DNA Import into Plant Mitochondria: Complex Approach for in organello and in vivo Studies
T. A. Tarasenko1#, V. I. Tarasenko1#, M. V. Koulintchenko1,a*, E. S. Klimenko1, and Yu. M. Konstantinov1
1Siberian Institute of Plant Physiology and Biochemistry, Siberian Branch of the Russian Academy of Sciences, 664033 Irkutsk, Russia# These authors contributed equally to this work.
* To whom correspondence should be addressed.
Received March 6, 2019; Revised April 2, 2019; Accepted April 10, 2019
Natural competence of mitochondria for DNA uptake has been known for the last 20 years. Until the present time, all studies of this process have been conducted exclusively in isolated mitochondria, as no system for investigation of the DNA transport into the mitochondria in intact cells has been available. The objective of this work was to improve and standardize the existing approaches for investigating DNA import into plant mitochondria in an in organello system. A method for detecting the import of fluorescently labeled DNA substrates has been developed. Based on the features of DNA import into the mitochondria, we suggested an efficient method for the evaluation of the DNA import efficiency by quantitative PCR. We also developed and characterized the in vivo system that allows to detect DNA transport from the cytoplasm to the mitochondrial matrix in Arabidopsis thaliana protoplasts. A combination of the proposed techniques for studying the DNA uptake by plant mitochondria might be useful for elucidating whether the properties of the mitochondrial DNA import established in the in organello system are preserved in vivo.
KEY WORDS: DNA import, plant mitochondria, fluorescent labeling, protoplasts transformation, Arabidopsis thalianaDOI: 10.1134/S0006297919070113
The majority of mitochondrial proteins are encoded by nuclear genes and imported into the mitochondria following their biosynthesis in the cytoplasm. Mitochondria have their own genetic system that provides these organelles with a set of proteins required for the formation of oxidative phosphorylation complexes and mitochondrial biogenesis. Compactly organized mammalian mitochondrial genome (16.5 kb) encodes 13 polypeptides of the electron transport chain, 22 tRNAs, and two rRNAs [1]. In comparison to the mammalian mitochondrial genomes, plant mitochondrial genomes are extremely large and can reach more than 1500 kb in size [2, 3]. Plant mtDNA contains 60 genes encoding three rRNAs, 15-20 tRNAs, and 30-35 known proteins. The functions of more than a half of sequences in the plant mitochondrial genome are unknown, and no homology has been observed between these sequences and sequences available in the databases [4].
Plant mitochondrial genomes demonstrate a surprisingly high frequency of the horizontal and intracellular gene transfer [5]. More than 5% nucleotide sequences in the plant mitochondrial genome are of the chloroplast, nuclear, or viral origin [4, 6]. It is very likely that the structure and dynamics of the plant mitochondrial genome are strongly affected by the natural competence of mitochondria, i.e., their ability to uptake DNA from the environment. The phenomenon of mitochondrial competence was first demonstrated in plants [7, 8], but later was discovered in mammals [9] and yeast [10]. Mitochondria can also import RNA (mostly, tRNA). The key player in this process is mitochondrial porin (VDAC); RNA import is also mediated by the components of the protein import machinery [11, 12].
Unlike the significance of the protein and tRNA import, the biological role and the molecular mechanisms of DNA import into the mitochondria are still poorly understood. According to the experimental data, VDAC is the main channel for the mitochondrial DNA import [7, 10, 13]. Recent studies identified other proteins that could be involved in this process [14], including precursor of the ATP synthase β-subunit. This protein is bound to the outer mitochondrial membrane (OMM), where it could interact with VDAC in the process of DNA binding. It was also shown that CuBPp, which is a complex I subunit acting as a receptor in the intermembrane space, participates in the import of long DNA fragments [14]. DNA transport through the inner mitochondrial membrane to the matrix is poorly studied and can occur in plants and mammals via different mechanisms. Thus, inhibitors of the inner membrane transporter protein adenine nucleotide translocator (ANT) block DNA import in plants, but not in mammalian mitochondria [9].
Previously, DNA import into the mitochondria has been investigated using radioactively labeled DNA substrates [7], which allowed visualization of the full-size molecules of imported DNA and assessment of the influence of various effectors on the rate of DNA import. However, radioactive labels have a number of disadvantages – the half-life of 32P is only 14 days; radioactivity is harmful for human health; experiments with radioactive labels require special, certified facilities.
Other methods used for studying DNA import into the mitochondria, such as transcription-mediated techniques [7] and homologous recombination in organello [15] have provided compelling evidence of DNA transport into the mitochondrial matrix. The import of genetic constructs into the mitochondria followed by analysis of their transcription by RT-PCR or homologous recombination in organello are the best approaches for investigating the processes associated with expression of exogenous DNA and its preservation in the content of mitochondrial genome. However, these methods are not suitable for the evaluation of the DNA import efficiency. To sum up, all the approaches used for investigating DNA import into the mitochondria have their limitations.
It is important to note that all studies of the DNA import into animal and plant mitochondria reported so far have been conducted in isolated mitochondria. It still remains unclear whether DNA import occurs in intact cells and whether the regularities of this process observed in organello are retained in vivo. The development of a system for investigating the mechanism of DNA transport from the cytoplasm to the mitochondrial matrix in cells, besides being important from the point of view of fundamental science, can be a starting point for the transformation of mitochondrial genomes of higher plants – an essential problem that has not been resolved so far.
Hence, there is a necessity to optimize and standardize conditions and approaches for investigating DNA import in organello and, most importantly, to develop a system for studying DNA transport into the mitochondria in vivo. In this work, we present a complex approach for studying DNA import into the mitochondria using two systems – in organello and in vivo.
MATERIALS AND METHODS
Plant material and growth conditions. Arabidopsis thaliana (L.) Heynh. ecotype Columbia (Col-0) plants were grown following stratification for 3 days at 4°C at 22°C in a KBW-720 growth chamber (Binder, Germany) in pots filled with a mixture of compost/peat moss/vermiculate (2 : 1 : 3) at the photosynthetic photon flux density (PPFD) of 150 μmol·m–2·s–1 and 16-h photoperiod. Etiolated corn seedlings (Zea mays, Kubanskii 250 MV variety) were grown at 29°C for 4 days in the dark. Potato tubers (Solanum tuberosum, Adretta variety) and Brassica rapa (Vnuchka variety) roots were used in the study. Corn seeds were obtained from NPO KOS-MAIS (Russia); potato plants were grown at the experimetal plots of the Siberian Institute of Plant Physiology and Biochemistry, Siberian Branch of the Russian Academy of Sciences; Brassica rapa seeds were purchased from commercial sources.
