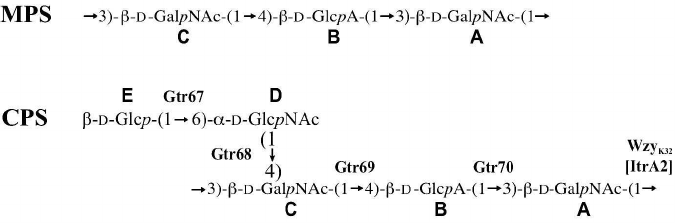Elucidation of the K32 Capsular Polysaccharide Structure and Characterization of the KL32 Gene Cluster of Acinetobacter baumannii LUH5549
S. M. Cahill1, N. P. Arbatsky2, A. S. Shashkov2, M. M. Shneider3, A. V. Popova4,5, R. M. Hall6#, J. J. Kenyon1#, and Y. A. Knirel2,a*#
1Institute of Health and Biomedical Innovation, School of Biomedical Sciences, Faculty of Health, Queensland University of Technology, QLD 4001 Brisbane, Australia2Zelinsky Institute of Organic Chemistry, Russian Academy of Sciences, 119991 Moscow, Russia
3Shemyakin and Ovchinnikov Institute of Bioorganic Chemistry, Russian Academy of Sciences, 117997 Moscow, Russia
4Moscow Institute of Physics and Technology, 141701 Dolgoprudny, Moscow Region, Russia
5State Research Center for Applied Microbiology and Biotechnology, 142279 Obolensk, Moscow Region, Russia
6School of Life and Environmental Sciences, The University of Sydney, Sydney, Australia
# These authors contributed equally to this work.
* To whom correspondence should be addressed.
Received October 29, 2019; Revised November 20, 2019; Accepted November 26, 2019
Capsular polysaccharide (CPS), isolated from Acinetobacter baumannii LUH5549 carrying the KL32 capsule biosynthesis gene cluster, was studied by sugar analysis, Smith degradation, and one- and two-dimensional 1H and 13C NMR spectroscopy. The K32 CPS was found to be composed of branched pentasaccharide repeats (K units) containing two residues of β-D-GalpNAc and one residue of β-D-GlcpA (β-D-glucuronic acid) in the main chain and one residue each of β-D-Glcp and α-D-GlcpNAc in the disaccharide side chain. Consistent with the established CPS structure, the KL32 gene cluster includes genes for a UDP-glucose 6-dehydrogenase (Ugd3) responsible for D-GlcA synthesis and four glycosyltransferases that were assigned to specific linkages. Genes encoding an acetyltransferase and an unknown protein product were not involved in CPS biosynthesis. Whilst the KL32 gene cluster has previously been found in the global clone 2 (GC2) lineage, LUH5549 belongs to the sequence type ST354, thus demonstrating horizontal gene transfer between these lineages.
KEY WORDS: Acinetobacter baumannii, capsule, K locus, polysaccharide structure, glucuronic acidDOI: 10.1134/S000629792002011X
Abbreviations: CPS, capsular polysaccharide; KL, chromosomal K locus; K unit, oligosaccharide repeat; MPS, modified polysaccharide; PSgc, polysaccharide gene cluster.
Capsular polysaccharide (CPS) is produced on the cell surface of many
bacterial pathogens including the nosocomial species, Acinetobacter
baumannii. It serves as a major pathogenicity determinant by
providing a layer protecting against host immune factors [1, 2]. It also protects cells from
desiccation and other environmental stressors [3,
4]. CPS is composed of a long chain of
oligosaccharide repeats (K units), which consist of two to six sugar
residues of various types joined by specific glycosidic linkages. The
biosynthesis and export of CPS in A. baumannii is mostly
determined by the genes at the chromosomal K locus (KL), which comes in
diverse forms [5].
The K locus includes an operon of genes (wza-wzb-wzc) responsible for the CPS export, and a divergently transcribed region for the synthesis of specific K units. The latter includes genes for sugar synthesis, initiation of K-unit synthesis (itr), glycosyl transfer (gtr), K-unit translocation (wzx) and polymerization (wzy), and occasionally for addition of acetyl or pyruvyl groups (atr/ptr).
