Chaperone and Immunoglobulin-Binding Activities of Skp Protein from Yersinia pseudotuberculosis
E. V. Sidorin1,a*, V. A. Khomenko1, N. Yu. Kim1, and T. F. Solov’eva1
1Elyakov Pacific Institute of Bioorganic Chemistry, Far-Eastern Branch of the Russian Academy of Sciences, 690022 Vladivostok, Russia* To whom correspondence should be addressed.
Received May 20, 2019; Revised August 20, 2019; Accepted September 10, 2019
Here, we determined qualitative and quantitative characteristics of the chaperone and immunoglobulin-binding activities of recombinant Skp protein (rSkp) from Yersinia pseudotuberculosis using the methods of dynamic light scattering and surface plasmon resonance. Commercial human polyclonal IgG and Fc and Fab fragments of human IgG were used as substrate proteins. The activity of rSkp strongly depended on the medium pH. The most stable low-molecular-weight complexes with a hydrodynamic radius up to 10 nm were formed by rSkp and protein substrates at acidic pH values. Under these conditions, rSkp exhibited the lowest propensity to self-association and the highest affinity for human IgG and its Fc and Fab fragments, as well as prevented their aggregation most efficiently (i.e., demonstrated the maximal chaperone activity). As the medium pH increased, the affinity of rSkp for IgG and its fragments decreased; rSkp was not able to completely prevent the aggregation of protein substrates, but significantly slowed it down. The obtained information may be of practical interest, since the stability of therapeutic IgG preparations affects their safety and efficacy in medical applications.
KEY WORDS: Skp chaperone, Yersinia pseudotuberculosis, human immunoglobulin G, Fc and Fab fragments of IgG, protein–protein interactions, dynamic light scattering, surface plasmon resonanceDOI: 10.1134/S0006297920010071
Abbreviations: DLS, dynamic light scattering; IgG, human immunoglobulin G; ka, rate constant of complex association; KD, kinetic constant of complex dissociation; kd, rate constant of complex dissociation; RH, hydrodynamic radius of particles (in distribution analysis); rSkp, recombinant Skp protein (17 kDa); RU, resonance unit; SPR, surface plasmon resonance; Z-average, average hydrodynamic radius of particles (in analysis of cumulants).
According to a large body of experimental data accumulated in modern
literature, many proteins have multiple activities. One of the striking
examples of multifunctional proteins is the chaperone Skp (17 kDa) that
is commonly found in enterobacteria. This protein belongs to the family
of homologous low-molecular-weight proteins (14-20 kDa) with clearly
pronounced basic properties (pI 9-10) [1-4]. The main substrates of the Skp chaperone activity
are proteins of the bacterial outer membrane. Skp selectively binds
unfolded outer membrane proteins after their translocation across the
plasma membrane into the periplasm and prevents their aggregation [5-7]. Moreover, Skp helps the
folding of soluble proteins in the bacterial periplasm [8]. Due to the chaperone activity, Skp co-expression
with target proteins, including full-size immunoglobulins (antibodies),
their Fab fragments, and numerous single-chain derivatives, increases
the yield and facilitates correct folding of the expressed proteins [9-11]. Along with the ability to
interact with the outer membrane proteins as a chaperone, Skp proteins
display other activities that can be biologically and physiologically
important. For example, they bind lipopolysaccharides and DNA [1, 2, 12, 13] and act as chemoattractants for monocytes and
polymorphonuclear leukocytes [14]. There is a
molecular mimicry between Skp and human histocompatibility antigen
HLA-B27: their amino acid sequences have homologous regions [15]. Therefore, it was suggested that these proteins
could have the properties of antigens that can immunologically
cross-react with HLA-B27. However, homologous regions in the Skp
sequence are not recognized in the humoral immune response in the
patients with HLA-B27-associated diseases [16].
We showed earlier that Skp from Yersinia pseudotuberculosis is able to bind as a monomer Skp [17] or homotrimer (Skp3) [18] to human and rabbit IgGs in a non-immune fashion (i.e., via bypassing antigen-binding sites of IgG). Using computer-aided modeling, spatial structures of Skp, Skp3, and Skp3 complexes with the Fc and Fab fragments of human IgG were calculated. Based on the obtained models, the most likely binding sites on the immunoglobulin molecule are located in the regions of its Fc and Fab fragments, and the IgG molecule can simultaneously bind two Skp3. We have shown Skp from Y. pseudotuberculosis binds to the pore-forming protein OmpF and phospholipase A of the bacterial outer membrane [19].
The purpose of this study was to determine qualitative and quantitative characteristics of the chaperone and immunoglobulin-binding activities of Skp toward human IgG and its Fc and Fab fragments at different pH values.
MATERIALS AND METHODS
Materials. Superdex 200 10/300 GL (GE Healthcare, USA); acrylamide (Serva, Germany); Amicon Ultra 3-kDa centrifugal filter units (Merck Millipore, Germany); SDS, 96-well ProteOn microplates, sensor chip GLC (Bio-Rad, USA); marker proteins (Fermentas, Lithuania); Coomassie R-250 (Sigma, USA); BSA, ovalbumin, chymotrypsinogen, myoglobin, cytochrome c, human IgG and Fc fragment of human IgG (ICN Biomedicals, USA); Fab fragment of human IgG (MP Biomedicals, USA); 0.45-μm cellulose membrane filters (Agilent, USA); and phosphate buffered saline (PBS), pH 7.4 (Helicon, Russia) were used. All other reagents (Reakhim, Russia) were of chemical purity grade and used without additional purification.
The following buffer systems were used: PBS (10 mM NaH2PO4, 137 mM NaCl, 2 mM KCl, pH 7.4); 50 or 100 mM CH3COONa/CH3COOH, pH 5.0 (buffer A); 50 mM Tris-HCl, pH 7.4 (buffer B); 50 or 100 mM Tris-HCl, pH 8.0 (buffer C); 50 mM CH3COONa/CH3COOH, pH 6.0 (buffer D).
