Complexes Formed via Bioconjugation of Genetically Modified TMV Particles with Conserved Influenza Antigen: Synthesis and Characterization
T. V. Gasanova1,a*, A. A. Koroleva1, E. V. Skurat1, and P. A. Ivanov1
1Lomonosov Moscow State University, Faculty of Biology, 119991 Moscow, Russia* To whom correspondence should be addressed.
Received September 30, 2019; Revised November 8, 2019; Accepted November 16, 2019
Recently we obtained complexes between genetically modified Tobacco Mosaic Virus (TMV) particles and proteins carrying conserved influenza antigen such as M2e epitope. Viral vector TMV-N-lys based on TMV-U1 genome was constructed by insertion of chemically active lysine into the exposed N-terminal part of the coat protein. Nicotiana benthamiana plants were agroinjected and TMV-N-lys virions were purified from non-inoculated leaves. Preparation was analyzed by SDS-PAGE/Coomassie staining; main protein with electrophoretic mobility of 21 kDa was detected. Electron microscopy confirmed the stability of modified particles. Chemical conjugation of TMV-N-lys virions and target influenza antigen M2e expressed in E. coli was performed using 5 mM 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide and 1 mM N-hydroxysuccinimide. The efficiency of chemical conjugation was confirmed by Western blotting. For additional characterization we used conventional electron microscopy. The diameter of the complexes did not differ significantly from the initial TMV-N-lys virions, but complexes formed highly organized and extensive network with dense “grains” on the surface. Dynamic light scattering demonstrated that the single peaks, reflecting the complexes TMV-N-lys/DHFR-M2e were significantly shifted relative to the control TMV-N-lys virions. The indirect enzyme-linked immunosorbent assay with TMV- and DHFR-M2e-specific antibodies showed that the complexes retain stability during overnight adsorption. Thus, the results allow using these complexes for immunization of animals with the subsequent preparation of a candidate universal vaccine against the influenza virus.
KEY WORDS: bioconjugation, tobacco mosaic virus (TMV), genetically modified particles, influenza A, multivalent nanovaccineDOI: 10.1134/S0006297920020091
Abbreviations: a.a., amino acid residue; ADFK, alanine–aspartic acid–phenylalanine–lysine sequence; CP, coat protein; DHFR, dihydrofolate reductase; EDC, 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide; M2e, conserved epitope of influenza A virus (25.5 kDa); NHS, N-hydroxysuccinimide; NP, nucleoprotein; TMV, tobacco mosaic virus.
Influenza A is one of the most dangerous respiratory diseases. The lack
of immunity to new seasonal and pandemic strains of this virus is a
serious problem that could lead to the development of severe
pathologies and death. Unfortunately, none of the existing vaccines can
completely prevent contracting influenza because of the high
variability of its major surface antigens [1].
Existing approaches for creating a universal influenza vaccine are based on using conserved proteins and peptides of the influenza virion. The efficacy of the suggested universal vaccine depends mainly on the induction of cross-reactive T cells [2]. Although this type of immunity cannot prevent infection entirely, it can considerably impede its development. For example, influenza virus nucleoprotein (NP) could be detected on the surface of infected cells at the early stages of infection and neutralized by antibodies against this protein [3].
The discovery of antibodies reacting with the conserved epitopes of the hemagglutinin (HA) stalk domain has stimulated the development of universal influenza vaccine that has been successfully tested in preclinical trials [1].
It was shown using chicken embryos that incorporation of NP into the virus-like particles (VLP) facilitated production of anti-NP antibodies and provided 100% protection from the lethal dose of heterologous influenza virus strain [4]. Matrix M1 protein is another conserved protein that stimulates effective T cell-mediated immune response similar to NP [4, 5]. The emergence of the wide-spectrum resistance in mice and birds was observed upon intracellular expression of the full-size NP and M1 genes employing vectors based on adeno-, baculo-, and poxviruses [6, 7]. The M2 protein tetramer forms an ionic transmembrane channel. The conserved M2e ectodomain of this protein (23 a.a.) exposed on the viral particle surface is also a promising target for designing the universal anti-influenza vaccine [8-10]. M2e epitope-based vaccines facilitate the development of resistance against influenza virus by inducing antibody-dependent cell-mediated cytotoxicity (ADCC) via T helper cells, natural killer cells, macrophages, and mast cells [2].
The objective of this study was investigation of the TMV conjugates as a platform to improve the universal subunit vaccine for influenza A by displaying conserved M2e antigen on the virion surface. TMV was chosen as a carrier because of its proven adjuvant [11] and immunogenic [12] properties, including the ability to stimulate stable humoral and cellular immune responses [13]. Rod-shaped TMV particles provide a convenient platform for displaying a large number (up to 2100 copies) of epitopes on their surface to ensure efficient immune response [13-15]. We investigated the possibility of using chemical conjugation for the generation of stable complexes between TMV particles containing reactive lysine on their surface and conserved M2e epitope of the influenza A virus. The TMV coat protein (CP), which has been characterized in detail by X-ray diffraction analysis, is capable of self-assembly. Moreover, it is sufficiently resistant to pH and temperature changes. According to the crystallography data, each CP subunit contains four sites exposed to the external environment that can be used for cloning target sequences. In addition to the N- and C-terminal regions, the protein also has two loops exposed at the virion outer surface (a.a. 59-65 and 152-156) [16]. Previously, conjugation with the CP N-terminus was achieved by introducing additional amino acid sequences containing lysine (K) residue with the reactive NH2-group. Out of these sequences, ADFK (alanine–aspartic acid–phenylalanine–lysine) proved to be the optimal one [15]. This approach allowed to produce complexes between stable TMV particles containing reactive lysine and epitopes from cytotoxic T lymphocytes associated with the melanoma antigens p15e or Trp2. Immunization with the modified viral particles containing both melanoma epitopes ensured more efficient stimulation of the anticancer activity in comparison with the use of the same peptides separately [13].