The arrangement of KL gene clusters carried by isolates from the Traub collection that were used to establish the original serotyping scheme for A. baumannii [6] had been examined previously [7]. However, these gene clusters were originally described as polysaccharide gene clusters (PSgc) and annotated using a traditional nomenclature system. The structures of CPSs produced by many of these isolates have now been established and, for all of them, their sugar content and configuration correlate perfectly with the genes in the gene cluster found at the K locus [8-12]. Accordingly, the annotations of these gene clusters have been revised [8-14] in accordance with the more transparent and widely adopted K locus nomenclature system for A. baumannii CPS biosynthesis gene clusters [5].
In this work, we establish for the first time the CPS structure of another strain from this collection, LUH5549, which carries the KL32 gene cluster, previously designated PSgc21.
MATERIALS AND METHODS
Bacterial strain and cultivation of bacteria. Acinetobacter baumannii LUH5549 was originally from the W. H. Traub collection at the Institut fur Medizinische Mikrobiologie und Hygiene, Universitat des Saarlandes (Saarland, Germany) and was kindly provided by Prof. Peter Reeves. The bacteria were cultivated in 2TY media overnight; the cells were harvested by centrifugation (10,000g, 20 min), washed with phosphate buffered saline, resuspended in aqueous 70% acetone, precipitated, and dried.
Isolation of CPS. CPS preparation was obtained by phenol–water extraction [15] of bacterial cells (1.1 g); the extract was dialyzed without layer separation and clarified from insoluble contaminants by centrifugation. The resulting solution was treated with cold (4°C) 50% aqueous CCl3COOH; after centrifugation, the supernatant was dialyzed against distilled water and freeze-dried to yield a crude CPS sample (370 mg). For CPS purification, this sample (120 mg) was heated with 2% aqueous AcOH at 100°C for 2 h, and CPS (27 mg) was purified by gel-permeation chromatography on a Sephadex G50 Superfine column 60 × 2.5 cm in 0.1% aqueous AcOH; eluted fractions were monitored with a differential refractometer (Knauer, Germany).
Monosaccharide analysis. Alditol acetates [16] were obtained by CPS hydrolysis with 2 M CF3COOH (120°C, 2 h) and analyzed by gas-liquid chromatography on an HP-5 capillary column with a Maestro (Agilent 7820) chromatograph (Interlab, Russia) using a temperature gradient of 160°C (1 min) to 290°C at 7°C/min. Glucuronic acid was identified by anion-exchange chromatography on a DA×8 resin column (7 × 0.4 cm) in 5 mM potassium phosphate buffer (pH 3) at 70°C; elution was monitored using bicinchoninic acid assay.
Smith degradation. CPS sample (12 mg) was oxidized with NaIO4 (29 mg in 1.5 ml water) at 20°C for 72 h in the dark and then reduced with NaBH4 (30 mg) for 24 h; water was evaporated and boric acid was removed by evaporation with 10% aqueous AcOH in methanol (three times). The degradation product was desalted on a Sephadex G-25 column (108 × 1.2 cm) in water; elution was monitored as described above. The obtained polymer was hydrolyzed with 2% aqueous AcOH (100°C, 2 h), and the products of hydrolysis were fractionated by gel-permeation chromatography on Sephadex G-25 as described above to yield modified polysaccharide (MPS) (2.6 mg).
NMR spectroscopy. The samples were deuterium-exchanged by freeze-drying from 99.9% D2O and then examined after dissolving in 99.95% D2O. The NMR spectra were recorded on an Avance II 600 MHz spectrometer (Bruker, Germany) at 60°C. Sodium 3-trimethylsilylpropanoate-2,2,3,3-d4 (δH 0, δC –1.6) was used as an internal reference for calibration. 2D NMR spectra were obtained using standard Bruker software; NMR data were acquired and processed with the Bruker TopSpin 2.1 program. A spin-lock time of 60 ms and a mixing time of 150 ms were used in TOCSY and ROESY experiments, respectively. A 60-ms delay was used for the evolution of long-range coupling to optimize 1H,13C HMBC experiments.