Preparation of human IgG, Fc and Fab fragments, and rSkp from Y. pseudotuberculosis. Commercially available proteins used as substrates for binding with Skp were first subjected to gel-filtration. A weighted portion of each lyophilized protein was dissolved in water; the resulting solution was filtered through a 0.45-µm filter and loaded (0.25 ml) on a Superdex 200 10/300 GL column equilibrated with PBS for gel-filtration. The proteins were eluted at 0.5 ml/min; protein-containing fractions (0.5-0.7 ml) were collected and their absorbance at 280 nm was determined. The fractions were combined according to the results of SDS-PAGE and concentrated using centrifugal filter units (cut-off, 3 kDa). The total protein concentration in the obtained samples of IgG and Fc and Fab fragments was determined by the Bradford method [20] using lysozyme as a calibration protein.
The recombinant Skp protein (rSkp) from Y. pseudotuberculosis was expressed in Escherichia coli and purified as described by us earlier [18]. The concentration of rSkp was determined from the absorbance at 280 nm using A0.1%1cm = 0.185 (Swiss-Prot, P31520 and Q667J8).
SDS-PAGE. Electrophoresis according to Laemmli [21] was performed in the presence of SDS. All samples for electrophoresis were prepared without heating at 100°C in the sample buffer containing no β-mercaptoethanol. Prestained proteins with molecular masses of 17, 24, 33, 40, 55, 72, 100, and 130 kDa were used as markers. The gels were stained with Coomassie R-250 solution in 10% acetic acid and 45% ethanol.
Dynamical light scattering (DLS). The size of particles formed by rSkp, substrate proteins, and complexes between rSkp and substrate proteins in solution at different incubation times and medium pH values (pH 5.1, 6.7, or 7.9) was determined by the DLS method using a ZetaSizer Nano ZS device (Malvern, Great Britain) equipped with a He-Ne-laser (λ = 633 nm, 4 mW) at the angle of 173°. The measurements were performed in a 10 × 10 mm cuvette at room temperature (20°C). The data were accumulated for 15-30 min. Before the measurement, the following buffer mixtures with the required pH values were prepared: 0.24 ml of PBS + 0.34 ml of 50 mM buffer A + 0.62 ml of either 100 mM buffer A (pH 5.1) or 50 mM buffer B (pH 6.7), or 100 mM buffer C (pH 7.9). Protein solutions used in the experiments were 0.15 mg/ml IgG in PBS, 0.12 mg/ml Fc fragment in PBS, 0.12 mg/ml Fab fragment in PBS, and 0.28 mg/ml rSkp in buffer A.
The Z-average (average particle hydrodynamic radius) and RH (hydrodynamic radius) values in the analysis of particle volume distribution were calculated with the software supplied with the ZetaSizer Nano ZS device (Malvern).
Circular dichroism (CD) spectra. CD spectra were recorded with a Chirascan-plus CD-spectrometer (Applied Photophysics, Great Britain) in quartz cuvettes with the optical pathlength of 0.1 or 1 cm for recording the spectra in the peptide bond and aromatic amino acid regions, respectively. In the peptide region of the CD spectrum (190-240 nm), the ellipticity [θ] of an average residue was calculated according to the following formula, assuming the molecular mass of the residue is 110 Da:
[θ] = [θ]obsS × 110/C·d (deg·m2·dmol–1),
where S is the sensitivity of the device scale; C is the protein concentration, mg/ml; d is the optical pathlength, cm. In the aromatic region of the CD spectrum (240-320 nm), the molar ellipticity [θ]M was calculated assuming that the protein molecular mass is 16.1 kDa. To record CD spectra of rSkp at different pH values, the protein was transferred to the buffer solution mixtures with pH 5.1, 6.7, and 7.9, respectively, prepared as described above.
Oligomeric structure of rSkp under native conditions was confirmed by gel filtration on a Superdex 200 10/300 GL column in buffer A using an FPLC system (Amersham Pharmacia Biotech, USA) as described by us in [18]. The column was preliminary calibrated using proteins with the known molecular masses: BSA (67 kDa), ovalbumin (45 kDa), chymotrypsinogen (24 kDa), myoglobin (18 kDa), and cytochrome c (13 kDa). The relative error in the determining the molecular mass was 7%.
Surface plasmon resonance (SPR). Quantitative characteristics of the rSkp interaction with human IgG and Fc and Fab fragments were obtained using the SPR method. The kinetics of rSkp (analyte) binding with the substrate proteins (ligands) immobilized on the surface of a GLS sensor chip was determined with a ProteOn XPR 36 matrix biosensor (Bio-Rad, USA). The ligands were covalently immobilized on the chip using Amine Coupling Kit reagents (Bio-Rad) according to the manufacturer’s instruction. The density of ligand binding was assessed from changes in the biosensor signal magnitude (RU, resonance unit). The density of ligand attachment was 1420, 690, and 1060 RU for IgG, Fc fragment, and Fab fragment, respectively. The channel with no immobilized protein was used as a control. The following buffer systems were used: 50 mM buffers A, B, C, and D. The analyte concentration was varied in the interval of 0.1-5.0 μM. Analyte association/ dissociation in each experiment was performed at a flow rate of 25 μl/min for 500 s. All experiments were conducted at 25°C. The surface of the sensor chip was regenerated with 50 mM NaOH at a flow rate of 30 μl/min for 60 s. To eliminate possible binding artifacts, the data obtained for the control surfaces (reference surfaces) and in the absence of protein (blank injections) were successively subtracted from the experimental data obtained for the ligand–analyte reaction surfaces. The sensograms were analyzed with the ProteOn Manager 3.0 software (Bio-Rad).