Insertion of reactive lysine also allows capsid biotinylation. As a result, the particles acquire the ability to bind targets conjugated with streptavidin. A fragment of the L2 structural protein (36 a.a.) of the dog papilloma virus located at the surface of such viral particles was significantly more immunogenic in comparison with the free L2 fragment [17]. Multivalent vaccines carrying two or more antigens can be generated using N-hydroxysuccinimide (NHS) and 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide (EDC) reagents by attaching these antigens to genetically modified TMV particles. It was demonstrated that simultaneous immunization of mice with a mixture of TMV particles carrying OmpA, DnaK, and Tul4 proteins from Francisella tularensis significantly promoted protective immune response to tularemia [18]. Similar results were obtained for the TMV particles containing reactive lysine and conjugated with hemagglutinin [11].
MATERIALS AND METHODS
Cloning of recombinant TMV viral vector containing reactive lysine at the CP N-terminus. Four codons encoding the ADFK sequence were inserted using the overlap extension PCR technique. The outer primers contained restriction sites for cloning into the intermediate construct pA4083, which contained 3′-terminal part of the TMV cDNA including the CP-coding sequence. PCR was carried out in a Tertsik amplifier (DNK Tekhnologiya, Russia) using Encyclo polymerase (Evrogen, Russia) with the exonuclease activity to decrease the probability of amplification errors. Two initial parallel PCRs were conducted with two pairs of primers: Pr Nco-mp-p (ggagggcccatggaac)/Pr ADFK-m (tactgtaagacttgaagtcagccatatttaaaacgaatccgattc) and Pr ADFK-p (gctgacttcaagtcttacagtatcactactccat)/Pr Apa-cp-m (tgggcccctaccgggggtaa). The pA4083 plasmid was used as a template. The resulting PCR products were purified and used in the second PCR with the terminal primers Pr Nco-mp-p and Pr Apa-cp-m.
The isolated PCR product was treated with NcoI and BstBI restriction endonucleases and cloned by same sites into the intermediate vector pA4083 with the generation of the pA4083-NLys plasmid. The presence of the insertion was confirmed by sequencing. Next, the BamHI/SalI fragment was cut out from the pA4083-NLys construct and cloned into the binary pBIN-TMV-wt vector previously designed in our laboratory (Fig. 1a). The final pBIN-TMV-N-lys construct contained the full-size TMV cDNA coding for the CP protein containing ADFK sequence in the CP N-fragment. The obtained cDNA was under control of the Actin 2 transcriptional promoter from Arabidopsis thaliana and contained transcriptional terminator from the nopaline synthase (nos) gene.
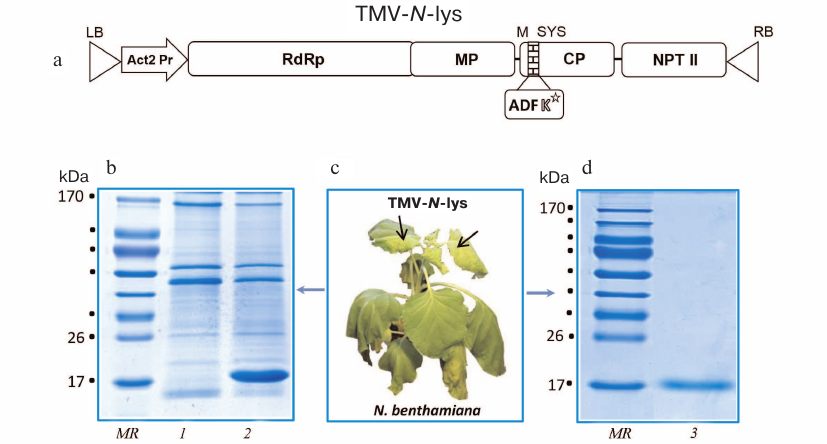
Fig. 1. Design and purification of genetically modified viral particles. a) pBIN-TMV-N-lys binary vector produced by insertion of the ADFK sequence into the binary vector pBIN-TMV-wt based on the pBin19 plasmid. RdRp, RNA-dependent RNA-polymerase (TMVU1) gene; Act2, Actin 2 gene promoter from A. thaliana; NPT, neomycin phosphotransferase gene (kanamycin-resistance gene); LB and RB, left and right borders; MP, TMV movement protein; CP, coat protein; ADFK, insertion to CP containing reactive lysine (K). b) Nicothiana benthamiana plant with systemic symptoms caused by the infection with recombinant TMV-N-lys 10 days post-inoculation. c) Electrophoretic analysis in 8-20% polyacrylamide gel of extracts from non-infected plants (1) and plants infected with TMV-N-lys (2). d) Electrophoretic analysis in 8-20% polyacrylamide gel of purified TMV-N-lys particles. MR, protein molecular weight markers (kDa); lane 3, TMV-N-lys, 10 μg.
Transformation of Agrobacterium tumefaciens cells. Agrobacterium tumefaciens GV 3101 cells were transformed with the produced binary vector pBIN-TMV-N-lys. Transformed bacterial cells were cultivated in LB liquid medium with the corresponding antibiotic at 28°C (optimal temperature for A. tumefaciens growth) overnight on a shaker (170 rpm). Next, aliquots (100 μl) of each overnight culture were mixed, plated on a Petri dish with agar without the antibiotic, and incubated at 28°C overnight. A series of 10-fold dilutions of the cells harvested by scrapping the bacterial lawn from the Petri dish was prepared; the dilutions were plated onto LB agar with selective antibiotics (50 μg/ml kanamycin, 50 μg/ml rifampicin, 25 μg/ml gentamycin) followed by cultivation for 48 h at 28°C. Agrobacterium tumefaciens colonies were tested by PCR using the following primers: PrCP-U1-PstI-p (actgctgcaggagtagacgacgcaacggtggccata), PrCP-U1-HindIII-m (actgaagcttcgcaccacgtgtgaattacggacacaat), PrTad23-p (gggaaaaatagtagtaatgatcggtcagtgccgaacaagaac), Pr CP-154-m (agaggtccaaaccaaaccag).