Bioinformatics analysis. Short reads for the LUH5549 genome sequence were obtained from the Sequence Read Archive (SRA number DRS005644) and assembled into contigs using SPAdes [17]. The KL32 gene cluster was identified between the fkpA and lldP genes and re-annotated using the nomenclature system described previously [5]. CAZy (http://www.cazy.org/) [18] and Pfam (https://pfam.xfam.org) [19] databases were used to assign encoded proteins to their respective biosynthesis roles. The assembled sequence and the newly assigned annotations are available in GenBank under accession number KC526897.2. The genome sequence was assigned to a sequence type (ST) using the Pasteur MLST scheme for A. baumannii.
RESULTS AND DISCUSSION
The sequence of the PSgc21 CPS biosynthesis gene cluster from A. baumannii LUH5549 (GenBank accession number KC526897.1) was found to be incomplete as it lacked the wza-wzb-wzc export genes, indicating a potential assembly issue. As the sequence had been previously assembled from short reads using Velvet 2.0 [7], these short reads (SRA number DRS005644) were reassembled into contigs using SPAdes. The complete sequence of the gene cluster between the conserved fkpA and lldP genes that flank the K locus was identified and found to be 99.56% identical to the KL32 gene cluster previously described for the A. baumannii Vietnamese isolate, BAL_058 (GenBank accession number KT359615.1) [20]. Hence, the LUH5549 gene cluster was renamed KL32 and its genes were reannotated according to the established nomenclature system [5]. The updated sequence and the annotations were deposited in the GenBank under accession number KC526897.2.
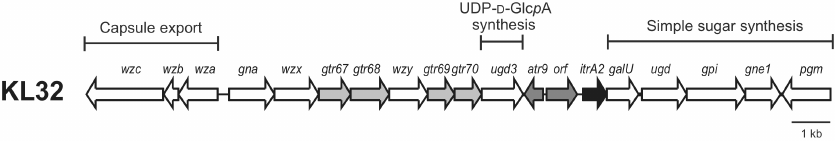
KL32 (Fig. 1) includes genes responsible for the K unit processing (wzy and wzx) and a galU-ugd-gpi-gne1-pgm gene module responsible for the synthesis of simple sugars, such as UDP-D-Glcp, UDP-D-GlcpNAc, and UDP-D-GalpNAc. Adjacent to galU, there is an itrA2 gene known to be responsible for initiating K unit synthesis by adding a D-GalpNAc residue as the first sugar of the K unit to the lipid carrier in the inner membrane [21]. The central region of KL32 also includes four predicted glycosyltransferase genes (gtr67, gtr68, gtr69, gtr70) for four internal K unit sugar linkages, an acetyltransferase gene (atr9), and a gene encoding an unidentified protein product (Orf).
Fig. 1. The KL32 capsule biosynthesis gene cluster from A. baumannii LUH5549. Arrows indicate the direction of transcription and are drawn to scale using the sequence from GenBank accession number KC526879.2. Genes for glycosyltransferases (gtr) are depicted in light grey; initiating transferase gene (itrA2) is shown in black. Genes shown in dark grey (atr9, orf) have no established role in the synthesis of the K32 CPS.
In the central region, another ugd gene, designated ugd3, is also present. Ugd3 (GenPept accession number AHB32286.1) is only 20% identical (74% coverage) to the product of the ugd gene (GenPept accession number AHB32291.1) located in the simple sugar synthesis gene module, which was previously predicted to be responsible for the conversion of UDP-D-Glcp to UDP-D-glucuronic acid (D-GlcpA) [5]. However, though ugd is a feature common to all A. baumannii KL gene clusters, its role in the CPS synthesis has never been established. Ugd3 is also 27% identical (71% coverage) to the second Ugd type (Ugd2) identified in the species, encoded in the central region of the KL20 and KL21 gene clusters. K20 and K21 CPSs contain D-GlcpA, which correlates with the presence of Ugd2 (GenPept accession numbers AUG44319.1 and AIT56461.1) [22]. The presence of ugd3 also in the central region of KL32 specific to the CPS synthesis suggests that the K32 CPS may also contain D-GlcpA.