Synthesis of DNA substrates. Taq polymerase (Thermo Scientific, USA) was used for DNA amplification according to the manufacturer’s instructions. Oligonucleotides with the Cy3 fluorescent group at the 5′-end (Evrogen, Russia) were used for the preparation of fluorescent DNA substrates of 269 bp, 852 bp, and 2.7 kb (Table 1). DNA amplification conditions for the 269- and 852-bp fragments were: 94°C – 3 min (1 cycle); 94°C – 30 s, 58°C – 45 s, 72°C – 45 s (30 cycles); 72°C – 1 min. The 2.7-kb fragment was produced under the following amplification conditions: 94°C – 3 min (1 cycle); 94°C – 30 s, 60°C – 45 s, 72°C – 180 s (30 cycles); 72°C – 5 min. The amplified DNA products were purified using a GeneJETTM PCR Purification Kit (Thermo Scientific) and their quality was evaluated by electrophoresis using a Gel Doc XR System (Bio-Rad, USA). DNA concentration was determined with a NanoPhotometer NP80 spectrophotometer (IMPLEN, Germany). The pCK/GFP/PRmt genetic construct [7] containing the GFP gene was used as a template for amplification.
Table 1. Primers used for the preparation of
fluorescent DNA substrates

Isolation of mitochondria. Mitochondria were isolated from A. thaliana plants [16], etiolated Z. mays seedlings [17], S. tuberosum tubers, and B. rapa roots [18] according to the procedures described previously. Protein content in the mitochondrial suspensions was measured by the Bradford method [19]. Purified mitochondrial fraction was examined with a Leo 906E transmission electron microscope (Zeiss, Germany) [20].
DNA import into the isolated mitochondria. DNA import was performed in the import buffer containing 0.4 M sucrose and 40 mM potassium phosphate (pH 7.0) at 25°C at constant shaking (350 rpm) for 30 min in a TS-100 thermo-shaker for microtubes and PCR plates (BioSan, Latvia) [7]; the reaction volume was 100-200 μl. Mitochondria were treated with DNase I (500 μg/ml; Sigma, USA) in the import buffer containing 10 mM MgCl2 for 20 min at 25°C. Further washing was carried out as described in [7]. To prepare mitoplasts, the OMM was disrupted by osmotic shock. For this, the mitochondrial pellet was resuspended in 1 ml of 5 mM potassium phosphate buffer (pH 7.5) and incubated on ice for 5 min; the suspension was then pelleted and washed with 10 mM potassium phosphate buffer (pH 7.2) containing 0.3 M sucrose and 1 mM EDTA.
Evaluation of respiratory control index and intactness of mitochondria. The functional activity of isolated mitochondria (100 μg protein) was assayed polarographically using a platinum oxygen electrode and an Oxytherm system (Hansatech, UK). Mitochondrial respiratory control index was evaluated according to the protocol described in [21]. The intactness of the OMM was evaluated as described in [22].
Protoplast preparation and DNA transfection. Protoplasts were produced from the leaves of 35-day-old A. thaliana plants according to the protocol [23] with modifications. Upper epidermis layer was removed using an adhesive tape. Pieces of the adhesive tape with the attached lower epidermis and mesophyll cells were placed on the surface of the protoplast isolation medium (PM) (0.4 M mannitol, 10 mM CaCl2, 20 mM KCl, 20 mM MES, pH 5.7, 0.1% BSA) containing 1% cellulase and 0.25% pectolyase (MP Biomedicals, USA) and incubated with gentle shaking at 23°C under light for 2 h. An aliquot (15 ml) of the washing medium (WM) containing 154 mM NaCl, 125 mM CaCl2, 5 mM KCl, 5 mM glucose, and 2 mM MES (pH 5.7) was added to the protoplast suspension, and the suspension was centrifuged for 3 min at 100g in an Allegra 64R centrifuge (Beckman Coulter, USA) at 20°C. The protoplast precipitate was resuspended in 15 ml of WM, centrifuged again, and resuspended in the medium containing 0.4 M mannitol, 15 mM MgCl2, 4 mM MES (pH 5.7) (300 μl per sample). A solution of DNA substrate (5 μg) was added to the protoplast suspension followed by addition of 300 μl of 20% PEG 2000 solution (w/v) containing 0.2 M mannitol and 100 mM CaCl2, and the mixture was incubated for 5 min. The protoplast suspension was subjected to three centrifugation cycles in 1.5 ml of WM for 1 min at 100g at 20°C. The protoplasts were then resuspended in WM and incubated in a 48-well plate (200 μl per well) at 22°C with low-intensity illumination for 20 h.
Isolation of mitochondria from A. thaliana protoplasts. Protoplast integrity was evaluated by light microscopy. The protoplast suspension was centrifuged for 1 min at 100g at 20°C. An aliquot (400 μl) of the mitochondria isolation medium (IM) (0.4 M sucrose, 50 mM Tris-HCl, pH 7.5, 5 mM MgCl2, 0.2% BSA, 2 mM DTT) was added to the precipitate. DNase I was added to the IM immediately prior to the use (1 unit enzyme per 50 μl IM). The protoplasts were disrupted with a Potter homogenizer. The IM medium was added to each sample up to 1 ml and the sample was centrifuged for 5 min at 3000g and then for 7 min at 15,000g at 4°C in an Allegra 64R centrifuge (Beckman Coulter). The resulting mitochondrial pellet was resuspended in 100 μl of the DNase treatment medium (0.4 M sucrose, 40 mM KH2PO4, pH 7.0) containing 0.2% BSA, 10 mM MgCl2, and DNase I (1 unit enzyme per 50 μl medium) and incubated for 20 min at 25°C. Following the incubation, the mitochondria were subjected to two centrifugation rounds for 7 min at 15,000g at 4°C in the IM containing 0.2% BSA, 10 mM EDTA, and 10 mM EGTA.
DNA isolation. DNA was extracted from the mitochondrial pellet with equal volumes of phenol and 10 mM Tris-HCl (pH 7.5) containing 1 mM EDTA and 1% SDS (w/v) and then precipitated with ethanol in the presence of 200 mM NaCl. DNA was eluted from the gel with a GeneJET Gel Extraction Kit (Thermo Scientific). To evaluate the efficiency of the import of fluorescently labeled DNA fragments into the mitochondria, the samples were analyzed by electrophoresis in 1% agarose gel; the gels were scanned with an Ettan™ DIGE Imager (GE Healthcare, Sweden).
Quantitative PCR. qPCR was carried out with the primers presented in Table 2 using a SYBR Select Master Mix (Applied Biosystems, USA) in a CFX96 cycler (Bio-Rad) according to the following protocol: 50°C – 2 min, 95°C – 3 min (1 cycle); 95°C – 20 s, 60°C – 30 s, 72°C – 30 s (40 cycles). The data were analyzed with the CFX Manager software (Bio-Rad). All experiments were repeated at least three times.