Agroinfiltration of Nicothiana benthamiana leaves. Agrobacterium tumefaciens cells carrying the pBIN-TMV-N-lys vector or binary vector expressing p19 with the anti-silencing activity from the tomato bushy stunt virus (TBSV) were cultivated overnight at 28°C. The cells were collected by centrifugation at 5000g for 5 min, resuspended in the buffer (10 mM MgSO4, 10 mM MES, pH 5.6) to the optical density OD600 = 0.5 and used for the agroinfiltration of third node leaves from the top of 1.5-month-old N. benthamiana plants. The agrobacterial mixture was infiltrated into the abaxial part of the leaf using a syringe without a needle.
Isolation of recombinant virus. Two to three weeks after infiltration, the leaves exhibited the signs of viral infection. Infected leaves were homogenized in two volumes of 0.1 M sodium phosphate buffer (NaH2PO4, Na2HPO4, pH 7.0, and 1% β-mercaptoethanol, v/v). Cell debris was removed by centrifugation in a preparative Beckman centrifuge for 10 min at 13,000g. The supernatant containing viral particles was clarified by vigorous shaking with chloroform (1/4 volume) for 20 min, and the sample was centrifuged again for 10 min at 13,000g for phase separation. The aqueous phase was mixed with PEG6000 and NaCl [final concentrations, 4% and 1% (w/v), respectively], and the virus was precipitated at 4°C overnight. The next day, the virus was centrifuged for 10 min at 13,000g, and the precipitate was resuspended in 0.01 M sodium phosphate buffer (NaH2PO4, Na2HPO4, pH 7.0). After dissolving the precipitate, the sample was centrifuged for 5 min at 5000g to remove insoluble components; the supernatant was collected and used in further experiments.
RNA isolation from N. benthamiana plants. RNA was isolated from 400 mg of plant material (upper leaves with pronounced symptoms of infection) according to the standard technique using RNeasy Mini-Kit (Qiagen, Germany). RNA was eluted from a column with RNase-free water provided with the kit; RNA amount was evaluated spectrophotometrically at OD260.
Western blot analysis. Proteins were separated by reducing SDS-PAGE (w/v) on 8-20% polyacrylamide gel and immobilized on a Hybond-P PVDF membrane (Amersham, USA) in the transfer buffer (25 mM Tris, 0.192 M glycine, 10% ethanol, v/v). Next, the membranes were blocked with 5% (w/v) nonfat dry milk (Difco, USA) for 1 h at room temperature in TBS-Tween buffer (TBS-T; 150 mM NaCl, 10 mM Tris-HCl, 0.1% Tween 20, v/v, pH 8.0). Following washing with TBS-T buffer, the membranes were incubated in the same buffer containing 2.5% (w/v) nonfat dry milk and mouse anti-TMV antibodies (generated at the Department of Virology, Moscow State University, via three intramuscular immunization with 500 μg protein per injection every 2 weeks; dilution, 1 : 5000), specific anti-TMV IgG (SRA 57400/1000; Agdia, USA) or mouse antibodies against M2e peptide (generated at the Department of Virology, Moscow State University, via three intramuscular immunization with 300 μg protein per injection weekly with adjuvant; dilution, 1 : 20,000 [9]). Following three 5-min washing of the membranes in TBS-T buffer, the membranes were incubated with secondary anti-mouse antibodies conjugated with horseradish peroxidase (Sigma, USA; dilution, 1 : 15,000) for 1 h. The membranes were washed three times in TBS-T buffer for 5 min and visualized with the ECL reagent kit (Amersham). Chemiluminescence signal was registered using an X-ray film.
Electron microscopy. Virus samples were applied onto a carbon-coated Copper EM grid (Ted Pella, USA), washed with MilliQ water, and treated with acidified solution of 2% uranyl acetate (w/v) for contrasting.
Indirect enzyme-linked immunoassay (ELISA). TMV-N-lys viral particles (200 ng) were incubated overnight at 4°C in microplate wells (Nunc MaxiSorb, Denmark) blocked with PBS-T containing 2% BSA (w/v) (1 h at room temperature). Subsequently, mouse antibodies against TMV or M2e in PBS-T buffer were added at a dilution 1 : 15,000 and 1 : 20,000, respectively, followed by 2-h incubation at room temperature. Incubation with the secondary anti-mouse antibodies conjugated with horseradish peroxidase (Sigma; A4416; dilution, 1 : 15,000) was carried out for 1 h at room temperature. ABTS substrate (MP Biomedicals, France; 0.04%, w/v) in 50 mM phosphate-citrate buffer containing 0.009% (v/v) hydrogen peroxide was added to each well, and the optical density at 405 nm was measured after 10, 20, and 40 min [9].
Conjugation of recombinant proteins and virus particles using EDC/NHS. Purified TMV-N-lys particles (10 µg/µl) were mixed with the dialyzed protein (20 μg/μl) in 0.1 M PBS buffer. EDC and NHS were added sequentially to the mixture to the final concentration of 5 and 1 mM, respectively. The conjugation reaction was carried out for 30 min at room temperature followed by 2-h incubation at 4°C with vigorous mixing [19].
Dynamic light scattering (DLS). The size of the viral particles and their complexes with the recombinant protein was determined by DLS based on the evaluation of particle hydrodynamic diameter from its diffusion coefficient (Stokes–Einstein equation) using a Zetasizer Nano ZS instrument (Malvern Instruments Ltd, United Kingdom) equipped with a helium-neon laser (633 nm). Each sample was tested 50 times according to the manufacturer’s protocol; the data were processed with a built-in Dispersion Technology Software (DTS) v. 5.10.