For the structure elucidation, CPS was isolated from the bacterial mass by phenol/water extraction followed by heating with 2% AcOH to remove contaminating short-chain lipopolysaccharides and then purified by gel-permeation chromatography on Sephadex G-50 Superfine. Sugar analysis using gas-liquid chromatography of alditol acetates derived after full acidic hydrolysis of CPS revealed the presence of Glc, GlcNAc, and GalNAc in the 1 : 1.6 : 1.8 ratio (detector response). In addition, GlcA was identified by anion-exchange chromatography. The absolute configuration of the monosaccharides was not determined chemically but inferred from the genetic data (see above).
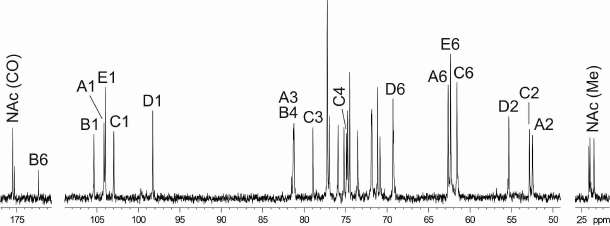
The 1H NMR and 13C NMR (Fig. 2) spectra of the CPS showed signals for five monosaccharide residues and three N-acetyl groups. Assignment of the spectra using two-dimensional 1H,1H COSY, 1H,1H TOCSY, 1H,1H ROESY, 1H,13C HSQC, and 1H,13C HMBC experiments revealed spin systems for one residue each of β-Glc (unit E), β-GlcA (unit B), α-GlcNAc (unit D), and two residues of β-GalNAc (units A and C), all being in the pyranose form (table). The α-gluco configuration of unit D, β-gluco configuration of units B and E, and β-galacto configuration of units A and C were inferred from the 3JH,H coupling constants for the ring protons, which were estimated from the one- and two-dimensional NMR spectra. The anomeric configuration of units A-C and E was confirmed by H-1/H-5 correlations in the ROESY spectrum.
Fig. 2. 13C NMR spectrum of the K32 CPS from A. baumannii LUH5549. Numbers refer to carbons in sugar residues denoted by letters as indicated in the table and Fig. 3.
1H and 13C NMR chemical shifts (ppm) of the K32
CPS from A. baumannii LUH5549
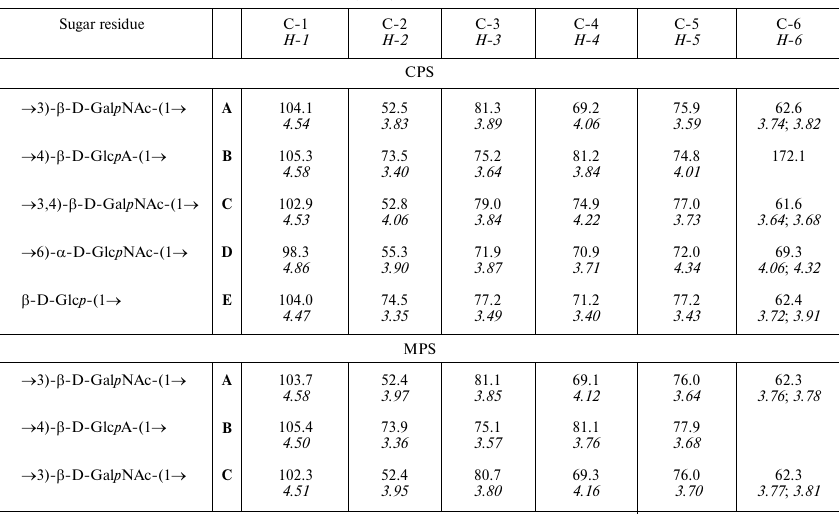
Notes: 1H NMR chemical shifts are italicized. Chemical shifts
for the N-acetyl groups are δH 1.96-2.06,
δC 23.3-24.0 (Me) and 175.3-175.5 (CO).
Low-field positions of signals for C-3 of units C and A at δ .0 and 81.3, C-4 of units C and B at δ 74.9 and 81.2, respectively, as well as C-6 of unit D at δ 69.3 (table), as compared with their positions in the corresponding non-substituted monosaccharides [23, 24], showed that the CPS is branched and allowed determination of the glycosylation pattern in the K unit. Accordingly, the 13C NMR chemical shifts for C-2,3,4,5,6 of the terminal unit E (table) were close to those of unsubstituted β-glucopyranose [23].
