The Role of LAM Genes in the Pheromone-Induced Cell Death of S. cerevisiae Yeast
S. S. Sokolov1,a*, K. V. Galkina1, E. A. Litvinova2, D. A. Knorre1,3, and F. F. Severin1
1Belozersky Institute of Physico-Chemical Biology, Lomonosov Moscow State University, 119991 Moscow, Russia2Lomonosov Moscow State University, Faculty of Biology, 119991 Moscow, Russia
3Sechenov First Moscow State Medical University, 119991 Moscow, Russia
* To whom correspondence should be addressed.
Received November 3, 2019; Revised January 21, 2020; Accepted January 21, 2020
Lam1-4 proteins perform non-vesicular transport of sterols from the plasma membrane to the endoplasmic reticulum. Disruption of their function leads to an increase in the content of sterols in the plasma membrane. In mammals, homologs of Lam proteins are responsible for the internalization of plasma cholesterol. The biological role of Lam proteins in yeast remains unclear, since the strains lacking individual LAM genes do not display any pronounced phenotype. Deletion of LAM1 (YSP1) gene inhibits the regulated death of Saccharomyces cerevisiae yeast cells induced by the mating pheromone. Here, we investigated whether LAM2 also plays a role in the cell death induced by the excess of mating pheromone and assessed genetic interactions between LAM2 and genes responsible for ergosterol biosynthesis. We have shown that LAM2 deletion partially prevents pheromone-induced death of yeast cells of the laboratory strain W303, while deletions of three other LAM genes – LAM1, LAM3, and LAM4 – does not provide any additional rescuing effect. The UPC2-1 mutation in the transcription factor UPC2 gene, which leads to the excessive accumulation of sterols in the cell, promotes cell survival in the presence of the pheromone and shows additivity with the LAM2 deletion. On the contrary, LAM2 deletion stimulates pheromone-induced cell death in the laboratory strain BY4741. We have found that the deletion of ergosterol biosynthesis genes ERG2 and ERG6 reduces the effect of LAM2 deletion. Deletion of LAM2 in the Δerg4 strain lacking the gene of the last step of ergosterol biosynthesis, significantly increased the proportion of dead cells and decreased the growth rate of the yeast suspension culture even in the absence of the pheromone. We suggest that the absence of the effect of LAM2 deletion in the Δerg6 and Δerg2 strains indicates the inability of Lam2p to transport some ergosterol biosynthesis intermediates, such as lanosterol. Taken together, our data suggest that the role of Lam proteins in the regulated death of yeast cells caused by the mating pheromone is due to their effect on the plasma membrane sterol composition.
KEY WORDS: Lam proteins, regulated cell death, yeast, sterols, YSP2, pheromoneDOI: 10.1134/S0006297920030050
Abbreviations: CFU, colony-forming unit; ER, endoplasmic reticulum; PI, propidium iodide; PM, plasma membrane; RCD, regulated cell death.
Programmed cell death (PCD) plays an important role in the organism
development, immunity, and maintenance of tissue homeostasis in
multicellular organisms [1, 2].
The physiological role of PCD in unicellular organisms is less clear.
In the case of well-studied model organism, such as baker’s yeast
Saccharomyces cerevisiae, there is a distinction between
regulated cell death (RCD) and accidental cell death (ACD) caused by
high-intensity stress factors and associated with the loss of the
plasma membrane (PM) barrier function [3]. RCD is
caused by mild stress and the action of external or internal signals.
It can be artificially prevented, e.g., by switching off expression of
certain genes. PCD is a natural (physiological) scenario of RCD. At
present, multiple inducers of RCD in yeast and other unicellular
organisms have been described, but the physiological role of RCD
remains unknown in most cases [4-8].
Earlier, we have shown that high concentrations (100 μg/ml) of the mating pheromone, α-factor, induce RCD in a-type yeast cells [9, 10]. The effect of the pheromone is accompanied by an increase in the cytoplasmic calcium concentration [11]. Therefore, it was suggested that calcium mediates the cell death cascade [10]. The antiarrhythmic drug amiodarone also causes an increase in the cytoplasmic calcium [12] and triggers a chain of events similar to those activated by 100 μg/ml pheromone [10]. It should be noted that even such high concentrations of α-factor lead to the death of less than half of the cells. Moreover, while the fraction of the dead cells depends on the strain, the further increase in the pheromone concentration does not increase the proportion of dead cells [13]. The mechanisms that limit the pheromone-induced cell death are still unknown. Certain mutations have been found to significantly upregulate pheromone-induced RCD, for example, a mutation in the calmodulin gene cmd1-6 [9] or the deletion of the yeast metacaspase gene MCA1 [13].
Earlier, we conducted two genetic screenings for positive regulators of pheromone- or amiodarone-induced RCD. As a result, LAM1 and LAM2 were identified as genes decreasing cell resistance to amiodarone [10, 14]. Originally, these genes had been named YSP1 and YSP2 (Yeast Suicidal Proteins), but later they were renamed as LAM1 and LAM2 (Lipid transfer protein Anchored at Membrane contact sites), respectively. Deletion of LAM1 was found to partially protect the cells from the pheromone-induced cell death [10].