RESULTS AND DISCUSSION
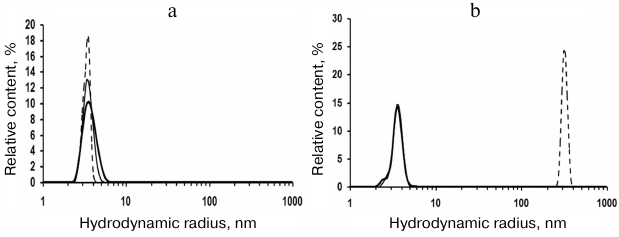
Effect of medium pH on Skp aggregation. We found that rSkp retained its immunoglobulin-binding activity for a long time if stored in buffer solutions with low pH values (4.0-5.0), but rapidly (within a week) aggregated and precipitated at neutral and basic pH values. Therefore, before studying the chaperone and immunoglobulin-binding activities of rSkp from Y. pseudotuberculosis by DLS, we investigated the behavior of rSkp incubated in solutions with different pH value for different periods of time (Fig. 1 and Table 1).
Fig. 1. Volume size distribution of rSkp particles: a) incubation at pH 5.0 for 6 days (solid thick line) and at pH 6.7 for 4 h (solid thin line) and 24 h (dashed line); b) incubation at pH 7.9 for 1 h (solid thick line), 4 h (solid thin line), and 24 h (dashed line).
Table 1. Average hydrodynamic radii
(Z-average and RH) of rSkp particles incubated in water
solutions at different pH values for varying periods of time
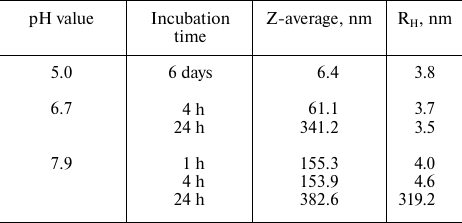
DLS showed that rSkp incubated in 50 mM sodium acetate (pH 5.0) for 6 days had a monomodal distribution with the predominant content of particles with RH and Z-average of 3.8 and 6.4 nm, respectively (Fig. 1a and Table 1). Similar distribution of RH for rSkp particles was also observed during the first 4 h of incubation in solutions with neutral and alkaline pH values, but in this case, the Z-average value of particles sharply increased (Table 1), which indicated the onset of protein self-association. Further incubation for 24 h led to further increase in the Z-average value and changes in the particle size distribution. Thus, protein solution with pH 6.7 contained mostly small particles (3.6 nm), whereas no particles with RH < 200 nm were observed in the buffer with pH 7.9 (Fig. 1b). Thus, we concluded that changes in the medium pH from acidic to alkaline induce self-association and aggregation of rSkp; both processes are accelerated with increase in pH up to the protein isoelectric point (pI 9.33).
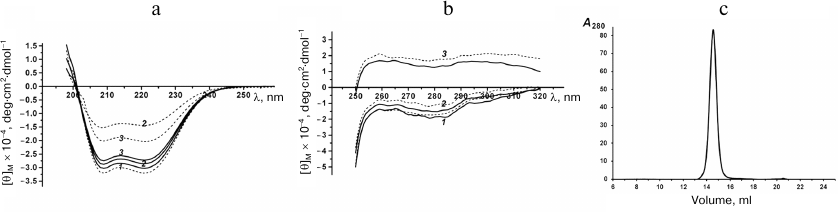
Spatial structure of rSkp. The effect of pH on the protein structure was studied using CD spectroscopy and DLS. The far-UV CD spectra of rSkp in the buffer solutions with pH 5.0, 6.7, and 7.9 were of the same intensity and shape with the minima at 208 and 222 nm, indicating the prevalence of α-helical structures (Fig. 2a). The spectra remained unchanged during protein incubation in the solution for the first 24 h (1, 4, and 24 h). Further incubation (72 h) at pH 5.0 did not change the characteristics of the spectrum, whereas at pH 6.7 and 7.9, the amplitudes of the spectra markedly decreased with the retention of the spectrum shape, which could be associated with the appearance of protein aggregates that could not be detected by CD spectroscopy [22].
Fig. 2. Far-UV (a) and near-UV (b) CD spectra of rSkp incubated at pH 5.0 (1), 6.7 (2) and 7.9 (3) for 1 h (solid line) and 72 h (dotted line). c) Gel-filtration of rSkp on a Superdex 200 HR column in 50 mM sodium acetate, pH 5.0.
Near-UV CD spectra of rSkp were used for detecting possible alterations in the protein tertiary structure caused by changes in the medium pH (Fig. 2b). It is known that the CD spectra in the region of 260-320 nm are generated by aromatic amino acids. Mature Skp has two Tyr and four Phe residues per monomer; some of these residues are located in the conformationally flexible region of the molecule (α-helical “tentacles”) [18, 23]. Figure 2b shows that the protein spectrum had the largest amplitude and the most pronounced fine structure (peaks at 260 and 270 nm characteristic for Phe and peaks at 278 and 285 nm characteristic for Tyr) at pH 5.0. At pH 6.7, some slight changes were observed in the spectrum, such as decreased signal intensity and peak heights. The pH increase to 7.9 resulted in the change of the spectrum sign, further lowering of its amplitude, and spectrum flattening with retention of its characteristic peaks. An increase in the incubation time from 1 to 72 h resulted in no essential changes in the observed spectra.
According to the DLS results, symmetrical monomodal distribution of rSkp particles with a small width and RH of 3.7 ± 0.3 nm was observed within the pH interval of 5.0 to 8.0 (Fig. 1). rSkp was eluted from the Superdex 200 10/300 GL column as a single symmetric peak (Fig. 2c) which corresponded to a protein with an apparent molecular mass of 45.2 ± 3.0 kDa and, therefore, represented the protein trimer [18].
Hence, pH increase from 5.0 to 7.9 did not affect the secondary and oligomeric structures of rSkp, but, presumably, slightly destabilized the tertiary structure of the protein.