Table 2. Primers used for the evaluation of
the amount of imported DNA substrates

RESULTS
DNA import into plant mitochondria in the in organello system. Here, we have developed an alternative method to analyze DNA import into the isolated plant mitochondria using fluorescently labeled DNA. We suggested that introduction of a non-radioactive fluorescent label would allow to overcome the disadvantages of radioactive labeling while preserving the main advantages of labeled DNA substrates, such as the possibility of testing the full-size imported DNA molecules after their extraction from the mitochondria, as well as a high sensitivity and specificity of their detection.
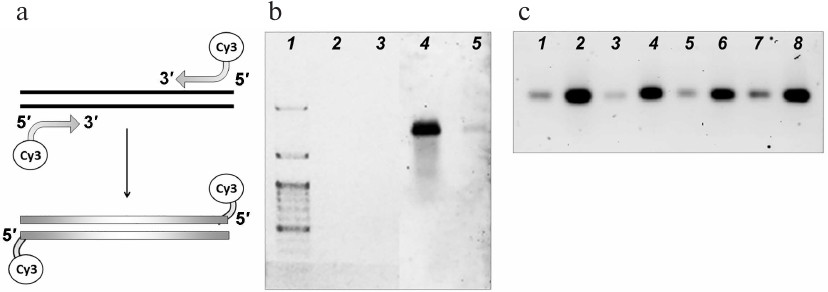
Fluorescent labels were incorporated into the DNA substrates by amplification with the primers carrying cyanine 3 (Cy3) fluorophore at the 5′-ends (Fig. 1a). Using this approach, we generated fluorescently labeled DNA substrates (Fig. 1b) and tested their stability after different storage times. The samples did not lose their fluorescence at least within one month of storage; the amount of the detected DNA (no less than 0.5 ng) remained the same (Fig. 1c).
Fig. 1. Fluorescent labeling of the DNA substrates for the import into the mitochondria. a) Introduction of the Cy3 fluorophore into DNA molecules. b) Visualization of the fluorescently labeled 852-bp DNA fragment: 2, 4) 5 ng; 3, 5) 0.5 ng. The fragment was visualized by ethidium bromide staining (1-3) or with a fluorescence scanner (4, 5). Lane 1, DNA size ladder. c) Visualization of samples of fluorescently labeled 852-bp DNA fragment after storage for 4 (1, 2), 3 (3, 4), 2 (5, 6), and 1 week (7, 8). The amounts of the samples loaded on the gels were 1 ng (1, 3, 5, 7) and 5 ng (2, 4, 6, 8).
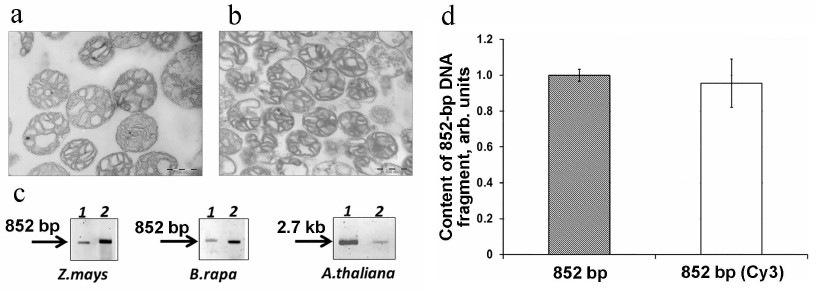
We analyzed the import of fluorescently labeled DNA substrates of various length into the mitochondria isolated from different plants (Z. mays, B. rapa, and A. thaliana) (Fig. 2). The mitochondria isolated from all three tested plants had a high degree of coupling between oxidation and phosphorylation (respiratory control for succinate oxidation was ~4), and their intactness was 85-90%. According to the electron microscopy, the isolated mitochondria were highly purified (Fig. 2, a and b).
Fig. 2. Import of Cy3-labeled DNA into plant mitochondria. Electron microscopy images of B. rapa (a) and Z. mays (b) mitochondria; scale bar, 500 nm. c) Fluorescently labeled 852-bp and 2.7-kb DNA fragments (0.5 μg) were incubated with the isolated mitochondria from Z. mays, B. rapa, and A. thaliana. Following the incubation, mtDNA was extracted and analyzed by electrophoresis in agarose gel. Lanes: 1) fluorescently labeled DNA substrate prior to the import, 3 ng; 2) the same fragment extracted from the mitochondria after its import. d) Effect of the presence of Cy3 at 5′-end of the 852-bp DNA fragment on the efficiency of the DNA fragment import into the mitochondria.
Analysis of fluorescent DNA fragments in the mtDNA samples isolated from all three tested plants showed the presence of the full-size DNA substrate molecules in the mitochondrial matrix (Fig. 2c). Hence, we demonstrated that fluorescently labeled DNA can be used in the experiments on the efficiency of mitochondrial import in organello.
However, the sensitivity of detection of fluorescently labeled DNA substrates after their import into the mitochondria was insufficient to solve a number of essential problems such as, for example, evaluation of DNA import into the mitochondria in vivo. At the same time, assessment of DNA import by quantitative PCR (qPCR) does not have this limitation, as it allows detection of significantly lower amounts of imported DNA. On the other hand, the fact that only small DNA fragments rather than the full-size DNA molecules are amplified in qPCR represents an obvious disadvantage of this method.
The evaluation of the import of non-labeled DNA substrates in organello by qPCR using TaqMan probes was for the first time performed by Klimenko et al. [24]. Here, we developed a standardized system for the qPCR analysis of DNA import using relatively fast and easy-to-use SYBR Green intercalating dye. To quantify the amount of imported DNA, we amplified a 185-bp fragment of the imported substrate. In parallel, each sample was analyzed for the amount of endogenous mitochondrial gene nad4, which was used as a reference gene.
We also confirmed that the presence of fluorophore at the 5′-end of the DNA substrates did not inhibit their transfer into the mitochondrial matrix: according to the qPCR results, both Cy3-labeled and non-labeled 852-bp DNA fragments were transported into A. thaliana mitochondria with an equal efficiency (Fig. 2d).