RESULTS
Design and purification of genetically modified viral particles containing reactive lysine residue. The obtained viral vector provided exposure of reactive lysine (K) on the surface of TMV particles; we introduced the ADFK (alanine, aspartic acid, phenylalanine, lysin) sequence into the N-terminus of CP using the full-size TMV cDNA [15]. The nucleotide sequence encoding the ADFK peptide was optimized for plant-based expression driven by the virus, according to the codon usage in A. thaliana, N. tabacum, and two viral CPs efficiently accumulated during infection of plants with tobacco mosaic virus (TMV-U1) and Alternanthera mosaic virus (AltMV-MU) [9]. The following codons sequences were selected: GCU for A; GAC for D; UUC for F; and AAG for K.
The sequence coding for ADFK in the N-terminus of the TMV CP was inserted by the overlap extension PCR. Next, the generated CPADFK fragment was ligated with the binary pBIN-TMV-wt vector, and the resulting pBIN-TMV-N-lys construct was used for the transformation of agrobacteria (Fig. 1a). The stability of the pBIN-TMV-N-lys binary vector in the agrobacteria was checked by PCR analysis of the formed colonies with specific primers. The presence of the foreign sequence in the PCR product was confirmed by sequencing. Two-week-old N. benthamiana plants were infiltrated with the produced agrobacterial culture.
The first pronounced symptoms of infection manifested as leaf yellowing (chlorosis) and curling of upper non-inoculated (systemic) leaves were observed 10 days post-inoculation, followed by deformation (bending) of the upper part of the stem (Fig. 1b). Systemic leaves were collected for further analysis on day 14 post inoculation. Electrophoresis of the plant extracts demonstrated the presence of a protein band with electrophoretic mobility corresponding to ~21 kDa, which was absent in the control non-inoculated plants (Fig. 1c). TMV-N-lys viral particles were isolated from the leaves of infected plants employing polyethylene glycol (PEG) precipitation followed by ultracentrifugation and electrophoretic analysis in a gradient polyacrylamide gel (Fig. 1d) that demonstrated the presence of a single protein band with electrophoretic mobility corresponding to ~21 kDa, which was in good agreement with the size of TMV-N-lys CP.
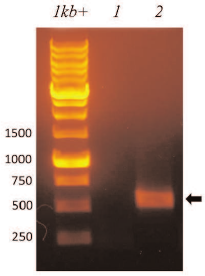
Viral RNA was isolated from the purified recombinant TMV-N-lys viral particles using RNeasy Mini-Kit (Qiagen, Germany) and used for reverse transcription-PCR (RT-PCR) with primers specific to the ADFK nucleotide sequence. Wild-type TMV particles (TMVwt) were used as a negative control. Analysis of the RT-PCR products by electrophoresis in 2% agarose gel confirmed the presence of the introduced sequence in the chimeric TMV-N-lys particles and genetic stability of the viral vector (Fig. 2).
Fig. 2. Electrophoretic analysis in 2% agarose gel of RT-PCR products obtained on isolated RNA samples. Lanes: 1 kb+, size markers (bp); 1) TMV U1, negative control for the presence of ADFK sequence; 2) TMV-N-lys particles; 600-bp band points to ADFK sequence (arrow).
The structural stability of the formed chimeric TMV-N-lys viral particles was evaluated by electron microscopy (Fig. 3b). It was found that the recombinant particles were similar in size to the particles of the wild-type TMV but differed morphologically due to a less compact structure resulting in the slightly curved shape of the virions.
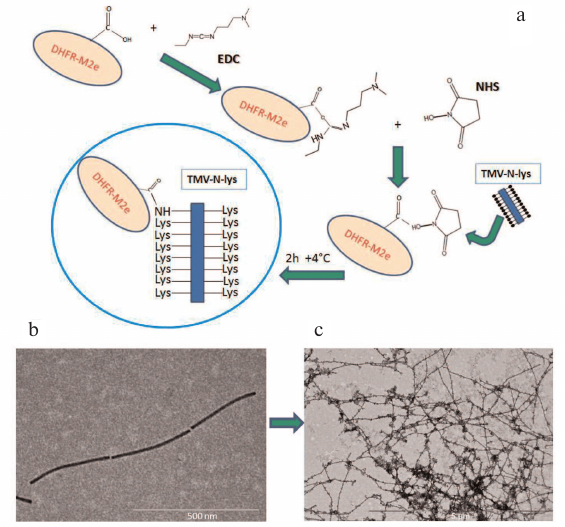
Fig. 3. Preparation and characterization of the virion–protein complexes. a) Conjugation of TMV-N-lys viral particles with DHFR-M2e using EDC and NHS reagents. The reaction was optimized based on the recommendation of NHS manufacturer and previously published technique [19]. b) Electron microphotograph of purified TMV-N-lys viral particles; magnification, 30,000×; negative contrast with 2% uranyl acetate. c) Electron microphotograph of the virion–protein complexes; magnification, 10,000×; negative contrast with 2% uranyl acetate.
Characterization of protein–virion complexes. Purified TMV-N-lys particles (10 μg/μl) containing reactive lysine were conjugated with the recombinant protein containing conserved M2e epitope of the influenza A virus. The recombinant protein (DHFR-M2e) composed of the mouse dihydrofolate reductase (DHFR) fused the M2e epitope (25.5 kDa) was isolated previously in our laboratory via expression in E. coli followed by purification on Ni-NTA agarose [9].