Effect of rSkp on the aggregation of IgG and its fragments at different pH values. Commercially available polyclonal IgG and Fc and Fab fragments were used as substrate proteins for binding with rSkp. Before the experiments, the proteins were additionally purified by gel-filtration on a Superdex 200 10/300 GL column equilibrated with PBS. DLS data showed that IgG in PBS had a monomodal volume size distribution with RH = 5.6 ± 0.5 nm, (which correlated with the size of IgG monomer reported earlier in [24-27]) and Z-average value of 17 nm. Fc and Fab fragments also had the monomodal volume size distribution by size with Z-average values of 14 and 11 nm, respectively; RH was 3.3 ± 0.3 nm for both proteins.
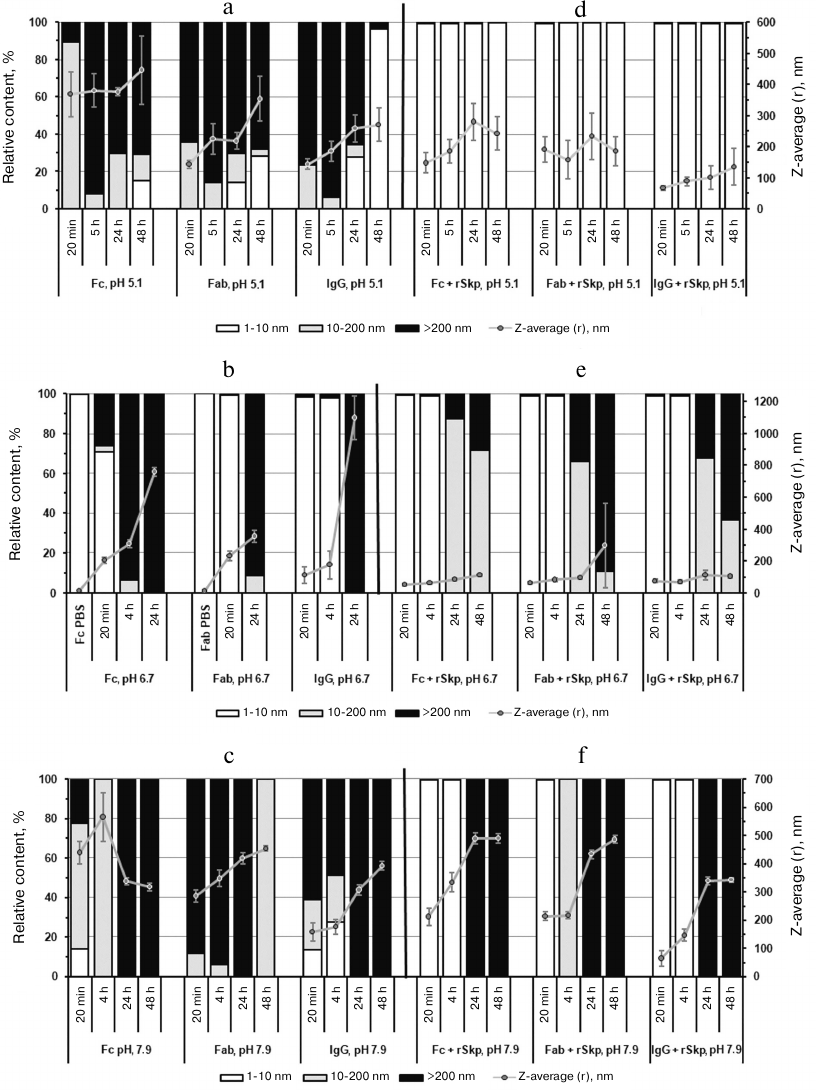
During the recent years, a considerable attention has been paid to the stability of biopharmaceutical preparations based on biologically active proteins. IgG is one of such proteins that is widely used in medicine. Studying the interaction and aggregation of IgG molecules [28-30], as well as factors affecting these processes [31-34], is of both fundamental and applied interest for the drug development. In this work, we observed aggregation of IgG molecules and Fc and Fab fragments upon their transfer from PBS into buffer solutions with lower ionic strength and with acidic, neutral, and alkaline pH values. For all the proteins, we observed increases in the Z-average, width of size distribution (multimodal distribution), and relative content (%) of particles with RH > 10 nm. Figure 3 shows the diagrams for the relative content (%) of particles at their volume size (RH) distribution and Z-average of particles produced by substrate proteins after their incubation at pH 5.1, 6.7, and 7.9 for different periods of time in the absence (Fig. 3, a-c) or presence (Fig. 3, d-f) of rSkp.
Fig. 3. The volume size (RH) distribution and Z-average values of particles produced by substrate proteins in solutions at different pH values and incubation times in the absence (a-c) or presence (d-f) of rSkp.
It can be seen from the diagrams that the aggregation rates of IgG and Fc and Fab fragments are different at different pH values. At acidic and alkaline pH values (Fig. 3, a and c), the aggregation rate was much higher than in the neutral medium (Fig. 3b); the highest aggregation rate was observed at acidic pH.
The growth of the relative content of small particles with RH < 10 and 200 nm with the increase in the incubation time (24 h and more) in acidic solutions (Fig. 3a) might be associated with the formation of large aggregates that precipitate from the solution and are not taken into account in the analysis.