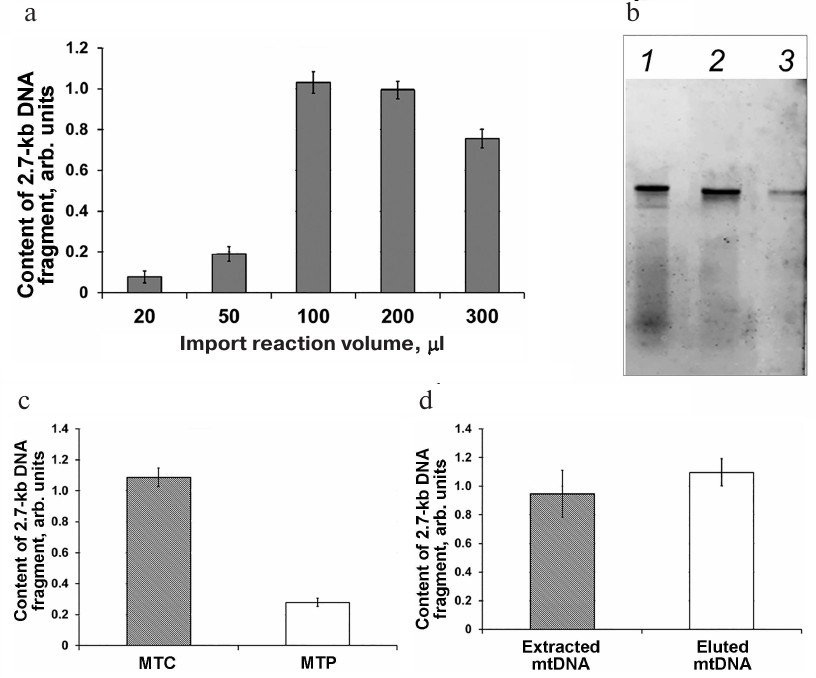
It should be emphasized that according to the accumulated experimental data, the validity of the results produced by qPCR depends on the conditions of DNA import into the mitochondria. The methods used for the DNA import differ significantly in the presence or absence of the mitoplast obtaining stage, reaction mixture volume, mtDNA extraction protocol, etc. [7, 9, 14, 24]. In this work, we determined the optimal volume of the import reaction mixture for the DNA uptake by the mitochondria. We analyzed the efficiency of DNA import in a series of samples differing in the reaction volume but containing the same amounts of mitochondrial protein (200 µg) and added DNA substrate (500 ng) (Fig. 3a). The efficiency of DNA import was low for the samples with the reaction volumes of 20 and 50 µl. It is likely that mitochondria aggregate in small volumes, which could cause the blockage of channels required for the macromolecule transfer. The reaction volumes of 100-300 μl was found to be optimal for the DNA transport into the mitochondria (Fig. 3a).
Fig. 3. Analysis of parameters affecting the import of non-labeled and fluorescently labeled 2.7-kb DNA fragments into the mitochondria isolated from S. tuberosum (a), A. thaliana (b, c), and B. rapa (d). a) Relative amounts of the 2.7-kb DNA fragment normalized to the content of the nad4 gene as determined by analysis of samples after DNA import into isolated mitochondria using different reaction volumes. b, c) The content of the imported fluorescently labeled DNA fragment after its extraction from the mitochondria (lane 1, MTC) or mitoplasts (lane 2, MTP); fluorescently labeled DNA substrate prior to the import, 1 ng (3). d) qPCR analysis of DNA fragments extracted after their import into the isolated mitochondria (extracted mtDNA) and eluted following separation in the agarose gel (eluted mtDNA).
Electrophoretic fractionation of fluorescently labeled DNA fragments extracted after their import into the mitochondria revealed that besides the full-size 852-bp fragment, the samples contained low-molecular-weight fragments (Fig. 3b, lane 1) that most probably emerged due to the degradation of non-imported DNA molecules. It is possible that some of these molecules were partially degraded by DNase I and retained bound to the OMM or in the intermembrane space after washing and then were extracted together with the mtDNA. The presence of these low-molecular-weight fragments could skew the results of the DNA import quantification. Removal of the OMM by osmotic shock (mitoplast preparation stage) after DNA import significantly reduced the content of the low-molecular-weight fragments (Fig. 3b, lane 2). At the same time, this procedure resulted in a significant decrease in the amount of DNA imported into the mitochondria as determined by qPCR (Fig. 3c). In order to find the contribution of the low-molecular-weight fraction to the results of quantification of the imported DNA, we examined by electrophoresis the DNA samples extracted after the import into the mitochondria and mitoplast preparation. mtDNA eluted from the agarose gel and a portion of the gel corresponding to the full-size imported DNA fragment were analyzed by qPCR (Fig. 3d). It was found that the efficiency of DNA import was virtually the same in both cases (i.e., when DNA extracted from the mitochondria directly after the DNA import or when DNA eluted from the gel and containing the full-size imported DNA and mtDNA were used as a template) (Fig. 3d).
These results demonstrated that qPCR analysis of the DNA isolated from the mitoplasts provides the same results as analysis of the sample containing the full-size fragment of the imported DNA, even if it was based on detection of only small amplified DNA fragment.
DNA import into the plant mitochondria in the in vivo system. Until recently, investigation of DNA import in animal and plant cells has been limited to the studies in isolated mitochondria. We were the first to develop a method for monitoring DNA transport from the cytoplasm to the mitochondria in A. thaliana protoplasts. This method includes (i) transformation of protoplasts with the DNA substrate; (ii) incubation of cells under optimal physiological conditions (temperature, illuminance) for 20 h; (iii) disruption of protoplasts and isolation of mitochondria by the micromethod; (iv) DNase treatment of isolated mitochondria in order to eliminate possible contamination by DNA molecules bound to the OMM; (v) lysis of the mitochondria and isolation of mtDNA; and (vi) qPCR evaluation of the amount of DNA imported to the mitochondria. The use of fluorescently labeled DNA substrates produced with Cy3-conjugated primers did not allow to achieve the level of DNA incorporation sufficient for detecting fluorescence in the samples of mitochondrial DNA isolated from the transformed protoplasts (data not shown). Hence, DNA quantification by qPCR was selected because of its higher sensitivity.
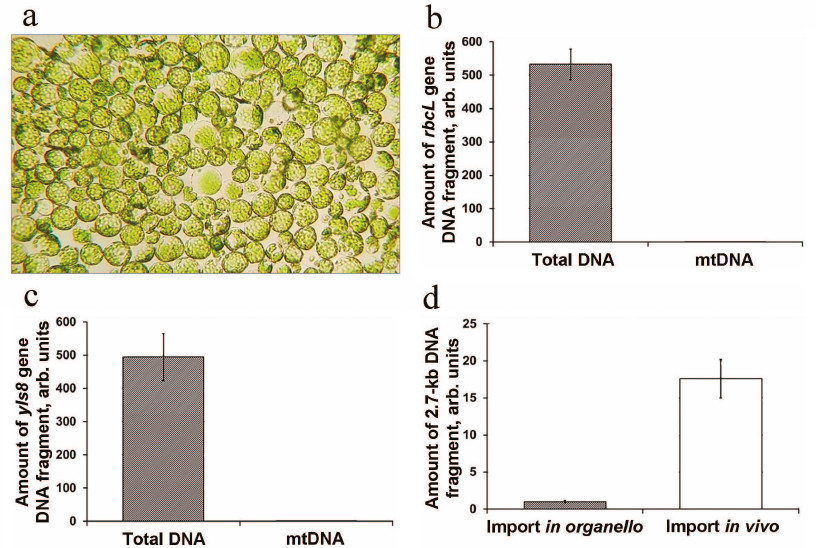
Protoplasts from A. thaliana leaves were prepared using the protocol developed for the experiments on the transient expression of genetic constructs in the cytoplasm [23]. We demonstrated that the produced protoplasts were free from contamination and maintained their integrity for at least 20 h after isolation and transfection with DNA (Fig. 4a).