Recently, it has been shown that LAM1 and LAM2 encode proteins belonging to the same family of membrane sterol-binding proteins [15]. The function of these proteins is the transport of sterols between the plasma membrane (PM) and the endoplasmic reticulum (ER). In mammals, such transport is necessary for the capture of cholesterol from the blood plasma [16]. Deletion of LAM1 in baker’s yeast decreased cell resistance to the antifungal drug amphotericin B [15]. Importantly, amphotericin B disrupts the functions only of ergosterol-containing membranes [17]. Therefore, an increase in the sensitivity of the Δlam1 and Δlam2 deletion mutants to amphotericin B indicates that Lam proteins transport ergosterol from the PM. We have shown recently that deletions of LAM family genes indeed lead to the increase in the sterol content in the PM and intracellular compartments [18].
In addition to LAM1 and LAM2, S. cerevisiae genome contains four more paralogous genes of the same family: LAM3, LAM4, LAM5, and LAM6. While Lam1, Lam2, Lam3, and Lam4 localize to the sites of PM and ER contacts, Lam5 and Lam6 are found at the ER–mitochondria junctions (see reviews [19, 20]).
The discovery of the role of Lam proteins in sterol transport has raised the question on how sterol composition of the membranes is linked to the yeast RCD. It is possible that the effects of LAM1 and LAM2 deletions on the RCD induced by the pheromone, amiodarone, or acetate are related to the changes in the PM ergosterol content. If this is the case, then other mechanisms affecting (either increasing or decreasing) the sterol content in the yeast cells are expected to affect the RCD. Alternatively, Lam proteins can play a direct role in the signal transduction leading to the cell death. Moreover, it is still unknown whether each of the LAM genes plays a unique role or their functions in the cell are redundant.
Here, we have studied RCD caused by high concentrations of the mating pheromone and amiodarone and evaluated the role in the effect of these compounds of LAM2 deletion and mutations increasing the sterol content (UPC2-1) or disrupting the terminal stages of ergosterol biosynthesis (Δerg2, Δerg3, Δerg4, Δerg6). We also compared the effects of a single (LAM2) and quadruple (LAM1-LAM4) gene deletions. Taken together with the previously published observations, our data suggest that the role of LAM genes in yeast RCD is indirect and mediated by changes in the PM sterol composition.
MATERIALS AND METHODS
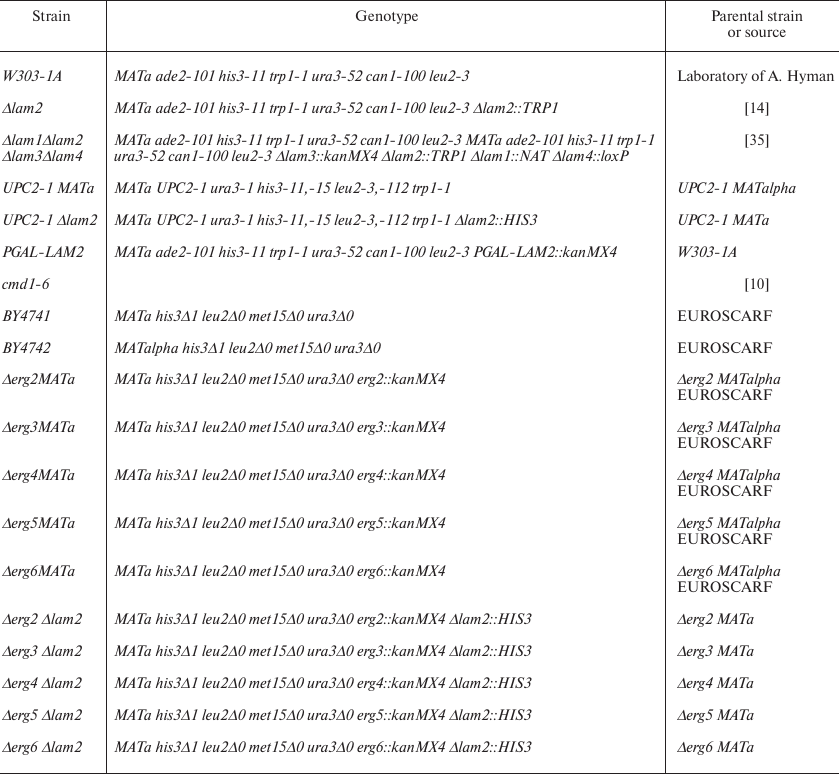
Yeast strains, cultivation conditions, and reagents used in the study. Amiodarone, α-factor, propidium iodide (PI), YNB synthetic medium, amino acid supplements, glucose, and galactose were from Sigma-Aldrich (USA). The cells were grown in YPD [1% yeast extract (Difco, USA), 2% peptone (Difco), 2% glucose (Helicon, Russia)] or YPGal [1% yeast extract (Difco), 2% peptone (Difco), 2% galactose (Sigma-Aldrich)]. Agar (Difco) was added up to 2% to solid media. The strains UPC2-1, Δlam1, Δlam2, Δlam3, Δlam4, and Δlam1Δlam2Δlam3Δlam4 are derivatives of the W303 strain. The strains Δerg2, Δerg2Δysp2, Δerg3, Δerg3Δysp2, Δerg4, Δerg4Δysp2, Δerg5, Δerg5Δysp2, Δerg6, Δerg6Δysp2 are derivatives of the BY4741 strain (table).