In order to study putative chaperone properties of rSkp toward IgG and its fragments (i.e., ability to prevent aggregation of the studied proteins via forming soluble complexes with them), rSkp was added to the protein solutions at different pH values (Skp3/IgG molar ratio, 8 : 1; Skp3/Fc fragment and Skp3/Fab fragment molar ratios, 3.5 : 1 using molecular mass of the Skp3 trimer in the calculations). According to the DLS results, the rates of self-association and aggregation of the substrate proteins markedly decreased in the presence of rSkp: the growth in the relative content (%) of particles with RH > 10 nm and Z-average values slowed down noticeably (Fig. 3, d-f). The chaperone activity of rSkp strongly depended on the medium pH. At acidic pH (conditions providing maximum rSkp stability), rSkp formed stable (up to 48 h) low-molecular-weight complexes with the substrate proteins with RH up to 10 nm (99.5-99.8%; Fig. 3d). In solutions with neutral and alkaline pH values (6.7 and 7.9), the chaperone activity of rSkp was noticeably decreased (Fig. 3, e and f). At these pH values, a high relative content (98.2-99.8%) of low-molecular-weight complexes (RH < 10 nm) produced during the first 20 min of incubation was retained for 4-5 h. The solution containing rSkp and Fab fragment at pH 7.9 was an exception: 100% of the resulting particles had RH in the range of 10-200 nm. Further incubation of the solutions for 24 and 48 h led to a considerable increase in the fraction of large particles (Fig. 3, e and f) with the size of 10-200 nm and over at pH 6.7 and over 200 nm (100%) at pH 7.9. These results show that at pH 6.7 and 7.9, rSkp was unable to fully prevent the self-association and aggregation of IgG and its fragments, but considerably slowed down these processes.
Recently, numerous studies have shown that disturbances in the protein structure caused by changes in the medium pH can affect chaperone activity of these proteins [35-37]. Our results clearly demonstrated the pH-dependent character of the chaperone activity of rSkp. The most stable low-molecular-weight complexes (RH < 10 nm) between rSkp and IgG, Fc fragments, or Fab fragments were produced only at acidic pH values. The chaperone activity of rSkp decreased with the increase in pH, and the aggregation of protein substrates somewhat slowed down. Based on our data, we propose a model of the inhibitory effect of rSkp on the pH-induced protein aggregation, according to which rSkp stably binds the substrate proteins at low pH values, thus preventing their irreversible aggregation. Further increase in the medium pH to neutral causes a slow release of the substrate proteins from their complexes with the chaperone.
In its chaperone properties, Skp resembles recently discovered homodimeric chaperones HdeA and HdeB that ensure E. coli resistance against acid stress [38, 39]. Similarly to Skp, these chaperones are located in the periplasm, have small molecular mass, are ATP-independent, and protect periplasm proteins from unfolding and aggregation under acidic conditions. They are activated at low pH values and function as partially unfolded monomers. At the same time, it was shown that Skp retains its native secondary and oligomeric structures at acidic pH values. In order to unambiguously elucidate the difference between Skp and the above-mentioned chaperones in their properties and mechanism of action, it is necessary to study the structure and chaperone activity of Skp at lower pH values (<5), at which HdeA and HdeB are active and function.
Kinetic characteristics of Skp binding to IgG and Fc and Fab fragments at different medium pH values. To study Skp binding with human IgG and its Fc and Fab fragments and to determine its quantitative characteristics at different pH values, the method of surface plasmon resonance (SPR) was used.
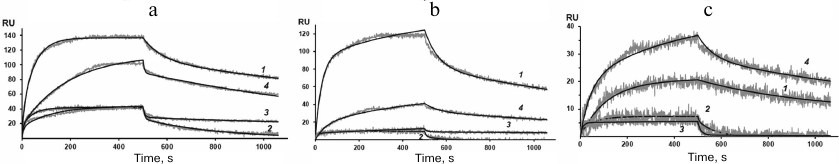
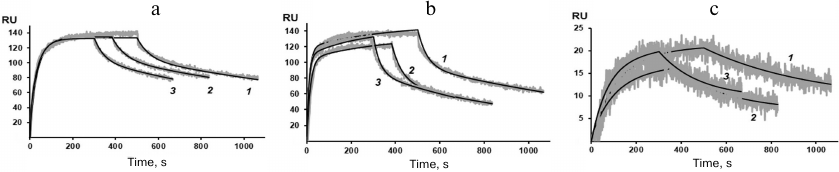
Figure 4 shows the sensograms (experimental and theoretical) of the rSkp binding with human IgG and its fragments at different pH values. Quantitative characteristics of binding, such as the kinetic constant of complex dissociation (KD), which is the main parameter that determines the strength of ligand–analyte binding, and the rate constants of association and dissociation of the complexes (ka and kd, respectively) are given in Table 2. The graphical data (Fig. 4) and the chi-test values (χ2; Table 2) confirmed that the models used to approximate the results were satisfactory for calculating quantitative characteristics of the ligand–analyte interactions. According to our calculations, rSkp binding with Fc and Fab fragments fitted the two-state model, and rSkp binding with IgG corresponded to the heterogeneous ligand model. Analysis of the rSkp binding to the substrate proteins at the same rSkp concentration and pH 5.0, but at different analyte flow rates across the cell (25, 35, and 45 μl/min; Fig. 5) indicated the absence of effects limiting the mass transfer in the ligand–analyte interaction.
Fig. 4. Sensograms of rSkp binding with IgG (a), Fc fragment (b), and Fab fragment (c) in buffer solutions with pH 5.0 (1), 6.0 (2), 7.4 (3) and 8.0 (4).
Fig. 5. Sensograms of rSkp binding with IgG (a), Fc fragment (b), and Fab fragment (c) in sodium acetate buffer (pH 5.0) at the analyte flow rates of 25 (1), 35 (2), and 45 μl/min (3).