The recombinant protein was dialyzed against water (MilliQ) and subjected to chemical modifications. Unlike previous publication [18], we performed sequential transformation of free COOH-groups in the recombinant protein into activated carboxylic acid esters using 5 mM N-cyclohexyl-N-(2-morpholinoethyl)carbodiimide-metho-p-toluenesulfonate (EDC analogue, further indicated in the text as EDC), followed by their transformation into intermediate NHS-ester using 1 mM N-hydroxysuccinimide (NHS). NHS reaction with carboxylic acid produced carboxylic acid activated esters that reacted with amino groups of lysine side chains on the surface of chimeric TMV-N-lys particles with the formation of carboxylic acid amides (see Fig. 3a for the reaction conditions). In order to develop the most efficient conjugation technique, we performed a series of test experiments with chemical reagents (EDC, NHS) and different buffer solutions; we found that the most effective protein binding occurred in PBS when both EDC and NHS were used. The obtained complexes were examined by electron microscopy (Fig. 3c) and SDS-PAGE.
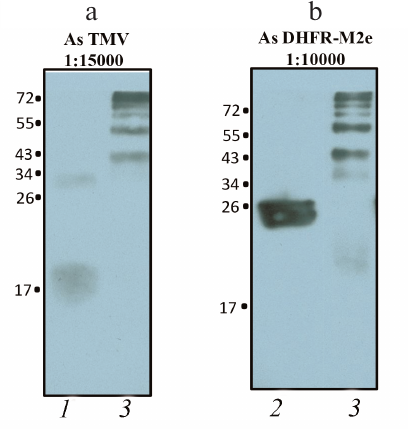
To confirm that the high molecular weight products of the conjugation reaction contained both TMV-N-lys particles and DHFR-M2e, they were analyzed by Western blotting using specific antibodies against TMV CP and DHFR-M2e produced in our laboratory [9]. The presence of appropriate bands with expected mobility confirmed successful conjugation of the protein with the viral particles (Fig. 4, a and b).
Fig. 4. Western blot analysis of virion–protein complexes produced by the DHFR-M2e conjugation with TMV-N-lys particles: 1) TMV CP (positive control); 2) DHFR-M2e recombinant protein; 3) TMV-N-lys complexes with DHFR-M2e. a) Staining with anti-TMV antibodies (dilution, 1 : 15,000); b) staining with anti-DHFR-M2e antibodies (dilution, 1 : 10,000).
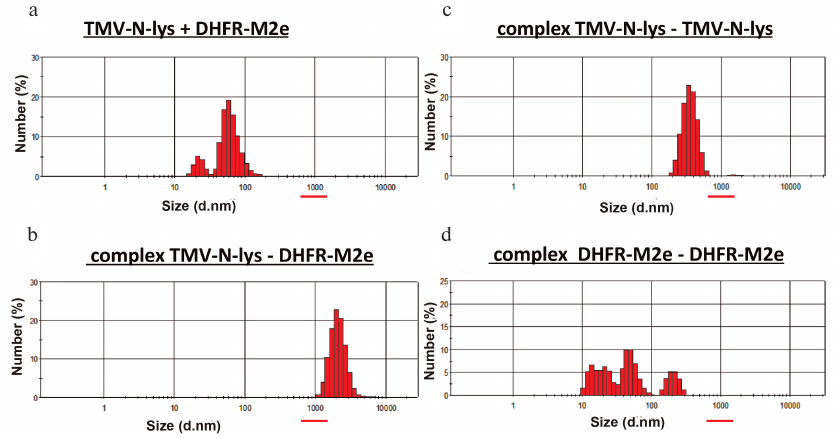
The stability of the obtained complex was studied by DLS. Each sample of the complex or control virus was examined 50 times. In order to avoid negative effects of NHS heterocycle, conjugation reactions were carried out in the presence of EDC at room temperature overnight. Analysis of the obtained data (Fig. 5) revealed that mixing the viral particles with the protein without conjugation produced complexes 10-150 nm in size, while the particles produced in the presence of EDC demonstrated a shift to the high-molecular-weight region with the size reaching ~2000 nm. In order to prove that the high-molecular-weight components represented the products of protein conjugation with viral particles, rather than association of each component to each other, we performed control experiments that showed that TMV-N-lys–TMV-N-lys particles ranged in size from 100 to 800 nm (with a peak size of 350 nm), while the size of the DHFR-M2e–DHFR-M2e complexes did not exceed 300 nm. Thus, we concluded that the TMV-N-lys–DHFR-M2e complexes were indeed formed as a result of conjugation reaction, since their size significantly differed from the size of complexes in the control experiments.
Fig. 5. DLS characterization of produced TMV-N-lys complexes with DHFR-M2e. a) Mixture of TMV-N-lys particles with DHFR-M2e before the conjugation reaction; b) complexes of TMV-N-lys particles with DHFR-M2e produced by conjugation; c) negative control (complexes of TMV-N-lys particles alone); d) negative control (complexes of DHFR-M2e protein alone).