Strains used in the study
Mating type change. To change the mating type, we transformed the strains with the pGAL-HO plasmid coding for the exonuclease HO under control of the galactose-regulated PGAL promoter [21]. The resulting strains were incubated overnight in YPGal medium (2% galactose) and then transferred onto solid YPD medium (2% glucose). The mating type of individual colonies was determined by crossing with a- and α-type strains. Colonies with the desired mating type were grown for 24 h in YPD liquid medium to lose the pGAL-HO plasmid; the loss of the pGAL-HO plasmid was confirmed by the inability to grow on YNB-leu selective medium (Sigma-Aldrich).
RCD induction by amiodarone and assessment of yeast cell viability. Yeast cells were grown for 12-16 h to the logarithmic growth stage and then diluted to 4·106 cells/ml with YPD containing 25 mM MES, pH 5.5. Amiodarone was added to the cell suspension to a final concentration of 60 μM; the cells were then incubated for 1 h at 30°C, diluted, and transferred onto solid YPD medium. After 24 h, the proportion of survived cells was determined as the ratio of colony-forming units (CFUs) after and before the stress.
Mating efficiency assay. Yeast cells of both mating types were grown for 12-16 h up to the logarithmic stage and then diluted in liquid YPD to ~2·106 cells/ml. Cells of the opposite mating types were mixed and incubated for 6 h at 30°C. After incubation, the cells were analyzed by microscopy and the proportion of zygotes (in 250-300 cells) was calculated.
Flow cytometry and RCD induction by the mating pheromone. Yeast cells were grown for 12-16 h in YPD liquid medium to 4·106 cells/ml. α-Factor was added to a final concentration of 100 μg/ml, and the cells were incubated for 4 h at 200 rpm at 30°C. Untreated cell suspension was used as a negative control; cell suspension incubated for 3 h at 50°C was used as a positive control (dead cells). After incubation, all samples were diluted 8 times with phosphate buffered saline (PBS; Gibco, USA) and stained with 0.5 μg/ml PI for 10 min. PI is a fluorescent probe that cannot penetrate into living cells and stains dead cells only [22]. Fluorescence of PI-stained cells was analyzed with a CytoFlex flow cytometer (Beckman, USA) and Beckman Coulter FC 500 at the excitation wavelength of 488 nm with the 690/50 nm emission filter. Each experiment was performed in at least three independent repeats on different days. In each experiment, at least 10,000 events (cells) were analyzed.
RESULTS
LAM2 deletion partially prevents cell death caused by the excess of mating pheromone and does not show additivity with the deletions of LAM1, LAM3, or LAM4 genes. We have previously identified LAM2 gene (YSP2) in genetic screening for deletions decreasing yeast sensitivity to amiodarone [14]. Here, we investigated whether deletion of LAM2 and additional deletions of the other three LAM genes coding for proteins of the PM–ER contact sites (LAM1, LAM3, and LAM4) inhibit pheromone-induced RCD. For this, we compared the ratio of live and dead cells in a suspension culture of yeast cells exposed to α-factor (100 μg/ml, 4 h).
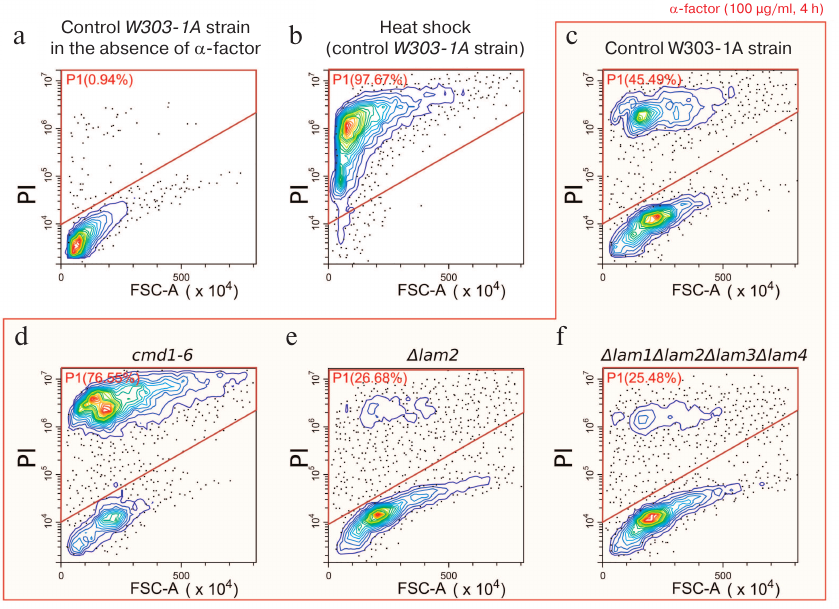
Live yeast cells were distinguished from the dead ones by flow cytometry based on PI accumulation (Fig. 1, a and b), because PI accumulates in dead cells only. Addition of α-factor induced the death of approximately 45% cells (Fig. 1c). In agreement with the published data [23], the cmd1-6 mutation increased the sensitivity of yeast cells to α-factor (Fig. 1d). We found that the deletion of LAM2 partially prevented cell death caused by α-factor (Fig. 1e). We also estimated linear sizes of yeast cells from light scattering. As shown in Fig. 1, α-factor caused a significant increase in the forward light scattering (FSC-A) that was most likely related to the formation of schmoos (cell protrusions characteristic for the mating process). Deletion of LAM2 gene did not prevent the increase in the linear light scattering, indicating that the conventional mating response was not impaired.