Table 2. Quantitative characteristics of
rSkp binding with human IgG, Fc and Fab fragments of IgG at different
pH values of the medium
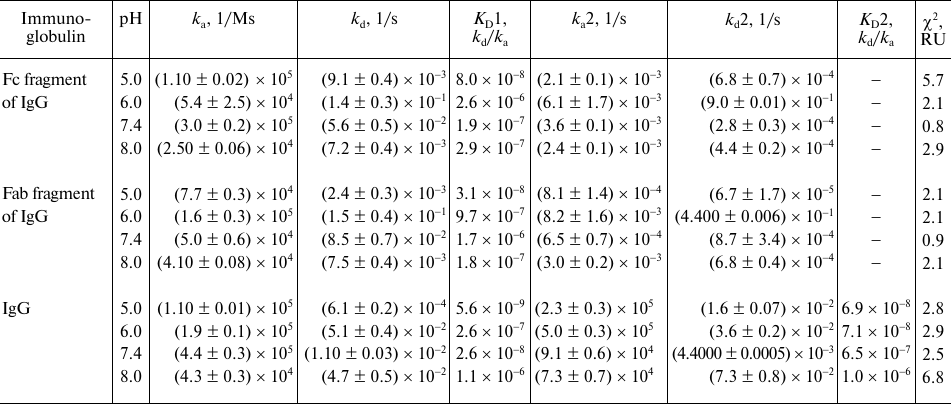
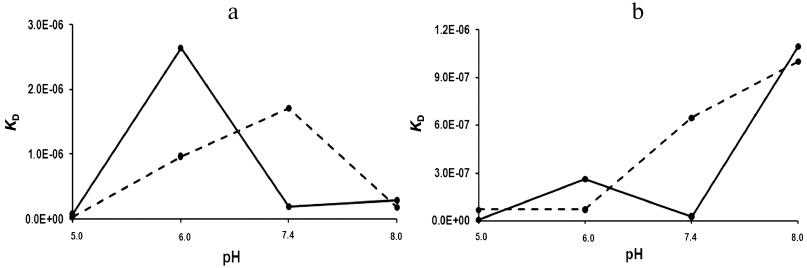
The dissociation constants of the studied complexes were low, which suggested specific interactions between the interaction participants. The strength of the rSkp binding with human IgG and its fragments strongly depended on the medium pH: at pH 5.0 the binding strength with all the tested substrate proteins was maximal and then decreased considerably with the increase in medium pH (Fig. 6 and Table 2). The decrease in the affinity of interaction with the increase in pH value was caused, first of all, by a considerable increase in the rate of the complex dissociation.
Fig. 6. pH-dependence of KD for rSkp complexes with IgG and Fc and Fab fragments. rSkp interactions: a) with Fc fragment (solid line) and Fab fragment (dashed line); b) with human IgG; KD1, solid line; KD2, dashed line.
At pH values close to neutral (6.0 and 7.4), the differences in the kinetics of formation and dissociation of rSkp complexes with protein ligands were most pronounced. With the increase in pH from 6.0 to 7.4, the changes in the kd values were similar for the complexes with Fc and Fab fragments (2.5- and 1.8-fold decrease, respectively). At the same time, ka for the Fc fragment increased 5.6 times, while ka for the Fab fragment decreased 3.2 times (Table 2). As a result, the binding affinity increased for the Fc fragment and decreased for the Fab fragment in this pH interval (Fig. 6a). These data correlate with the DLS results according to which the chaperone activity of rSkp toward the Fc fragment at pH 6.7 was higher than toward the Fab fragment (Fig. 3e). It is possible that the differences in the interaction of the Fc and Fab fragments with rSkp are caused by the fact the pI of Fc fragment in the pH interval of 6.0-7.4 is lower than pI of the Fab fragment [40]. In the case of IgG, the pH-dependence of the dissociation constants in the pH interval of 6.0 to 7.4 for the site with KD1 was similar to that for the Fc fragment and the pH dependence for the site with KD2 coincided with that for the Fab fragment (Fig. 6b). Hence, it may be assumed that KD1 and KD2 relate to the rSkp binding with the Fc and Fab fragments, respectively, on the immobilized human IgG.
The affinity of rSkp for the Fc fragment remained unchanged, while its affinity for the Fab fragment increased with the increase in the medium pH from 7.4 to 8.0 (Fig. 6a and Table 2). These results contradict the DLS data, according to which rSkp has the lowest chaperone activity at pH 7.9 (Fig. 3f). This discrepancy might result from a decrease in the KD values due to fast self-association of rSkp in the alkaline medium with pH close to the protein isoelectric point (Table 1). An apparent increase in the affinity recorded by the SPR method with a growth in the size of aggregates involved in the ligand–receptor interaction was described earlier in [41]. Both binding sites of the IgG molecule demonstrated the lowest interaction affinity at pH 8.0 (Fig. 6b and Table 2), which is in a good agreement with the low chaperone activity of rSkp recorded by the DLS method at pH 7.9 (Fig. 3f). However, this does not exclude the possibility that the KD values can be underestimated due to the self-association of rSkp.
Our study showed that the maximal immunoglobulin-binding and chaperone activities of rSkp from Y. pseudotuberculosis were manifested in solution with acidic pH in the pH range of 5.0 to 8.0, at which the protein was most stable. Slow self-association and aggregation of rSkp molecules was observed with the increase in medium pH together with a noticeable decrease in the binding properties of this protein.
The obtained information can be of practical interest because immunoglobulins are widely used in medicine as pharmaceutical preparations for the treatment of various autoimmune diseases, cancer, etc. IgG aggregates formed during the long-term storage can increase the immunogenicity of the preparations and change their physical properties, first of all, viscosity. Controlling protein aggregation is a key issue in the production of stable biopharmaceutical preparations of high quality.
Conflict of interest. The authors declare no conflict of interest.
Ethical norm compliance. This article does not contain description of studies with participation of humans or animals performed by any of the authors.
REFERENCES
1.Holck, A., Lossius, I., Aasland, R., Haarr, L., and
Kleppe, K. (1987) DNA- and RNA-binding proteins of chromatin from
Escherichia coli, Biochim. Biophys. Acta,
908, 188-199, doi: 10.1016/0167-4781(87)90058-3.
2.Holck, A., and Kleppe, K. (1988) Cloning and
sequencing of the gene for the DNA-binding 17K protein of
Escherichia coli, Gene, 67, 117-124, doi:
10.1016/0378-1119(88)90014-5.
3.Koski, P., Rhen, M., Kantele, J., and Vaara, M.
(1989) Isolation, cloning, and primary structure of a cationic 16-kDa
outer membrane protein of Salmonella typhimurium, J. Biol.
Chem., 264, 18973-18980.