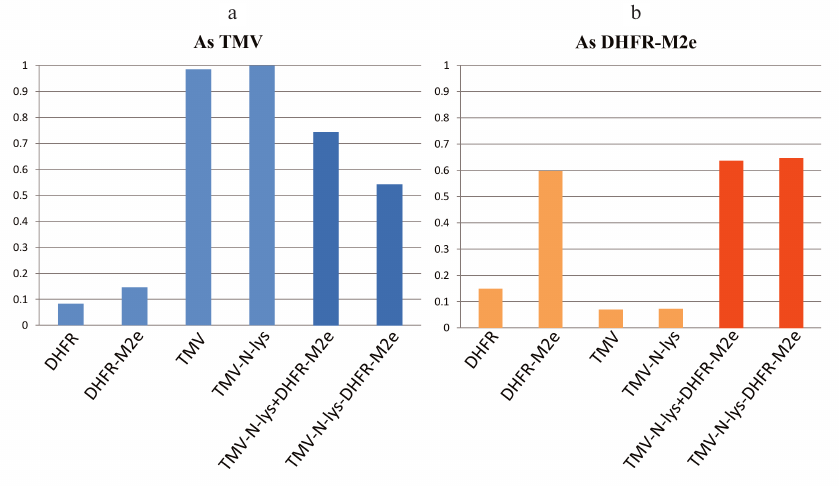
The results of indirect enzyme-linked immunoassay (ELISA) provided additional evidence for the formation of stable virion–protein complexes (Fig. 6). Microplates loaded with the samples (in triplicate) were incubated overnight at 4°C. On the next day, antibodies against TMV or DHFR-M2e were added to the wells and further steps were conducted according to the standard indirect ELISA protocol. The use of secondary antibodies and ABTS substrate allowed visualization of the enzyme reaction producing color reaction product. The amount of the product was checked with a Multiskan FC microbiological analyzer (Thermo Scientific, USA) at 405 nm after 10, 20, and 40 min of incubation. It was found that the signal from the TMV-N-lys–DHFR-M2e complexes incubated with the anti-TMV antibodies was 1.5 times weaker than the signal from the virion–protein mixture without conjugation. This indicated that the virions were almost entirely coated with the DHFR-M2e protein and the binding of the anti-TMV antibodies to them was less efficient than in the case of simple mixing of the two components (virions and protein). The signals from the samples incubated with the anti-DHFR-M2e antibodies were virtually identical to the conjugated complexes and virion–protein mixtures, which implied uniform coating of viral particles with the protein.
Fig. 6. Characterization of complexes between TMV-N-lys particles and DHFR-M2e by indirect ELISA (results from three independent experiments): a) samples incubated with anti-TMV antibodies; b) samples incubated with anti-DHFR-M2e antibodies; TMV-N-lys + DHFR-M2e, mixture of equimolar amounts of viral particles and protein.
Therefore, we concluded that the produced complexes of TMV-N-lys particles with the surface-exposed epitope M2e of the influenza A virus are stable and can be used for animal immunization in order to further improve the existing universal vaccine candidates against influenza A virus [9, 20].
DISCUSSION
According to the published data, the use of TMV as a safe platform provides a number of advantages for developing multivalent vaccines based on the virion–protein biocomplexes against a broad spectrum of human and animal diseases.
The immunogenic properties of the wild-type TMV have been investigated in detail for many years [12]. It was established recently that TMV virions can serve as a successful adjuvant, in particular, for repeated immunization [11]. The usage of TMV particles as a carrier of antigenic proteins and peptides results in the emergence of respective antibodies in the blood. It should be emphasized that these are not “junk” antibodies. According to the data reported by Liu et al., the presence of antibodies against TMV in the blood of smokers positively correlates with the decreased risk of developing Parkinson’s and Alzheimer diseases [21]. Moreover, the presence of these antibodies practically does not affect the results of further vaccinations [11]. Thus, a vaccine based on the virion–protein biocomplexes will be multifunctional, as the antibodies against the target peptide will be supplemented with functional antibodies against the carrier.
The shape of particles used for immunization is important. Thus, synthetic polymer nanoparticles with a micelle-like shape circulate longer in an organism in comparison with similar spherical particles. It was suggested that this effect could be associated with less efficient capture of elongated particles by mononuclear phagocytes [22]. Similar results were reported for gold nanoparticles [23]. Immunization of mice with the TMV particles (300 nm in length) and spherical particles (diameter, ~50 nm) produced by thermal modification of TMV virions showed that the clearance rate for the spherical particles was higher than for the rod-shaped ones [14].
Recombinant subunit vaccines are apparently safer comparing to inactivated or live attenuated vaccines. Current studies on the development of subunit vaccine against tularemia revealed several Francisella tularensis antigens capable of inducing partial protective immune response [24, 25]. It was shown that the protective potential of vaccine increased when multiple antigens were present in its composition [26]. One of the essential problems in the development of multivalent subunit vaccines is efficient delivery of such antigens across the mucous membranes. On the contrary, induction of reliable immune response without adjuvants is a significant advantage for developing new vaccination strategies based on TMV conjugation.
Two different approaches were used for evaluating the potential for developing a multivalent vaccine: (i) vaccine design targeting all proteins to the same TMV virion (monoconjugate TMV vaccine); or (ii) using a mixture of target proteins independently conjugated to their “own” TMV particles (multiconjugate TMV vaccine). Both vaccine types induce formation of antibodies against all recombinant proteins; moreover, purification and/or conjugation procedures do not change the conformation of the native epitopes. Comparison of the immune responses to the mono- and multiconjugate vaccines demonstrated that immobilization of all proteins on the same particle is less advantageous, while separate binding of each antigen to TMV in equimolar amounts ensures efficient delivery of multiple antigens [18]. TMV-conjugated vaccine is safe and can be administered to mice in multiple doses without side effects [27]. In order to design an efficient multiconjugate vaccine, various conserved antigens of the influenza virus can be used, including M1 and NP proteins together with conserved hemagglutinin stalk domain.
Another approach to creating multivalent subunit vaccine may be associated with a combination of genetic mutations and chemical conjugation in the same TMV particle. To develop chimeric particles based on the TMV genome, a foreign sequence can be cloned into the open reading frame of the CP gene. It was demonstrated previously that such particles could provide protection against influenza A [9, 20], papilloma [28], and foot-and-mouth disease [29] viruses, as well as be used in anticancer therapy [30, 31]. In particular, TMV-M2e-ala and TMV-M2e-ser particles generated previously in our laboratory contained up to 90% of recombinant protein in the purified preparations. The ratio of antibodies against the epitope and the carrier was 5 : 1 in the case of intraperitoneal immunization of mice, which indicated stability of these particles in the animal post injection; this ratio was significantly higher than the one reported previously (approximately 1 : 1) [30, 32]. The TMV-M2e-ala nanovaccine provided protection against five lethal doses of homologous and heterologous strains of influenza A virus. This degree of protection is considered very high for this type of anti-influenza vaccines. We believe that the use of conjugated protein–virion complexes generated and described in this study will increase the efficiency of vaccination due to the induction of strong humoral and cell-mediated immune responses. Furthermore, it is possible to use various combinations of chimeric particles and obtained complexes in order to increase the efficacy of multivalent universal vaccines against influenza [9].