Fig. 1. Distribution of linear cell sizes (FSC-A) and PI signal intensity. a) Control W303-1A cells in the absence of stress (negative control for dead cells); b) W303-1A cells subjected to heat shock at 50°C for 3 h (positive control for dead cells); c-f) cells of different strains exposed to α-factor (100 μg/ml, 4 h): c) parental strain; d) cmd1-6 strain with mutation in the calmodulin gene; e) strain with LAM2 deletion; f) strain with deletion of all four LAM genes (LAM1-4). The cells within the indicated area of the graphs were counted as dead; the percentage of dead cells is indicated in each graph. The data from representative experiments (10,000 events in each experiment) are presented.
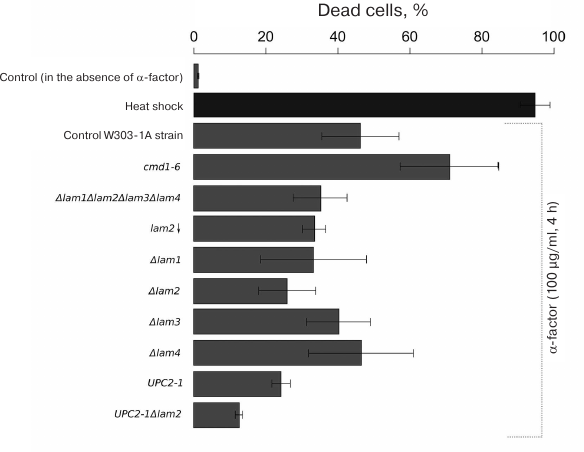
Figure 2 shows that LAM2 deletion had the highest effect on the cell resistance to α-factor, while additional deletions of the other three LAM genes did not increase the cell survival. Similar results were obtained in the experiments with amiodarone (Fig. 3), which kills yeast by the mechanism analogous to that triggered by the mating pheromone [10]. Similarly to the α-factor-induced death, LAM2 disruption had the maximum effect on the cell resistance to amiodarone. No significant differences in the resistance to this compound were observed between the Δlam2 strain and the quadruple deletion mutant (Fig. 3a).
Fig. 2. RCD induced by α-factor (100 μg/ml) in yeast cells with impaired transport and accumulation of ergosterol. The ratio of dead to live cells was evaluated by flow cytometry using PI staining. The data are presented as mean ± standard deviation (n = 3).
Fig. 3. Cell survival in the presence of yeast RCD inducer amiodarone (60 μM, 1-h incubation); 100% corresponds to the number of CFUs prior to amiodarone addition.
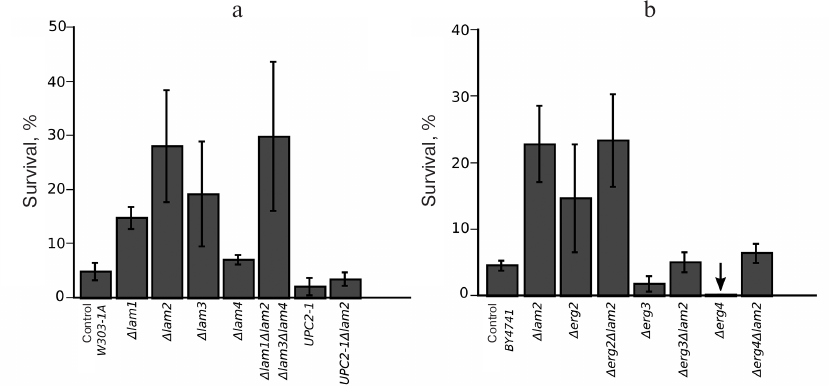
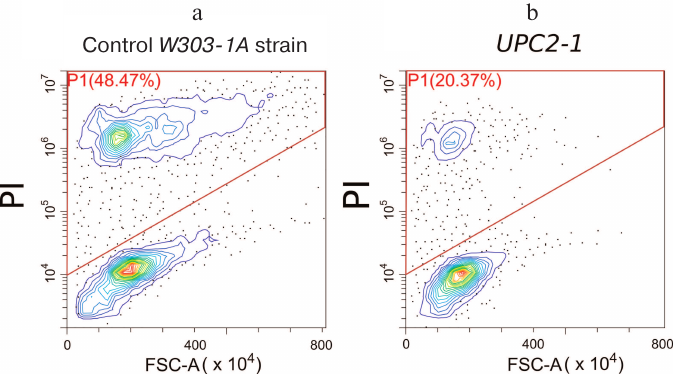
α-Factor-induced RCD is reduced in UPC2-1 strain. The strain with the partially dominant UPC2-1 allele demonstrated an increased rate of sterol uptake by the cells [24] and upregulated transcription of ERG genes, leading to the increase in total sterol content in the cell and sterol content in the PM in particular [25]. Deletion of LAM genes also leads to the increase in the concentration of ergosterol in the PM [15, 18]. If the resistance of the LAM deletion strains to the mating pheromone is related to the changes in the ergosterol concentration in the PM, then the UPC2-1 allele should also prevent α-factor-induced cell death. To test this hypothesis, we investigated the death of the UPC2-1 and wild-type yeast strains in the presence of 100 μg/ml α-factor. As shown in Fig. 4, UPC2-1 mutation resulted in the decrease in a relative number of PI-stained cells, i.e., this mutation suppressed cell death caused by the α-factor (also see Fig. 2). By contrast, the same strain showed increased sensitivity to amiodarone (Fig. 3a). Comparison of mating efficiency for the crosses UPC2-1 MATa × UPC2-1 MATalpha (2.2% zygotes), Δlam1Δlam2Δlam3Δlam4 MATa × Δlam1Δlam2Δlam3Δlam4 MATalpha (1.8%), and wild-type BY4741 (MATa) × BY4742 (MATalpha) (2.3%) revealed no pronounced differences between them. Therefore, an increased resistance of UPC2-1 and Δlam1Δlam2Δlam3Δlam4 strains to α-factor was not associated with partial inhibition of conventional mating response.