Fig. 4. DNA import into the mitochondria from A. thaliana protoplasts. a) Protoplasts after transformation with the DNA fragment and incubation for 20 h; b) analysis of mitochondria isolated from the protoplasts for contamination with chloroplasts; c) analysis of mitochondria isolated from the protoplasts for contamination with nuclear DNA. The amounts of the rbcL (chloroplast DNA) and yls8 (nuclear DNA) gene fragments in the DNA samples obtained from the protoplasts and isolated mitochondria are normalized to the amount of the nad4 gene (mtDNA); d) analysis of DNA import in isolated organelles (in organello) and A. thaliana protoplasts (in vivo) .
Mitochondria were isolated using the method developed for the preparation of mitochondria from the protoplasts of A. thaliana suspension cell culture [25] that we adapted for small samples. In order to eliminate the possibility of exogenous DNA uptake by the mitochondria during isolation, MgCl2 and DNase I were added to the isolation medium. We evaluated the contamination of the DNA extracted from the isolated mitochondria with DNA sequences of a non-mitochondrial origin. For this purpose, the contents of the chloroplast gene rbcL and nuclear gene yls8 were estimated by qPCR analysis. We found that the content of both chloroplast and nuclear DNA in the obtained mitochondrial sample was reduced approximately 500-fold in comparison with its content in the total DNA sample from the protoplasts (Fig. 4, b and c). Hence, the mitochondria isolated using this method were sufficiently free from contamination.
In preliminary experiments, the protoplasts were transformed with the 2.7-kb DNA substrate. Quantification of the imported DNA fragment by qPCR following 20-h incubation of the protoplasts, isolation of mitochondria, and mtDNA extraction showed a sufficiently high content of the imported DNA fragment. In parallel, the content of the same DNA substrate imported into the isolated A. thaliana mitochondria was analyzed. It was demonstrated that the content of the imported DNA substrate was significantly higher in vivo in comparison with the in organello import (Fig. 4d).
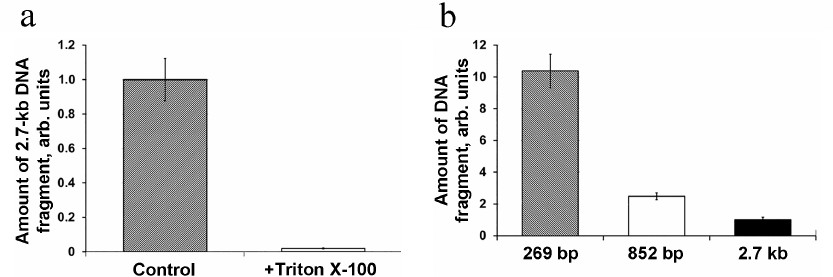
In order to confirm the intramitochondrial localization of the detected DNA, the mitochondria isolated from A. thaliana protoplasts were treated with the non-ionic detergent Triton X-100 that disrupts the mitochondrial membranes. Prior to the DNase I treatment, the isolated mitochondria were incubated in the medium containing 0.5% Triton X-100 (v/v) for 10 min, which resulted in virtually complete absence of DNA import (Fig. 5a). This implied that the detected fragment was located in the mitochondria matrix rather than associated with the OMM.
Fig. 5. a) Import of 2.7-kb DNA into the mitochondria in Arabidopsis protoplasts followed by the treatment of isolated mitochondria with Triton X-100. b) Import of DNA fragments of different size in Arabidopsis protoplasts; the content of gene fragments was normalized to the content of the nad4 gene.
Next, the efficiency of DNA import was evaluated for the DNA substrates of varying sizes – 269 bp, 852 bp, and 2.7 kb. Equal DNA amounts (5 μg) were used for the protoplast transfection with each of the three DNA substrates. It can be seen in Fig. 5b that the amount of DNA molecules imported into mitochondria in vivo decreases with the increase of the imported molecule size. Similar dependence was observed previously for the import of DNA fragments of different sizes into isolated mitochondria [7], which supports the suggestion that the observed regularities of the DNA import in organello reflect the processes occurring in vivo. Hence, we have shown for the first time that DNA can be actively transported from the plant cell cytoplasm into the mitochondria.
DISCUSSION
Taking into consideration the properties of the plant mitochondrial genome structural organization such as (i) large size in comparison with the mitochondrial genomes of other eukaryotic organisms, (ii) large number of sequences of foreign origin, (iii) presence of autonomous replicons, and (iv) capacity for recombination, it seems very likely that plant mitochondria should display a high propensity for uptake and integration of foreign sequences into their genome. Indeed, we have shown the existence of DNA import into the mitochondria of higher plants using both in organello and in vivo approaches. At present, it is difficult to establish the significance of this process for the functioning of mitochondria in the cells. One cannot rule out that this phenomenon is inherited from the bacterial ancestors of mitochondria, since the ability of bacteria to uptake foreign genetic material is well known and plays a vital role in their evolution [26, 27]. At the same time, participation of mitochondrial channels in the uptake of viral DNA associated with the initiation of mitochondria-mediated cell defense mechanisms is also possible.
The pathways of DNA transport into the nuclei of plant and animal cells are well known. Viral DNA can be transported across the nuclear membrane by the mechanism involving the import of nucleus-targeted cell proteins [28]. In dicotyledons, the transport of plasmid DNA into the nucleus during plant infection with agrobacteria (agroinfection) is mediated by two bacterial proteins, VirD2 and VirE2. Furthermore, the possibility of non-specific import of DNA molecules up to several kb in size into the nuclei of mammalian cells was demonstrated using microinjection of DNA fragments into the cytoplasm [29] and cell permeabilization with digitonin [30]. During the course of evolution, numerous eubacterial genes have migrated into nucleus from the cell organelles via intracellular gene transfer [31]. Thus, almost entire copy of the chloroplast genome is present in the plant cell nucleus [32, 33], as well as copies of many mitochondrial genes encoding polypeptides [34] and tRNAs [35]. Nuclear mobile genetic elements have been acquired mostly by the horizontal gene transfer [36]. Interestingly enough, the events of genetic material transfer into the chloroplast genome are extremely rare [37]; the capacity for active DNA uptake is untypical for the chloroplasts [38]. Hence, there exists a clear correlation between the ability of cell organelles to uptake DNA and the direction of the intracellular gene transfer in the process of evolution. We suggest that the natural competence of mitochondria for the DNA uptake could be one of the mechanisms facilitating migration of the genetic material within the cell. In this work, we present modern approaches for investigating the phenomenon of DNA import into the plant mitochondria.