Conflict of interest. The authors declare no conflict of interest in financial or any other sphere.
Ethical approval. This article does not contain description of studies with human participants or animals performed by any of the authors.
REFERENCES
1.Krammer, F. (2016) Novel universal influenza virus
vaccine approaches, Cur. Opin. Virol., 17, 95-103; doi:
10.1016/j.coviro.2016.02.002.
2.Deng, L., Cho, K. J., Fiers, W., and Saelens, X.
(2015) M2e-based universal influenza A vaccines, Vaccines,
3, 105-136; doi: 10.3390/vaccines3010105.
3.Virelizier, J. L., Allison, A. C., Oxford, J. S.,
and Schild, G. C. (1977) Early presence of ribonucleoprotein antigen on
surface of influenza virus-infected cells, Nature, 266,
52-54; doi: 10.1038/266052a0.
4.Xue, C., Tian, G., Chen, X., Liu, Q., Ma, J., Xu,
S., Li, X., Chen, H., and Cao, Y. (2015) Incorporation of conserved
nucleoprotein into influenza virus-like particles could provoke a broad
protective immune response in BALB/c mice and chickens, Virus
Res., 195, 35-42; doi: 10.1016/j.virusres.2014.09.018.
5.Gotch, F., McMichael, A., Smith, G., and Moss, B.
(1987) Identification of viral molecules recognized by influenza
specific human cytotoxic T lymphocytes, J. Exp. Med.,
165, 401-416; doi: 10.1084/jem.165.2.408.
6.Boyd, A. C., Ruiz-Hernandez, R., Peroval, M. Y.,
Carsona, C., Balkissoonb, D., Staines, K., Turner, A. V., Hill, A. V.
S., Gilbert, S. C., and Butter, C. (2013) Towards a universal vaccine
for avian influenza: protective efficacy of modified vaccinia virus
Ankara and Adenovirus vaccines expressing conserved influenza antigens
in chickens challenged with low pathogenic avian influenza virus,
Vaccine, 31, 670-675; doi:
10.1016/j.vaccine.2012.11.047.
7.Pushko, P., Pearce, M. B., Ahmad, A., and
Tretyakova, I. (2011) Influenza virus-like particle can accommodate
multiple subtypes of hemagglutinin and protect from multiple influenza
types and subtypes, Vaccine, 29, 5911-5918; doi:
10.1016/j.vaccine.2011.06.068.
8.Fiers, W., De Filette, M., Bakkouri, K., Schepens,
B., Roose, K., Schotsaert, M., Birkett, A., and Saelens, X. (2009)
M2e-based universal influenza A vaccine, Vaccine, 27,
6280-6283; doi: 10.1016/j.vaccine.2009.07.007.
9.Petukhova, N. V., Gasanova, T. V., Stepanova, L.
A., Rusova, O. A., Potapchuk, M. V., Korotkov, A. V., Skurat, E. V.,
Tsybalova, L. M., Kiselev, O. I., Ivanov, P. A., and Atabekov, J. G.
(2013) Immunogenicity and protective efficacy of candidate universal
influenza A nanovaccines produced in plants by tobacco mosaic
virus-based vectors, Curr. Pharm. Des., 19, 5587-5600;
doi: 10.2174/13816128113199990337.
10.Stepanova, L. A., Kotlyarov, R. Y., Kovaleva, A.
A., Potapchuk, M. V., Korotkov, A. V., Sergeeva, M. V., Kasianenko, M.
A., Kuprianov, V. V., Ravin, N. V., Tsybalova, L. M., Skryabin, K. G.,
and Kiselev, O. I. (2015) Protection against multiple influenza A virus
strains induced by candidate recombinant vaccine based on heterologous
M2e peptides linked to flagellin, PloS One, 10, e0119520;
doi: 10.1371/journal.pone.0119520.
11.Mallajosyula, J. K., Hiatt, E., Hume, S.,
Johnson, A., Jeevan, T., Chikwamba, R., Pogue, G. P., Bratcher, B.,
Haydon, H., Webby, R. J., and McCormick, A. A. (2014) Single-dose
monomeric HA subunit vaccine generates full protection from influenza
challenge, Hum. Vaccin. Immunother., 10, 586-595; doi:
10.4161/hv.27567.
12.Van Regenmortel, M. H. (1999) The antigenicity of
tobacco mosaic virus, Philos. Trans. R Soc. Lond. B Biol.
Sci., 354, 559-568; doi: 10.1098/rstb.1999.0407.
13.McCormick, A. A., Corbo, T. A., Wykoff-Clary, S.,
Palmer, K. E., and Pogue, G. P. (2006) Chemical conjugate TMV-peptide
bivalent fusion vaccines improve cellular immunity and tumor
protection, Bioconj. Chem., 17, 1330-1338; doi:
10.1021/bc060124m.
14.Bruckman, M. A., Randolph, L. N., VanMeter, A.,
Hern, S., Shoffstall, A. J., Taurog, R. E., and Steinmetz, N. F. (2014)
Biodistribution, pharmacokinetics, and blood compatibility of native
and PEGylated tobacco mosaic virus nano-rods and -spheres in mice,
Virology, 449, 163-173; doi:
10.1016/j.virol.2013.10.035.
15.Smith, M. L., Lindbo, J. A., Dillad-Telm, S.,
Brosio, P. M., Lasnik, A. B., McCormick, A. A., Nguyen, L. V., and
Palmer, K. E. (2006) Modified tobacco mosaic virus particles as
scaffolds for display of protein antigens for vaccine applications,
Virology, 348, 475-488; doi:
10.1016/j.virol.2005.12.039.