Fig. 4. Distribution of linear size (FSC-A) of yeast cells and PI signal intensity in wild-type yeast (a) and mutant UPC2-1 strain (b). The cells were incubated for 4 h in the presence of 100 μg/ml α-factor. The data of representative experiments are shown (10,000 events in each experiment).
Genetic interactions between LAM2 and ergosterol biosynthesis genes. In the following series of experiments, we examined RCD caused by α-factor and amiodarone in yeast cells with deletions of ERG2-6 genes. These genes encode enzymes catalyzing the downstream steps of ergosterol biosynthesis. Disruption of ergosterol biosynthesis is usually lethal [26], however, deletion of genes coding for the enzymes of the late biosynthesis stages, from lanosterol to ergosterol, does not lead to the loss of cell viability [26]. We excluded the ERG5 gene from our analysis, since the Δerg5 strain loses mitochondrial DNA at a high frequency (up to 80%), which significantly reduces its sensitivity to α-factor [9, 10]. It should be also mentioned that in this series of experiments, we used the parental BY4741 strain, while experiments with UPC2-1 were performed with the strains derived from the laboratory W303 strain, because these strains showed drastically different sensitivity to the pheromone-induced RCD.
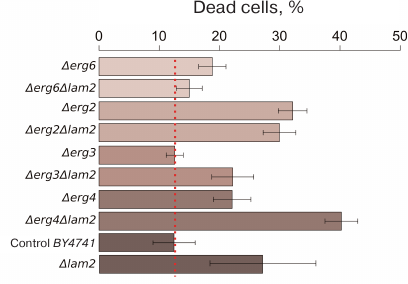
We estimated the percentage of dead cells in the wild-type strain and in the strains lacking the ERG6, ERG2, ERG3, or ERG4 genes (Fig. 5). Figure 5 shows the results arranged to reflect the order of ergosterol biosynthesis reactions catalyzed by the encoded enzymes. Deletions of the ERG genes did not decrease, and sometimes even increased the fraction of dead cells after incubation with the α-factor (Fig. 5). Moreover, additional deletion of LAM2 in the Δerg6 and Δerg2 strains produced no significant effect on the pheromone-dependent RCD (Fig. 5). At the same time, LAM2 deletion did not suppress RCD in the strains with impaired ergosterol synthesis. Deletion of LAM2 in the Δerg4 strain and in the control BY4741 strain led to a significant increase in the proportion of dead cells in the presence of α-factor.
Fig. 5. RCD of yeast cells with mutations in the ergosterol biosynthesis genes induced by α-factor (100 μg/ml). The ratio of dead to live cells was evaluated by flow cytometry using PI staining. The data are presented as mean ± standard deviation (n = 3).
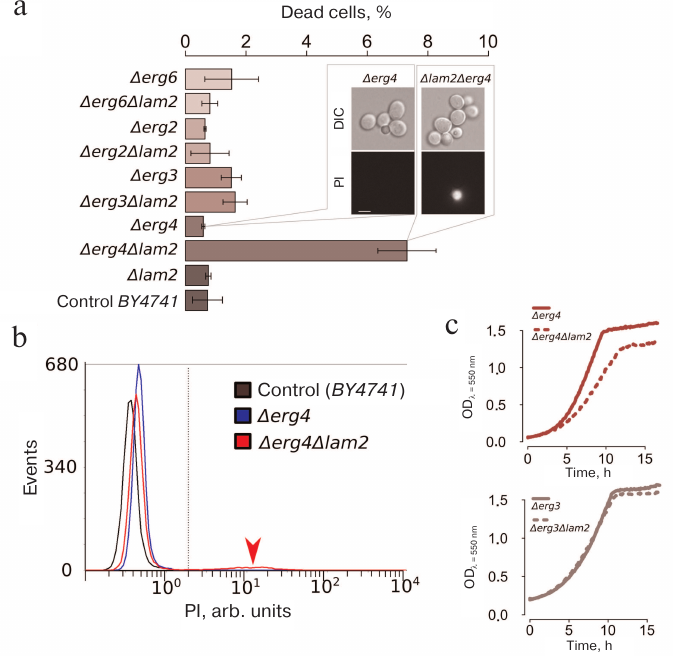
We noticed that, even in the absence of the mating pheromone, the number of dead (PI-positive) cells in the Δerg4Δlam2 strain was significantly higher than in the parental strain and all other double and single mutants (Fig. 6, a and b). Deletion of LAM2 significantly reduces the growth rate of the Δerg4 strain, but not of the Δerg3 strain. Importantly, Erg3p acts one step upstream of Erg4p (Fig. 6c). Therefore, our data indicate epistasis between LAM2 and ERG4 genes.