4.Koski, P., Hirvas, L., and Vaara, M. (1990)
Complete sequence of the ompH gene encoding the 16-kDa cationic
outer membrane protein of Salmonella typhimurium, Gene,
88, 117-120, doi: 10.1016/0378-1119(90)90068-3.
5.De Cock, Y., Schafer, U., Potgeter, M., Demel, R.,
Muller, M., and Tommassen, J. (1999) Affinity of the periplasmic
chaperone Skp of Escherichia coli for phospholipids,
lipopolysaccharides and non-native outer membrane proteins. Role of Skp
in the biogenesis of outer membrane protein, Eur. J. Biochem.,
259, 96-103, doi: 10.1046/j.1432-1327.1999.00010.x.
6.Harms, N., Koningstein, G., Dontje, W., Muller, M.,
Oudega, B., Luirink, J., and de Cock, H. (2001) The early interaction
of the outer membrane protein PhoE with the periplasmic chaperone Skp
occurs at cytoplasmic membrane, J. Biol. Chem.,
276, 18804-18811, doi: 10.1074/jbc.M011194200.
7.Solov’eva, T. F., Novikova, O. D., and
Portnyagina, O. Y. (2012) Biogenesis of β-barrel integral proteins
of bacterial outer membrane, Biochemistry (Moscow),
77, 1221-1236.
8.Entzminger, K. C., Chang, C., Myhre, R. O.,
McCallum, K. C., and Maynard, J. A. (2012) The Skp chaperone helps fold
soluble proteins in vitro by inhibiting aggregation,
Biochemistry, 51, 4822-4834, doi: 10.1021/bi300412y.
9.Mazor, Y., Van Blarcom, T., Mabry, R., Iverson, B.
L., and Georgiou, G. (2007) Isolation of engineered, full-length
antibodies from libraries expressed in Escherichia coli, Nat.
Biotechnol., 25, 563-565, doi: 10.1038/nbt1296.
10.Levy, R., Weiss, R., Chen, G., Iverson, B. L.,
and Georgiou, G. (2001) Production of correctly folded Fab antibody
fragment in the cytoplasm of Escherichia coli trxB gor mutants
via the co-expression of molecular chaperones protein expression and
purification, Protein Expr. Purif., 23, 338-347, doi:
10.1006/prep.2001.1520.
11.Sonoda, H., Kumada, Y., Katsuda, T., and Yamaji,
H. (2011) Effects of cytoplasmic and periplasmic chaperones on
secretory production of single-chain Fv antibody in Escherichia
coli, J. Biosci. Bioeng., 111, 465-470, doi:
10.1016/j.jbiosc.2010.12.015.
12.Geyer, R., Galanos, C., Westphal, O., and
Golecki, J. (1979) A lipopolysaccharide-binding cell-surface protein
from Salmonella minnesota. Isolation, partial characterization
and occurrence in different Enterobacteriaceae, Eur. J.
Biochem., 98, 27-38, doi:
10.1111/j.1432-1033.1979.tb13156.x.
13.Holck, A., Lossius, I., Aasland, R., and Kleppe,
K. (1987) Purification and characterization of the 17 K protein, a
DNA-binding protein from Escherichia coli, Biochim. Biophys.
Acta, 914, 49-54, doi: 10.1016/0167-4838(87)90160-9.
14.Shrestha, A., Shi, L., Tanase, S., Tsukamoto, M.,
Nishino, N., Tokita, K., and Yamamoto, T. (2004) Bacterial chaperone
protein, Skp, induces leukocyte chemotaxis via C5a receptor, Am. J.
Pathol., 164, 763-772, doi:
10.1016/S0002-9440(10)63164-1.
15.Vuorio, R., Hirvas, L., Raybourne, R. B., Yu, D.
T. Y., and Vaara, M. (1991) The nucleotide and deduced amino acid
sequence of the cationic 19 kDa outer membrane protein OmpH of
Yersinia pseudotuberculosis, Biochim. Biophys. Acta,
1129, 124-126, doi: 10.1016/0167-4781(91)90226-C.
16.Lahesmaa, R., Skurnik, M., Vaara, M.,
Leirisalo-Repo, M., Nissila, M., Granfors, K., and Toivanen, P. (1991)
Molecular mimicry between HLA B27 and Yersinia,
Salmonella, Shigella and Klebsiella within the
same region of HLA α1-helix, Clin. Exp.
Immunol., 86, 399-404, doi:
10.1111/j.1365-2249.1991.tb02944.x.
17.Sidorin, E. V., Ziganshin, R. Kh., Naberezhnykh,
G. A., Likhatskaya, G. N., Trifonov, E. V., Anastiuk, S. D., Chernikov,
O. V., and Solov’eva, T. F. (2009) Chaperone Skp from Yersinia
pseudotuberculosis exhibits immunoglobulin G binding activity,
Biochemistry (Moscow), 74, 406-415.
18.Sidorin, E. V., Tishchenko, N. M., Khomenko, V.
A., Isaeva, M. P., Dmitrenok, P. S., Kim, N. Y., Likhatskaya, G. N.,
and Solov’eva, T. F. (2012) Molecular cloning, isolation, and
properties of chaperone Skp from Yersinia pseudotuberculosis,
Biochemistry (Moscow), 77, 1315-1325.
19.Sidorin, E. V., Sidoorova, O. V., Tishchenko, N.
M., Khomenko, V. A., Novikova, O. D., and Solov’eva, T. F. (2015)
Chaperone activity of immunoglobulin-binding protein from Yersinia
pseudotuberculosis, Biol. Membr. (Moscow), 32,
217-220, doi: 10.7868/S0233475515030081.
20.Bradford, M. M. (1976) A rapid and sensitive
method for the quantitation of microgram quantities of protein
utilizing the principle of protein–dye binding, Anal.
Biochem., 72, 248-254, doi: 10.1006/abio.1976.9999.