The 1H,13C HMBC spectrum of the CPS [Fig. S1; see Supplement to this paper on the web sites of the journal (http://protein.bio.msu.ru) and Springer (Link.springer.com)] showed correlations between the anomeric protons and linkage carbons at δ 4.47/69.3, 4.53/81.2, 4.54/79.0, 4.58/81.3, and 4.86/74.9, which were assigned to the E H-1/D C-6, C H-1/B C-4, A H-1/C C-3, B H-1/A C-3, and D H-1/C C-4 correlations, respectively. The glycosylation pattern of the CPS was confirmed by correlations of the anomeric protons and anomeric carbons with protons at the linkage carbons in the 1H,1H ROESY (see Fig. S2 in the Supplement) and 1H,13C HMBC spectra, respectively.
Therefore, the K32 CPS of A. baumannii LUH5549 has the structure shown in Fig. 3, which, to our knowledge, is unique among the known bacterial polysaccharide structures deposited in the Bacterial Carbohydrate Structure Database (BCSD; http://csdb.glycoscience.ru/bacterial/) [25]. This structure was confirmed by Smith degradation, which yielded a modified polysaccharide (MPS). Its structure (Fig. 3) was established by one- and two-dimensional NMR spectroscopy as described above for the CPS (see the table for assigned 1H and 13C NMR chemical shifts of the MPS).
Fig. 3. Structures of the K32 CPS from A. baumannii LUH5549 and MPS derived by Smith degradation of the CPS. Glycosyltransferases, Wzy polymerase, and ItrA2 initiating transferase are shown near the linkage they presumably form.
The K32 CPS was found to be composed of pentasaccharide K units that include the common sugars, D-GalpNAc, D-GlcpNAc, and D-Glcp, as well as D-GlcpA, as predicted. We previously suggested that the ugd gene present in the module for the simple sugar synthesis may be redundant with respect to the CPS production [5], and that D-GlcpA can only be a component of the A. baumannii CPS when a second ugd gene is present in the corresponding KL gene cluster [22]. Indeed, KL32 contains a ugd3 gene in the central region, and as a result, K32 CPS contains D-GlcpA.
As the CPS structure determined includes two D-GalpNAc residues (Fig. 3), it was unclear which residue was the first monosaccharide of the K32 unit that is added by ItrA2 (GenPept accession number AHB32289.2). However, the identity of the first sugar was determined by examining the linkage formed by the WzyK32 polymerase responsible for joining K units together into the CPS chain, as this linkage would connect the first and the last sugars in the chain of K units. WzyK32 (GenPept accession number AHB32283.1) was found to be 28% identical to WzyK116 encoded by A. baumannii KL116 (GenBank accession number MK399425.1), which forms the β-D-GalpNAc-(1→3)-D-GalpNAc linkage [26]. As an identical linkage is present in the K32 unit, WzyK32 was assigned to it. Therefore, the structure of the K32 unit begins with the monosubstituted D-GalpNAc residue as drawn in Fig. 3.
The assembly of this K unit would therefore require three inverting glycosyltransferases to form the three β linkages, and one glycosyltransferase with a retaining mechanism for the only α linkage. Only one glycosyltransferase encoded by KL32, Gtr68K32 (GenPept accession number AHB32282.2), was identified as a retaining enzyme belonging to the GT4 family of retaining glycosyltransferases in the CAZy database [18]. Therefore, Gtr68K32 was assigned to the α-D-GlcpNAc-(1→4)-D-GalpNAc linkage.