Fig. 6. Double deletion Δerg4Δlam2 strain contains a subpopulation of dead cells stained with PI and exhibits a reduced growth rate. a) Cell survival under control conditions (mean ± standard deviation, n = 3). The inset shows a photograph of Δerg4Δlam2 and Δerg4 cells stained with PI. DIC, differential interference contrast. b) Representative histogram of the flow cytometry experiment. c) Comparison of the growth rates (estimated by light scattering, wavelength λ = 550 nm) of Δerg4 and Δerg4Δlam2 strains (top) and Δerg3 and Δerg3Δlam2 strains (bottom).
Deletion of LAM2 increased cell resistance to amiodarone and promoted cell survival in all the mutants tested (Δerg2, Δerg3, Δerg4, and UPC2-1) (Fig. 3).
DISCUSSION
Studying LAM genes in yeast is difficult because these genes are represented by paralogous pairs [20]. Indeed, very few phenotypes have been described so far for the strains harboring deletions of individual LAM genes. However, we have previously shown that the deletion of LAM2 gene leads to an increase in the fragmentation of mitochondrial network in the cells at the stationary growth phase [27]. We have also shown resistance to amiodarone in the Δlam1 and Δlam2 strains [10, 14] and resistance to α-factor in the Δlam1 strain [10]. In this work, we demonstrated that LAM2 deletion also decreases the sensitivity of S. cerevisiae W303 strain to the RCD induced by α-factor (Figs. 1 and 2). Using this phenotype for studying genetic interactions, we examined the pheromone-induced RCD in the Δlam1Δlam2Δlam3Δlam4 strain and found that the deletions of additional LAM genes did not lead to the decrease in the sensitivity to α-factor compared to the Δlam2 strain. This result suggests that out of the four Lam proteins of the PM–ER contacts, Lam2 is the major contributor to the pheromone-induced cell death. Another possible explanation is that Lam proteins exist as heterodimers and that the presence of Lam2p is necessary for their functioning. The latter explanation seems unlikely since the strains with single deletions (Δlam3 and Δlam4) exhibited resistance to α-factor (Fig. 2). The data of large-scale mass-spectrometric proteomic analysis show that Lam2p is present in the cells in higher quantities than the other Lam proteins [28, 29]. Therefore, one can speculate that the deletion of LAM3 and LAM4 genes does not affect the pheromone-induced death because of the difference in the concentration of these proteins. However, in these studies, the amount of Lam2p did not exceed the total amount of its three paralogs. In another study, that used a simplified protocol for preparing protein samples for mass spectrometry, Lam1p was found in even larger quantities than Lam2p [30]. Hence, we believe that the absence of the effect of the triple deletion of LAM1, LAM3, and LAM4 in the Δlam2 strain is not associated with the low concentration of these proteins in the cells but rather reflects their functional relationship. The effect of the deletions of LAM genes on the amiodarone resistance was similar – LAM2 deletion promoted the resistance to this compound, while additional disruption of the LAM1, LAM3, and LAM4 genes provided no further increase in the cell resistance.
The BY4741 strain was significantly more resistant to the α-factor-induced RCD than the W303 strain. Moreover, deletion of the LAM2 gene did not decrease, but, on the contrary, increased the proportion of dead cells in our experimental model of the mating pheromone-induced RCD (Fig. 5). At the same time, the effect of LAM2 deletion on the resistance to amiodarone did not depend on the genetic background (Fig. 4). It is believed that amiodarone induces the same cascade of events as the mating pheromone (i.e., stimulates calcium entry to the cytoplasm), but at later stages [10, 12]. This might be due to the hyperpolarization of PM caused by the dysfunction of ion channels [31]. The simplest explanation is that the increase in the PM ergosterol content caused by the LAM2 deletion affects the activity of PM ion channels and pheromone receptors, and that the pheromone-to-calcium part of the cascade is highly dependent on the genetic background.