21.Laemmli, U. K. (1970) Cleavage of structural
proteins during the assembly of the head of bacteriophage T4,
Nature, 227, 680-685, doi: 10.1038/227680a0.
22.Ioannou, J. C., Donald, A. M., and Tromp, R. H.
(2015) Characterizing the secondary structure changes occurring in high
density systems of BLG dissolved in aqueous pH 3 buffer, Food
Hydrocolloids, 46, 216-225, doi:
10.1016/j.foodhyd.2014.12.027.
23.Walton, T. A., and Sousa, M. C. (2004) Crystal
structure of Skp, a prefoldin-like chaperone that protects soluble and
membrane proteins from aggregation, Mol. Cell, 15,
367-374, doi: 10.1016/j.molcel.2004.07.023.
24.Hawea, A., Kasperb, J. C., Friessb, W., and
Jiskoot, W. (2009) Structural properties of monoclonal antibody
aggregates induced by freeze–thawing and thermal stress, Eur.
J. Pharm. Sci., 38, 79-87, doi:
10.1016/j.ejps.2009.06.001.
25.Arosio, P., Barolo, G., Muller-Spath, T., Wu, H.,
and Morbidelli, M. (2011) Aggregation stability of a monoclonal
antibody during downstream processing, Pharm. Res., 28,
1884-1894, doi: 10.1007/s11095-011-0416-7.
26.Amani, S., Nasim, F., Khan, T. A., Fazili, N. A.,
Furkan, M., Bhat, I. A., Khan, J. M., Khan, R. H., and Naeem, A. (2014)
Detergent induces the formation of IgG aggregates: a
multi-methodological approach, Spectrochim. Acta A Mol. Biomol.
Spectrosc., 120, 151-160, doi:
10.1016/j.saa.2013.09.141.
27.Esfandiary, R., Parupudi, A., Casas-Finet, J.,
Gadre, D., and Sathish, H. (2015) Mechanism of reversible
self-association of a monoclonal antibody: role of electrostatic and
hydrophobic interactions, J. Pharm. Sci., 104, 577-586,
doi: 10.1002/jps.24237.
28.Nezlin, R. (2010) Interactions between
immunoglobulin G molecules, Immunol. Lett., 132, 1-5,
doi: 10.1016/j.imlet.2010.06.006.
29.Joubert, M. K., Luo, Q., Nashed-Samuel, Y.,
Wypych, J., and Narhi, L. O. (2011) Classification and characterization
of therapeutic antibody aggregates, J. Biol. Chem.,
286, 25118-25133, doi: 10.1074/jbc.M110.160457.
30.Luo, Q., Joubert, M. K., Stevenson, R., Ketchem,
R. R., Narhi, L. O., and Wypych, J. (2011) Chemical modifications in
therapeutic protein aggregates generated under different stress
conditions, J. Biol. Chem., 286, 25134-25144, doi:
10.1074/jbc.M110.160440.
31.Wang, W. (2005) Protein aggregation and its
inhibition in biopharmaceutics, Intern. J. Pharm., 289,
1-30, doi: 10.1016/j.ijpharm.2004.11.014.
32.Arosio, P., Rima, S., and Morbidelli, M. (2013)
Aggregation mechanism of an IgG2 and two IgG1 monoclonal antibodies at
low pH: from oligomers to larger aggregates, Pharm. Res.,
30, 641-654, doi: 10.1007/s11095-012-0885-3.
33.Gil, D., and Schrum, A. G. (2013) Strategies to
stabilize compact folding and minimize aggregation of antibody-based
fragments, Adv. Biosci. Biotechnol., 4, 73-84, doi:
10.4236/abb.2013.44A011.
34.Roberts, C. J. (2014) Therapeutic protein
aggregation: mechanisms, design, and control, Trends
Biotechnol., 32, 372-380, doi:
10.1016/j.tibtech.2014.05.005.
35.Poon, S., Rybchyn, M. S., Easterbrook-Smith, S.
B., Carver, J. A., Pankhurst, G. J., and Wilson, M. R. (2002) Mildly
acidic pH activates the extracellular molecular chaperone clusterin,
J. Biol. Chem., 277, 39532-39540, doi:
10.1074/jbc.M204855200.
36.Tapley, T. L., Franzmann, T. M., Chakraborty, S.,
Jakob, U., and Bardwell, J. C. A. (2010) Protein refolding by
pH-triggered chaperone binding and release, PNAS, 107,
1071-1076, doi: 10.1073/pnas.0911610107.
37.Bose, D., Patra, M., and Chakraborty, A. (2017)
Effect of pH on stability, conformation, and chaperone activity of
erythroid and non-erythroid spectrin, Biochim. Biophys. Acta
Proteins Proteom., 1865, 694-702, doi:
10.1016/j.bbapap.2017.03.012.
38.Malki, A., Le, H.-T., Milles, S., Kern, R.,
Caldas, T., Abdallah, J., and Richarme, G. (2008) Solubilization of
protein aggregates by the acid stress chaperones HdeA and HdeB, J.
Biol. Chem., 283, 13679-13687, doi:
10.1074/jbc.M800869200.
39.Tapley, T. L., Korner, J. L., Bargea, M. T.,
Hupfelda, J., Schauertec, J. A., Gafnic, A., Jakoba, U., and Bardwella,
J. C. A. (2009) Structural plasticity of an acid-activated chaperone
allows promiscuous substrate binding, PNAS, 106,
5557-5562, doi: 10.1073/pnas.0811811106.
40.Coleman, L., and Mahler, S. M. (2003)
Purification of Fab fragments from a monoclonal antibody papain digest
by Gradiflow electrophoresis, Protein Expres. Purif., 32,
246-251, doi: 10.1016/j.pep.2003.07.005.
41.Luo, Y., Lu, Z., Raso, S. W., Entrican, C., and
Tangarone, B. (2009) Dimers and multimers of monoclonal IgG1 exhibit
higher in vitro binding affinities to Fcγ receptors,
mAbs, 1, 491-504.