The remaining glycosyltransferases were assigned to linkages in the K32 unit based on their homology to enzymes with known or predicted activities as follows. Gtr70K32 (GenPept accession number AHB32285.1) is 54% identical to WdbN from the Escherichia coli O143 O-antigen gene cluster (GenPept accession number STM86374.1), and the O143 structure contains the β-D-GlcpA-(1→3)-D-GlcpNAc linkage [27]. A similar linkage, β-D-GlcpA-(1→3)-D-GalpNAc, is present in K32 (Fig. 3), and therefore Gtr70K32 was assigned to this linkage. Gtr67K32 (GenPept accession number AHB32281.1) was found to be 27% identical to Gtr75K37 encoded by A. baumannii KL37 gene cluster (GenBank accession number KX712115.1). The structure of K37 is known [26], and Gtr75K37 was predicted to catalyze formation of the β-D-Glcp-(1→6)-D-GalpNAc linkage. Hence, the β-D-Glcp-(1→6)-D-GlcpNAc side branch of K32 would be formed by Gtr67K32. Finally, Gtr69K32 (GenPept accession number AHB32284.1) that falls into the glycos_transf_2 family (Pfam PF00535) of glycosyltransferases, returns no significant hits to other proteins in BLASTp. However, as there was only one linkage left to be assigned in the K32 structure, Gtr69K32 was assigned to the β-D-GalpNAc-(1→4)-D-GlcpA linkage (Fig. 3).
Since the K32 unit contains no acetyl or other acyl groups, and all its structural features were attributed to the presence of other genes encoded by KL32, a role for Atr9 (GenPept accession number AHB32287.2) and Orf (GenPept accession number AHB32288.2) in the synthesis of the CPS could not be established.
Previously, the KL32 gene cluster had been identified in the widely disseminated clonal group, Global Clone 2 (GC2; equivalent to sequence type ST2 in the Pasteur MLST scheme) [20]. We found that A. baumannii LUH5549 belongs to another type, ST354, suggesting that the KL32 gene cluster is distributed amongst multiple distinct clonal lineages.
Funding. This work was supported by the Russian Science Foundation (project no. 19-14-00273) to YAK and Australian Research Council (ARC) DECRA Fellowship 180101563 to JJK.
Conflict of interest. The authors declare no conflict of interest in financial nor in any other area.
Ethical approval. This article does not contain any studies with human participants or animals performed by any of the authors.
REFERENCES
1.Russo, T., Luke N., Beanan, J., Olson, R.,
Sauberan, S., MacDonald, U., Schultz, L., Umland, T., and Campagnari,
A. (2010) The K1 capsular polysaccharide of Acinetobacter
baumannii strain 307-0294 is a major virulence factor, Infect.
Immun., 78, 3993-4000.
2.Singh, J. K., Adams, F. G., and Brown, M. H. (2018)
Diversity and function of capsular polysaccharide in Acinetobacter
baumannii, Front. Microbiol., 9, 3301.
3.Tipton, K. A., Chin, C. Y., Farokhyfar, M., Weiss,
D. S., and Rather, P. N. (2018) Role of capsule in resistance to
disinfectants, host antimicrobials, and desiccation in Acinetobacter
baumannii, Antimicrob. Agents Chemother., 62,
e01188-18.
4.Geisinger, E., Huo, W., Hernandez-Bird, J., and
Isberg, R. R. (2019) Acinetobacter baumannii: envelope
determinants that control drug resistance, virulence, and surface
variability, Ann. Rev. Microbiol., 73, 481-506,
doi: 10.1146/annurev-micro-020518-115714.
5.Kenyon, J. J., and Hall, R. M. (2013) Variation in
the complex carbohydrate biosynthesis loci of Acinetobacter
baumannii genomes, PLoS One, 8, e62160.
6.Traub, W. H. (1989) Acinetobacter baumannii
serotyping for delineation of outbreaks of nosocomial cross-infection,
J. Clin. Microbiol., 27, 2713-2716.
7.Hu, D., Liu, B., Dijkshoorn, L., Wang, L., and
Reeves, P. R. (2013) Diversity in the major polysaccharide antigen of
Acinetobacter baumannii assessed by DNA sequencing, and
development of a molecular serotyping scheme, PLoS One,
8, e70329, doi: 10.1371/journal.pone.0070329.
8.Senchenkova, S. N., Kenyon, J. J., Jia, T., Popova,
A. V., Shneider, M. M., Kasimova, A. A., Shashkov, A. S., Liu, B.,
Hall, R. M., and Knirel, Y. A. (2019) The K90 capsular polysaccharide
produced by Acinetobacter baumannii LUH5553 contains
di-N-acetylpseudaminic acid and is structurally related to the K7
polysaccharide from A. baumannii LUH5533, Carbohydr.