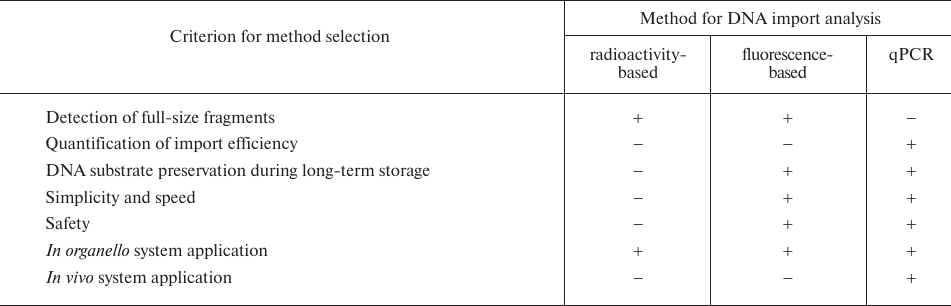
Previously, the studies on the DNA import into mitochondria have used mostly radioactively labeled substrates [7, 9, 10, 14]. Since the use of radioactively-labeled DNA has several significant drawbacks (Table 3), new alternative approaches for detection and analysis of the imported DNA are required.
Table 3. Advantages and drawbacks of the
methods for the DNA import analysis
Here, we developed a method for detection of the DNA imported into the plant mitochondria using fluorescent labeling. Previously, the only study of DNA import that used fluorescently labeled DNA substrates was performed in isolated human mitochondria [39]. DNA imported into the mitochondria was visualized by confocal microscopy. It was shown that the labeled DNA was preserved following the treatment of the mitochondria with DNase I, thereby establishing its localization to the mitochondrial matrix. However, this method did not include removal of the OMM, i.e., failed to guarantee elimination of DNA molecules associated with the OMM. It also did not allow to distinguish between fluorescence of the full-size DNA fragments transported into the mitochondria from the fluorescence of their degradation products.
We investigated the import of fluorescently labeled DNA fragments of different sizes into the mitochondria isolated from three plant species (A. thaliana, B. rapa, and Z. mays). The presence of the fluorophore did not interfere with the transport of the full-size DNA molecules into the mitochondrial matrix. However, this method did not allow quantification of the imported DNA, which is required in the experiments on the mechanisms of DNA transport and its kinetic characteristics. These limitations can be overcome by employing qPCR analysis. We optimized our approach by selecting conditions for the DNA import in the in organello system and DNA extraction for qPCR analysis. The reaction volume of 100-300 μl was shown to be optimal for the DNA import. The use of DNA eluted after the import as a template for qPCR confirmed that amplification of small (typical for qPCR) amplicons in the context of imported DNA substrates produced reliable results. Removal of the OMM ensured elimination of the bound DNA molecules that have not penetrated into the matrix.
The optimized in organello approach allows the use of intact mitochondria for investigating the mechanisms of DNA import and the role of mitochondrial membrane transport proteins in it; however, this system does not take into consideration numerous cellular factors that could affect the DNA transport.
The natural competence of animal and plant mitochondria for DNA uptake has not been investigated in vivo. At the same time, the search for the methods for the transformation of mammalian mitochondrial genome has been continuing for many years. Numerous attempts have been made to transform mitochondria using DNA conjugated with mitochondria-targeting peptides or recombinant mitochondrial TFAM protein, bacterial conjugation, delivery with the help of lipophilic cationic compounds, adenovirus-mediated gene transfer, and other approaches [40, 41]. None of these methods resulted in stable transformation. No successful transformation of mitochondria from higher plants has been reported [12, 40]. Meanwhile, such studies are of considerable interest, as they will make it possible, in particular, to generate transgenic plants with maternally inherited foreign genetic material.
Protoplast transformation is widely used for the introduction of transiently expressed constructs into the cells in the studies of intracellular protein targeting [42] and promoter activity of regulatory sequences [43]. Electroporation-mediated protoplast transformation was used in the studies of the in vivo import of tRNA into S. tuberosum mitochondria [44]. However, protoplast transformation has not been used previously for studying the mechanisms of DNA import.
We developed an approach for investigating the DNA import in vivo based on PEG-mediated transformation of A. thaliana protoplasts [23]. Using this method, the 270-2700-bp DNA fragments transfected into the protoplasts were imported from the cytoplasm into the mitochondria with a sufficiently high efficiency. The developed approach might be useful in elucidating whether the regularities of DNA import demonstrated in isolated mitochondria (dependence of the import efficiency on the size and structural features of the DNA molecule, effect of inactivation of certain mitochondrial membrane proteins on the import efficiency) are preserved at the cell level. In addition to studying the mechanisms of DNA transport in vivo, this approach could be used for developing the system for the transformation of plant mitochondrial genome, including protoplast transfection with constructs containing sequences facilitating DNA incorporation into the mitochondrial genome [15], transcription of target sequences [7], cell wall regeneration, selection of transformed cells, and generation of whole plants from them.
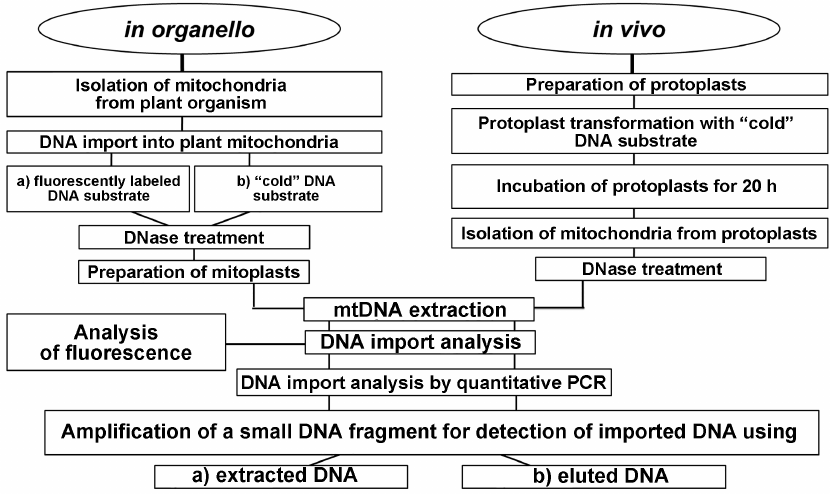
In conclusion, we developed a complex approach for investigating the DNA import into plant mitochondria that included analysis both in organello and in vivo (Fig. 6). The application of this approach will make it possible to elucidate whether the regularities of DNA import established in organello are preserved in vivo.
Fig. 6. Schematic representation of the complex approach based on the use of in organello and in vivo systems for investigating DNA import into plant mitochondria.