16.Gasanova, T. V., Petukhova, N. V., and Ivanov, P.
A. (2016) Chimeric particles of tobacco mosaic virus as a platform for
the development of next-generation nanovaccines, Nanotechnologies in
Russia, 11, 227-236; doi: 10.1134/S1995078016020051.
17.Lee, S. Y., Royston, E., Culver, J. N., and
Harris, M. T. (2005) Improved metal cluster deposition on a genetically
engineered tobacco mosaic virus template, Nanotechnology,
16, 435-441; doi: 10.1088/0957-4484/16/7/019.
18.Banik, S., Mansour, A. A., Suresh, R. V.,
Wykoff-Clary, S., Malik, M., McCormick, A. A., and Bakshi, C. S. (2015)
Development of a multivalent subunit vaccine against tularemia using
tobacco mosaic virus (TMV) based delivery system, PloS One,
10, e0130858; doi: 10.1371/journal.pone.0130858.
19.Narain, R. (2014) Chemistry of Bioconjugates:
Synthesis, Characterization, and Biomedical
Applications, John Wiley & Sons, Hoboken; doi:
10.1002/9781118775882.
20.Petukhova, N. V., Gasanova, T. V., Ivanov, P. A.,
and Atabekov, J. G. (2014) High-level systemic expression of conserved
influenza epitope in plants on the surface of rod-shaped chimeric
particles, Viruses, 6, 1789-1800; doi:
10.3390/v6041789.
21.Liu, R., Vaishnav, R. A., Roberts, A. M., and
Friedland, R. P. (2013) Humans have antibodies against a plant virus:
evidence from tobacco mosaic virus, PLoS One, 8, e60621;
doi: 10.1371/journal.pone.0060621.
22.Geng, Y., Dalhaimer, P., Cai, S. S., Tsai, R.,
Tewari, M., Minko, T., and Discher, D. E. (2007) Shape effects of
filaments versus spherical particles in flow and drug delivery, Nat.
Nanotechnol., 2, 249-255; doi: 10.1038/nnano.2007.70.
23.Arnida, Janat-Amsbury, M. M., Ray, A., Peterson,
C. M., and Ghandehari, H. (2011) Geometry and surface characteristics
of gold nano particles influence their biodistribution and uptake by
macrophages, Eur. J. Pharm. Biopharm., 77, 417-423; doi:
10.1016/j.ejpb.2010.11.010.
24.Huntley, J. F., Conley, P. G., Rasko, D. A,
Hagman, K. E., Apicella, M. A., and Norgard, M. V. (2008) Native outer
membrane proteins protect mice against pulmonary challenge with
virulent type A Francisella tularensis, Infect. Immun.,
76, 3664-3671; doi: 10.1128/IAI.00374-08.
25.Apicella, M. A., Post, D. M., Fowler, A. C.,
Jones, B. D., Rasmussen, J. A., Hunt, J. R., Imagawa, S., Choudhury,
B., Inzana, T. J., Maier, T. M., Frank, D. W., Zahrt, T. C., Chaloner,
K., Jennings, M. P., McLendon, M. K., and Gibson, B. W. (2010)
Identification, characterization and immunogenicity of an O-antigen
capsular polysaccharide of Francisella tularensis, PLoS
One, 5, e11060; doi: 10.1371/journal.pone.0011060.
26.Huntley, J. F., Conley, P. G., Hagman, K. E., and
Norgard, M. V. (2007) Characterization of Francisella
tularensis outer membrane proteins, J. Bacteriol.,
189, 561-574; doi: 10.1128/JB.01505-06.
27.Mallajosyula, J. K., Jeevan, T., Chikwamba, R.,
Webby, R. J., and McCormick, A. A. (2016) A single dose TMV-HA vaccine
protects mice from H5N1 influenza challenge, Int. J. Vaccine
Res., 1, 6; doi: 10.15226/2473-2176/1/2/00106.
28.Palmer, K. E., Benko, A., Doucette, S. A.,
Cameron, T. I., Foster, T., Hanley, K. M., McCormick, A. A., McCulloch,
M., Pogue, G. P., Smith, M. L., and Christensen, N. D. (2006)
Protection of rabbits against cutaneous papillomavirus infection using
recombinant tobacco mosaic virus containing L2 capsid epitopes,
Vaccine, 24, 5516-5525; doi:
10.1016/j.vaccine.2006.04.058.
29.Jiang, L., Li, Q., Li, M., Zhou, Z., Wu, L., Fan,
J., Zhang, Q., Zhu, H., and Xu, Z. (2006) A modified TMV-based vector
facilitates the expression of longer foreign epitopes in tobacco,
Vaccine, 24, 109-115; doi:
10.1016/j.vaccine.2005.09.060.
30.Fitchen, J., Beachy, R. N., and Hein, M. B.
(1995) Plant virus expressing hybrid coat protein with added murine
epitope elicits autoantibody response, Vaccine, 13,
1051-1057; doi: 10.1016/0264-410x(95)00075-c.
31.Frolova, O. Y., Petrunia, I. V., Komarova, T. V.,
Kosorukov, V. S., Sheval, E. V., Gleba, Y. Y., and Dorokhov, Y. L.
(2010) Trastuzumab-binding peptide display by tobacco mosaic virus,
Virology, 407, 7-13; doi:
10.1016/j.virol.2010.08.005.
32.Koo, M., Bendahmane, M., Lettieri, G. A.,
Paoletti, A. D., Lane, T. E., Fitchen, J. H., Buchmeier, M. J., and
Beachy, R. N. (1999) Protective immunity against murine hepatitis virus
(MHV) induced by intranasal or subcutaneous administration of hybrids
of tobacco mosaic virus that carries an MHV epitope, Proc.
Natl. Acad. Sci. USA, 96, 7774-7779; doi:
10.1073/pnas.96.14.7774.