Res., 479, 1-5, doi: 10.1016/j.carres.2019.04.008.
9.Shashkov, A. S., Senchenkova, S. N., Popova, A. V.,
Mei, Z., Shneider, M. M., Liu, B., Miroshnikov, K. A., Volozhantsev, N.
V., and Knirel, Y. A. (2015) Revised structure of the capsular
polysaccharide of Acinetobacter baumannii LUH5533 (serogroup O1)
containing di-N-acetyllegionaminic acid, Russ. Chem. Bull.
Int. Ed., 64, 1196-1199, doi: 10.1007/s11172-015-1000-9.
10.Senchenkova, S. N., Popova, A. V., Shashkov, A.
S., Shneider, M. M., Mei, Z., Arbatsky, N. P., Liu, B., Miroshinikov,
K. A., Volozhantsev, N. V., and Knirel, Y. A. (2015) Structure of a new
pseudaminic acid-containing capsular polysaccharide of Acinetobacter
baumannii LUH5550 having the KL42 capsule biosynthesis locus,
Carbohydr. Res., 407, 154-157, doi:
10.1016/j.carres.2015.02.006.
11.Kasimova, A. A., Kenyon, J. J., Arbatsky, N. P.,
Shashkov, A. S., Popova, A. V., Knirel, Y. A., and Hall, R. M. (2018)
Structure of the K82 capsular polysaccharide from Acinetobacter
baumannii LUH5534 containing a D-galactose 4,6-pyruvic acid acetal,
Biochemistry (Moscow), 83, 831-835, doi:
10.1134/S0006297918070064.
12.Shashkov, A. S., Liu, B., Kenyon, J. J., Popova,
A. V., Shneider, M. M., Senchenkova, S. N., Arbatsky, N. P.,
Miroshnikov, K. A., Wang, L., and Knirel, Y. A. (2017) Structures of
the K35 and K15 capsular polysaccharides of Acinetobacter
baumannii LUH5535 and LUH5554 containing amino and diamino uronic
acids, Carbohydr. Res., 448, 28-34, doi:
10.1016/j.carres.2017.05.017.
13.Kenyon, J. J., Shashkov, A. S., Senchenkova, S.
N., Shneider, M. M., Liu, B., Popova, A. V., Arbatsky, N. P.,
Miroshnikov, K. A., Wang, L., Knirel, Y. A., and Hall, R. M. (2017)
Acinetobacter baumannii K11 and K83 capsular polysaccharides
have the same 6-deoxy-L-talose-containing pentasaccharide K units but
different linkages between the K units, Int. J. Biol. Macromol.,
103, 648-655, doi: 10.1016/j.ijbiomac.2017.05.082.
14.Shashkov, A. S., Kenyon, J. J., Arbatsky, N. P.,
Shneider, M. M., Popova, A. V., Miroshnikov, K. A., Hall, R. M., and
Knirel, Y. A. (2016) Related structures of neutral capsular
polysaccharides of Acinetobacter baumannii isolates that carry
related capsule gene clusters KL43, KL47, and KL88, Carbohydr.
Res., 435, 173-179, doi: 10.1016/j.carres.2016.10.007.
15.Westphal, O., and Jann, K. (1965) Bacterial
lipopolysaccharides: extraction with phenol–water and further
applications of the procedure, in Methods in Carbohydrate
Chemistry (Whistler, R., ed.) Academic Press, New York, pp.
83-91.
16.Sawardeker, J. S., Sloneker, J. H., and Jeanes,
A. (1965) Quantitative determination of monosaccharides as their
alditol acetates by gas liquid chromatography, Anal. Chem.,
37, 1602-1604, doi: 10.1021/ac60231a048.
17.Bankevich, A., Nurk, S., Antipov, D., Gurevich,
A. A., Dvorkin, M., Kulikov, A. S., Lesin, V. M., Nikolenko, S. I.,
Pham, S., Prjibelski, A. D., Pyshkin, A. V., Sirotkin, A. V., Vyahhi,
N., Tesler, G., Alekseyev, M. A., and Pevzner, P. A. (2012) SPAdes: a
new genome assembly algorithm and its applications to single-cell
sequencing, J. Comput. Biol., 19, 455-477, doi:
10.1089/cmb.2012.0021.