We have found that the effects of the deletions of LAM2 and ERG4 genes were additive (Figs. 5 and 6). ERG4 encodes C-24(28) sterol reductase, which catalyzes the last reaction in the metabolic pathway of ergosterol biosynthesis in yeast [32]. Importantly, in the Δerg3 strain lacking the enzyme catalyzing one before the last reaction of the same pathway, the effect of LAM2 deletion was lower in magnitude. No changes in the pheromone resistance were observed when LAM2 was deleted in the strains lacking ERG2 or ERG6 genes. Deletion of ergosterol biosynthesis genes changes the sterol composition of cell membranes, including ergosterol replacement by the intermediates of its biosynthesis [33]. Deletion of ERG6 and ERG2 interrupts ergosterol biosynthesis at the levels of zymosterol and fecosterol, respectively [34]. To our knowledge, no quantitative analysis of sterol biosynthesis intermediates in the deletion strains has been performed yet. It is possible that the absence of effect of LAM2 deletion in the Δerg2 and Δerg6 strains can be explained by Lam2 inability to transport early ergosterol biosynthesis intermediates (zymosterol, fecosterol, and lanosterol). If this is the case, the deletion of LAM2 gene would not affect cell phenotype. At the same time, according to our data, Lam2p does transport late ergosterol biosynthesis intermediates (and ergosterol).
The fact that LAM deletion reduces the death of yeast cells induced by high concentrations of the mating pheromone or amiodarone has been known for more than 10 years [10, 14]. However, the molecular mechanism of the involvement of Lam proteins in this type of RCD has remained unclear. While planning our study, we considered two possibilities: (1) deletion of LAM genes affects RCD indirectly, via changes in the PM ergosterol concentration; (2) Lam proteins directly interact with the RCD signal transduction intermediates. Our data point to the first possibility, since all other tested interventions in the sterol homeostasis also influenced cell resistance to this type of RCD. Deletion of LAM genes likely increases the content of PM ergosterol, as follows from the data on the LAM2 deletion strain sensitivity to amphotericin B and from direct measurements of PM ergosterol content with the sterol-sensitive fluorescent dye filipin [15, 18, 35].
How can elevated PM ergosterol possibly affect RCD? High ergosterol content is known to prevent Upc2 from relocation to the nucleus. Nuclear Upc2 activates transcription of at least two genes of the MAP kinase cascade, also initiated by the pheromone [36]. Is it possible that in LAM mutants, increased PM ergosterol prevents RCD by decreasing the activity of Upc2? According to our data, this seems unlikely. Upc2 hyperactivation (UPC2-1 mutation) does not increase, but rather reduces the pheromone-dependent RCD (Figs. 1 and 2). Since Upc2 is the main activator of ergosterol synthesis, it can be assumed that an increase in the PM ergosterol concentration affects ion fluxes through the PM, and this leads to the pheromone resistance. Indeed, it is known that the mutations in ergosterol biosynthesis genes can induce dysfunction of the PM transporters [37].
An increased sensitivity of UPC2-1 cells to amiodarone is also in good agreement with the previously published data. Amiodarone causes an increase in the cytoplasmic calcium concentration [12], which results in a downstream cascade of events leading to the cell death [10]. Moreover, an increase in the ergosterol levels affects vacuoles [38]. Normal functioning of vacuoles, in turn, provides resistance to amiodarone, because vacuoles are able to remove excessive Ca2+ from the cytoplasm. Since the toxicity of amiodarone is mediated by the rise in Ca2+ [39], this vacuolar function can be crucial for the resistance to amiodarone.
Combined with the previous observations, our data indicate that the effects of LAM deletions are associated with the changes in ergosterol content in the PM, which, in turn, affect the activity of membrane proteins and ion fluxes across the PM. In our opinion, the most likely explanation for the role of LAM genes in the pheromone-induced RCD is their indirect involvement in the signaling processes.
Funding. This work was supported by the Russian Science Foundation (project 18-14-00151) (Figs. 1, 2, and 4-6) and Russian Foundation for Basic Research (project 18-04-01183) (Fig. 3).
Acknowledgements. We thank Vasilina Efimova for participating in the generation of the double ΔergΔlam2 mutants.
Conflict of interest. The authors declare no conflict of interest in financial or any other sphere.
Ethical approval. This article does not contain description of studies involving humans or animals as research subjects performed by any of the authors.
REFERENCES
1.Nagata, S. (2018) Apoptosis and clearance of
apoptotic cells, Annu. Rev. Immunol., 36, 489-517; doi:
10.1146/annurev-immunol-042617-053010.
2.Fuchs, Y., and Steller, H. (2015) Live to die
another way: modes of programmed cell death and the signals emanating
from dying cells, Nat. Rev. Mol. Cell Biol., 16, 329-344; doi:
10.1038/nrm3999.
3.Carmona-Gutierrez, D., Bauer, M. A., Zimmermann,
A., Aguilera, A., Austriaco, N., et al. (2018) Guidelines and
recommendations on yeast cell death nomenclature, Microb. Cell
Fact., 5, 4-31; doi: 10.15698/mic2018.01.607.
4.Gordeeva, A. V., Labas, Y.A., and Zvyagilskaya, R.
A. (2004) Apoptosis in unicellular organisms: mechanisms and evolution,
Biochemistry, 69, 1055-1066; doi:
10.1023/b:biry.0000046879.54211.ab.
5.Severin, F. F., Meer, M. V., Smirnova, E. A.,
Knorre, D. A., and Skulachev, V. P. (2008) Natural causes of programmed
death of yeast Saccharomyces cerevisiae, Biochim. Biophys. Acta Mol.
