-70.
23.Sun, L., Blair, H. C., Peng, Y., Zaidi, N.,
Adebanjo, O. A., Wu, X. B., Wu, X. Y., Iqbal, J., Epstein, S., and Abe,
E. (2005) Calcineurin regulates bone formation by the osteoblast,
Proc. Natl. Acad. Sci. USA, 102, 17130-17135.