18.Lombard, V., Golaconda Ramulu, H., Drula, E.,
Coutinho, P. M., and Henrissat, B. (2014) The carbohydrate-active
enzymes database (CAZy) in 2013, Nucleic Acids Res., 42,
D490-D495, doi: 10.1093/nar/gkt1178.
19.Finn, R. D., Coggill, P., Eberhardt, R. Y., Eddy,
S. R., Mistry, J., Mitchell, A. L., Potter, S. C., Punta, M., Qureshi,
M., Sangrador-Vegas, A., Salazar, G. A., Tate, J., and Bateman, A.
(2016) The Pfam protein families database: towards a more sustainable
future, Nucleic Acids Res., 44, D279-D285, doi:
10.1093/nar/gkv1344.
20.Schultz, M. B., Thanh, D. P., Hoan, N. T. D.,
Wick, R. R., Ingle, D. J., Hawkey, J., Edwards, D. J., Kenyon, J. J.,
Lan, N. P. H., Campbell, J. I., Thwaites, G., Nhu, N. T. K., Hall, R.
M., Fournier-Level, A., Baker, S., and Holt, K. E. (2016) Repeated
local emergence of carbapenem-resistant Acinetobacter baumannii
in a single hospital ward, Microb. Genom., 2, e000050,
doi: 10.1099/mgen.0.000050.
21.Kenyon, J. J., Marzaioli, A. M., Hall, R. M., and
De Castro, C. (2014) Structure of the K2 capsule associated with the
KL2 gene cluster of Acinetobacter baumannii,
Glycobiology, 24, 554-563, doi:
10.1093/glycob/cwu024.
22.Kasimova, A. A., Kenyon, J. J., Arbatsky, N. P.,
Shashkov, A. S., Popova, A. V., Shneider, M. M., Knirel, Y. A., and
Hall, R. M. (2018) Acinetobacter baumannii K20 and K21 capsular
polysaccharide structures establish roles for UDP-glucose dehydrogenase
Ugd2, pyruvyl transferase Ptr2 and two glycosyltransferases,
Glycobiology, 28, 876-884, doi:
10.1093/glycob/cwy074.
23.Lipkind, G. M., Shashkov, A. S., Knirel, Y. A.,
Vinogradov, E. V., and Kochetkov, N. K. (1988) A computer-assisted
structural analysis of regular polysaccharides on the basis of
13C NMR data, Carbohydr. Res., 175, 59-75,
doi: 10.1016/0008-6215(88)80156-3.
24.Jansson, P. E., Kenne, L., and Schweda, E. (1987)
Nuclear magnetic resonance and conformational studies on monoacetylated
methyl D-gluco- and D-galacto-pyranosides, J. Chem.
Soc. Perkin Trans., 1, 377-383, doi:
10.1039/P19870000377.
25.Toukach, P. (2011) Bacterial Carbohydrate
Structure Database 3: principles and realization, J. Chem. Inf.
Model, 51, 159-170, doi: 10.1021/ci100150d.
26.Shashkov, A. S., Cahill, S. M., Arbatsky, N. P.,
Westacott, A. C., Kasimova, A. A., Shneider, M. M., Popova, A. V.,
Shagin, D. A., Shelenkov, A. A., Mikhailova, Y. V., Yanushevich, Y. G.,
Edelstein, M. V., Kenyon, J. J., and Knirel, Y. A. (2019)
Acinetobacter baumannii K116 capsular polysaccharide structure
is a hybrid of the K14 and revised K37 structures, Carbohydr.
Res., 484, 107774, doi: 10.1016/j.carres.2019.107774.
27.Landersjo, C., Weintraub, A., and Widmalm, G.
(1996) Structure determination of the O-antigen polysaccharide from the
enteroinvasive Escherichia coli (EIEC) O143 by component
analysis and NMR spectroscopy, Carbohydr. Res., 291,
209-216, doi: 10.1016/s0008-6215(96)00168-1.
Supplementary Figures S1 & S2 (PDF)