Funding. This work was financially supported by the Russian Foundation for Basic Research (project 18-04-00603).
Acknowledgements. The authors are grateful to E. V. Klimenkov for performing microscopic examination and N. E. Korotaeva for help in performing fluorescence analysis. Equipment of the Bioanalitika Center for Collective Use, Siberian Institute of Plant Physiology and Biochemistry, Siberian Branch of the Russian Academy of Sciences, Irkutsk, was used in this study.
Conflict of interest. The authors declare no conflict of interest in financial or any other sphere.
Ethical approval. This article does not contain any studies with human participants or animals performed by any of the authors.
REFERENCES
1.Saccone, C., De Giorgi, C, Gissi, C., Pesole, G.,
and Reyes, A. (1999) Evolutionary genomics in Metazoa: the
mitochondrial DNA as a model system, Gene, 238, 195-209;
doi: 10.1016/S0378-1119(99)00270-X.
2.Alverson, A. J., Rice, D. W., Dickinson, S., Barry,
K., and Palmer, J. D. (2011) Origins and recombination of the
bacterial-sized multichromosomal mitochondrial genome of cucumber,
Plant Cell, 23, 2499-2513; doi:
10.1105/tpc.111.087189.
3.Kubo, T., and Newton, K. J. (2008) Angiosperm
mitochondrial genomes and mutations, Mitochondrion, 8,
5-14; doi: 10.1016/j.mito.2007.10.006.
4.Unseld, M., Marienfeld, J. R., Brandt, P., and
Brennicke, A. (1997) The mitochondrial genome of Arabidopsis
thaliana contains 57 genes in 366,924 nucleotides, Nat.
Genet., 15, 57-61; doi: 10.1038/ng0197-57.
5.Sanchez-Puerta, M. V. (2014) Involvement of
plastid, mitochondrial and nuclear genomes in plant-to-plant horizontal
gene transfer, Acta Soc. Bot. Pol., 83, 317-323; doi:
10.5586/asbp.2014.041.
6.Kubo, T., Nishizawa, S., Sugawara, A., Itchoda, N.,
Estiati, A., and Mikami, T. (2000) The complete nucleotide sequence of
the mitochondrial genome of sugar beet (Beta vulgaris L.)
reveals a novel gene for tRNAcys(GCA), Nucleic Acids Res.,
28, 2571-2576; doi: 10.1093/nar/28.13.2571.
7.Koulintchenko, M., Konstantinov, Y., and Dietrich,
A. (2003) Plant mitochondria actively import DNA via the permeability
transition pore complex, EMBO J., 22, 1245-1254; doi:
10.1093/emboj/cdg128.
8.Konstantinov, Y. M., Dietrich, A., Weber-Lotfi, F.,
Ibrahim, N., Klimenko, E. S., Tarasenko, V. I., Bolotova, T. A., and
Koulintchenko, M. V. (2016) DNA import into mitochondria,
Biochemistry (Moscow), 81, 1044-1056; doi:
10.1134/S0006297916100035.
9.Koulintchenko, M., Temperley, R. J., Mason, P. A.,
Dietrich, A., and Lightowlers, R. N. (2006) Natural competence of
mammalian mitochondria allows the molecular investigation of
mitochondrial gene expression, Hum. Mol. Genet., 15,
143-154; doi: 10.1093/hmg/ddi435.
10.Weber-Lotfi, F., Ibrahim, N., Boesch P., Cosset,
A., Konstantinov, Y., Lightowlers, R. N., and Dietrich, A. (2009)
Developing a genetic approach to investigate the mechanism of
mitochondrial competence for DNA import, Biochim. Biophys. Acta,
1787, 320-327; doi: 10.1016/j.bbabio.2008.11.001.
11.Campo, M. L., Peixoto, P. M., and
Martinez-Caballero, S. (2017) Revisiting trends on mitochondrial
mega-channels for the import of proteins and nucleic acids, J.
Bioenerg. Biomembr., 49, 75-99; doi:
10.1007/s10863-016-9662-z.
12.Verechshagina, N. A., Konstantinov, Y. M.,
Kamenski, P. A., and Mazunin, I. O. (2018) Import of proteins and
nucleic acids into mitochondria, Biochemistry (Moscow),
83, 643-661; doi: 10.1134/S0006297918060032.
13.Delage, L., Duchene, A. M., Zaepfel, M., and
Marechal-Drouard, L. (2003) The anticodon and the D-domain sequences
are essential determinants for plant cytosolic tRNAVal
import into mitochondria, Plant J., 34, 623-633; doi:
10.1046/j.1365-313X.2003.01752.x.
14.Weber-Lotfi, F., Koulintchenko, M. V., Ibrahim,
N., Hammann, P., Mileshina, D. V., Konstantinov, Y. M., and Dietrich,
A. (2015) Nucleic acid import into mitochondria: new insights into the
translocation pathways, Biochim. Biophys. Acta, 1853,
3165-3181; doi: 10.1016/j.bbamcr.2015.09.011.
15.Mileshina, D., Koulintchenko, M., Konstantinov,
Yu., and Dietrich, A. (2011) Transfection of plant mitochondria and
in organello gene integration, Nucleic Acids Res.,
39, e115; doi: 10.1093/nar/gkr517.
16.Sweetlove, L. J., Taylor, N. L., and Leaver, C.
J. (2007) Isolation of intact, functional mitochondria from the model
plant Arabidopsis thaliana, Methods Mol. Biol.,
372, 125-136; doi: 10.1007/978-1-59745-365-3_9.
17.Newton, K. J., and Walbot, V. (1985) Maize
mitochondria synthesize organ-specific polypeptides, PNAS,
82, 6879-6883; doi: 10.1073/pnas.82.20.6879.
18.Neuburger, M., Journet, E. P., Bligny, R., Carde
J.-P., and Douce, R. (1982) Purification of plant mitochondria by
isopycnic centrifugation in density gradients of Percoll, Arch.
Biochem. Biophys., 217, 312-323; doi:
10.1016/0003-9861(82)90507-0.
19.Bradford, M. M. (1976) A rapid and sensitive
method for the quantitation of microgram quantities of protein
utilizing the principle of protein–dye binding, Anal.
Biochem., 72, 248-254; doi:
10.1016/0003-2697(76)90527-3.
20.Layton, B. E., Sastry, A. M., Lastoskie, C. M.,
Philbert, M. A., Miller, T. J., Sullivan, K. A., Feldman, E. L., and
Wang, C.-W. (2004) In situ imaging of mitochondrial outer
membrane pores using atomic force microscopy, Biotechniques,
37, 564-573; doi: 10.2144/04374BI01.
21.Douce, R., and Neuburger, M. (1989) The
uniqueness of plant mitochondria, Annu. Rev. Plant Physiol. Plant
Mol. Biol., 40, 371-414; doi:
10.1146/annurev.pp.40.060189.002103.
22.Grabel’nykh, O. I., Kirichenko, K. A.,
Pobezhimova, T. P., Borovik, O. A., Pavlovskaya, N. S., Lyubushkina, I.
V., Koroleva, N. A., and Voinikov, V. K. (2014) Effect of cold shock on
fatty acid composition and functional state of mitochondria in
stratified and non-stratified seedlings of winter wheat, Biol.
Membr. (Moscow), 31, 204-217; doi:
10.7868/S0233475514020029.
23.Wu, F.-H., Shen, S.-C., Lee, L.-Y., Chan, M.-T.,
and Lin, C.-S. (2009) Tape–Arabidopsis sandwich – a
simpler Arabidopsis protoplast isolation method, Plant
Methods, 5, 1-10; doi: 10.1186/1746-4811-5-16.
24.Klimenko, E. S., Mileiko, V. A., Morozkin, E. S.,
Laktionov, P. P., and Konstantinov, Yu. M. (2011) Study of DNA import
and export in potato (Solanum tuberosum) mitochondria using
quantitative PCR, Biochemistry (Moscow), Suppl. Ser. A,
5, 170-176; doi: 10.1134/S1990747811030044.
25.Meyer, E. H., and Millar, A. H. (2008) Isolation
of mitochondria from plant cell culture, Methods Mol. Biol.,
425, 163-169; doi: 10.1007/978-1-60327-210-0_15.
26.Doolittle, W. F., Boucher, Y., Nesbo, C. L.,
Douady, C. J., Andersson, J. O., and Roger, A. J. (2003) How big is the
iceberg of which organellar genes in nuclear genomes are but the tip?
Philos. Trans. R. Soc. Lond. B Biol. Sci., 358, 39-57;
doi: 10.1098/rstb.2002.1185.
27.Nakamura, Y., Itoh, T., Matsuda, H., and
Gojobori, T. (2004) Biased biological functions of horizontally
transferred genes in prokaryotic genomes, Nat. Genet.,
36, 760-766; doi: 10.1038/ng1381.
28.Bai, H., Schiralli Lester, G. M., Petishnok, L.
C., and Dean, D. A. (2017) Cytoplasmic transport and nuclear import of
plasmid DNA, Biosci. Rep., 37, BSR20160616; doi:
10.1042/BSR20160616.
29.Dowty, M. E., Williams, P., Zhang, G., Hagstrom,
J. E., and Wolff, J. A. (1995) Plasmid DNA entry into postmitotic
nuclei of primary rat myotubes, Proc. Natl. Acad. Sci. USA,
92, 4572-4576; doi: 10.1073/pnas.92.10.4572.
30.Hagstrom, J. E., Ludtke, J. J., Bassik, M. C.,
Sebestyen, M. G., Adam, S. A., and Wolff, J. A. (1997) Nuclear import
of DNA in digitonin-permeabilized cells, J. Cell Sci.,
110, 2323-2331.
31.Johnston, I. G., and Williams, B. P. (2016)
Evolutionary inference across eukaryotes identifies specific pressures
favoring mitochondrial gene retention, Cell Syst., 2,
101-111; doi: 10.1016/j.cels.2016.01.013.
32.Pichersky, E., Logsdon, J. M., Jr., McGrath, J.
M., and Stasys, R. A. (1991) Fragments of plastid DNA in the nuclear
genome of tomato: prevalence, chromosomal location, and possible
mechanism of integration, Mol. Gen. Genet., 225, 453-458;
doi: 10.1007/BF00261687.
33.Ayliffe, M. A., and Timmis, J. N. (1992) Plastid
DNA sequence homologies in the tobacco nuclear genome, Mol. Gen.
Genet., 236, 105-112; doi: 10.1007/BF00279648.
34.Mower, J. P., Jain, K., and Hepburn, N. J. (2012)
The role of horizontal transfer in shaping the plant mitochondrial
genome, Adv. Botan. Res., 63, 41-69; doi:
10.1016/B978-0-12-394279-1.00003-X.
35.Marechal-Drouard, L., Small, I., Weil, J. H., and
Dietrich, A. (1995) Transfer RNA import into plant mitochondria,
Methods Enzymol., 260, 310-327; doi:
10.1016/0076-6879(95)60148-1.
36.Diao, X. M., Freeling, M., and Lisch, D. (2006)
Horizontal transfer of a plant transposon, Plos Biol., 4,
e5; doi: 10.1371/journal.pbio.0040005.
37.Rice, D. W., and Palmer, J. D. (2006) An
exceptional horizontal gene transfer in plastids: gene replacement by a
distant bacterial paralog and evidence that haptophyte and cryptophyte
plastids are sisters, BMC Biol., 4, 31; doi:
10.1186/1741-7007-4-31.
38.Richardson, A. O., and Palmer, J. D. (2007)
Horizontal gene transfer in plants, J. Exp. Bot., 58,
1-9; doi: 10.1093/jxb/erl148.
39.Jackson, C. B., Zbindena, C., Gallati, S., and
Schaller, A. (2014) Heterologous expression from the human D-loop in
organello, Mitochondrion, 17, 67-75; doi:
10.1016/j.mito.2014.05.011.
40.Remacle, C., Larosa, V., Salinas, T., Hamel, P.,
Subrahmanian, N., Bonnefoy, N., and Kempken, F. (2012) in Genomics
of Chloroplasts and Mitochondria, Advances in Photosynthesis and
Respiration (Bock, R., and Knoop, V., eds.) Springer Science,
Belgium; doi: 10.1007/978-94-007-2920-9.
41.Niazi, A. K., Mileshina, D., Cosset, A., Val, R.,
Weber-Lotfi, F., and Dietrich, A. (2013) Targeting nucleic acids into
mitochondria: progress and prospects, Mitochondrion, 13,
548-558; doi: 10.1016/j.mito.2012.05.004.
42.Bhushan, S., Pavlov, P. F., Rudhe, C., and
Glaser, E. (2007) In vitro and in vivo methods to study
protein import into plant mitochondria, Methods Mol. Biol.,
390, 131-150; doi: 10.1007/978-1-59745-466-7_9.
43.Wehner, N., Hartmann, L., Ehlert, A., Bottner,
S., Onate-Sanchez, L., and Droge-Laser, W. (2011) High-throughput
protoplast transactivation (PTA) system for the analysis of
Arabidopsis transcription factor function, Plant J.,
68, 560-569; doi: 10.1111/j.1365-313X.2011.04704.x.
44.Wintz, H., and Dietrich, A. (1996)
Electroporation of small RNAs into plant protoplasts: mitochondrial
uptake of transfer RNAs, Biochem. Biophys. Res. Commun.,
223, 204-210; doi: 10.1006/bbrc.1996.0870.