Cell Res., 1783, 1350-1353; doi: 10.1016/j.bbamcr.2008.02.001.
6.Carmona-Gutierrez, D., Eisenberg, T., Buttner, S.,
Meisinger, C., Kroemer, G., and Madeo, F. (2010) Apoptosis in yeast:
triggers, pathways, subroutines, Cell Death Differ., 17,
763-773; doi: 10.1038/cdd.2009.219.
7.Sukhanova, E. I., Rogov, A. G., Severin, F. F., and
Zvyagilskaya, R. A. (2012) Phenoptosis in yeasts, Biochemistry, 77,
761-775; doi: 10.1134/S0006297912070097.
8.Aouacheria, A., Cunningham, K. W., Hardwick, J. M.,
Palkova, Z., Powers, T., Severin, F. F., and Vachova, L. (2018) Comment
on “Sterilizing immunity in the lung relies on targeting fungal
apoptosis-like programmed cell death”, Science, 360; doi:
10.1126/science.aar6910.
9.Severin, F. F., and Hyman, A. A. (2002) Pheromone
induces programmed cell death in S. cerevisiae, Curr. Biol., 12,
R233-235; doi: 10.1016/j.cellbi.2005.10.023.
10.Pozniakovsky, A. I., Knorre, D. A., Markova, O.
V., Hyman, A. A., Skulachev, V. P., and Severin, F. F. (2005) Role of
mitochondria in the pheromone- and amiodarone-induced programmed death
of yeast, J. Cell Biol., 168, 257-269; doi:
10.1083/jcb.200408145.
11.Ohsumi, Y., and Anraku, Y. (1985) Specific
induction of Ca2+transport activity in MATa cells of Saccharomyces
cerevisiaeby a mating pheromone, alpha factor, J. Biol. Chem.,
260, 10482-10486.
12.Gupta, S. S., Ton, V.-K., Beaudry, V., Rulli, S.,
Cunningham, K., and Rao, R. (2003) Antifungal activity of amiodarone is
mediated by disruption of calcium homeostasis, J. Biol. Chem.,
278, 28831-28839; doi: 10.1074/jbc.M303300200.
13.Zhang, N.-N., Dudgeon, D. D., Paliwal, S.,
Levchenko, A., Grote, E., and Cunningham, K. W. (2006) Multiple
signaling pathways regulate yeast cell death during the response to
mating pheromones, Mol. Biol. Cell, 17, 3409-3422; doi:
10.1091/mbc.e06-03-0177.
14.Sokolov, S., Knorre, D., Smirnova, E., Markova,
O., Pozniakovsky, A., Skulachev, V., and Severin, F. (2006) Ysp2
mediates death of yeast induced by amiodarone or intracellular
acidification, Biochim. Biophys. Acta, 1757, 1366-1370; doi:
org/10.1016/j.bbabio.2006.07.005.
15.Gatta, A. T., Wong, L. H., Sere, Y. Y.,
Calderon-Norena, D. M., Cockcroft, S., Menon, A. K., and Levine, T. P.
(2015) A new family of StART domain proteins at membrane contact sites
has a role in ER–PM sterol transport, Elife, 4; doi:
org/10.7554/eLife.07253.
16.Sandhu, J., Li, S., Fairall, L., Pfisterer, S.
G., Gurnett, J. E., Xiao, X., Weston, T. A., Vashi, D., Ferrari, A.,
Orozco, J. L., Hartman, C. L., Strugatsky, D., Lee, S. D., He, C.,
Hong, C., Jiang, H., Bentolila, L. A., Gatta, A. T., Levine, T. P.,
Ferng, A., Lee, R., Ford, D. A., Young, S. G., Ikonen, E., Schwabe, J.
W. R., and Tontonoz, P. (2018) Aster proteins facilitate nonvesicular
plasma membrane to ER cholesterol transport in mammalian cells, Cell,
175, 514-529.e20; doi: 10.1016/j.cell.2018.08.033.
17.Huang, W., Zhang, Z., Han, X., Tang, J., Wang,
J., Dong, S., and Wang, E. (2002) Ion channel behavior of amphotericin
B in sterol-free and cholesterol- or ergosterol-containing supported
phosphatidylcholine bilayer model membranes investigated by
electrochemistry and spectroscopy, Biophys. J., 83, 3245-3255;
doi: 10.1016/S0006-3495(02)75326-5.
18.Sokolov, S., Vorobeva, M., Smirnova, E.,
Smirnova, A., Trushina, N., Galkina, K., Severin, F., and Knorre, D.
(2020) LAM genes contribute to environmental stress tolerance but
sensibilise yeast cells to azoles, Front. Microbiol., 11;
doi: 10.3389/fmicb.2020.00038.
19.Alli-Balogun, G. O., and Levine, T. P. (2019)
Regulation of targeting determinants in interorganelle communication,
Curr. Opin. Cell Biol., 57, 106-114; doi:
10.1016/j.ceb.2018.12.010.
20.Sokolov, S. S., Trushina, N. I., Severin, F. F.,
and Knorre, D. A. (2019) Ergosterol turnover in yeast: an interplay
between biosynthesis and transport, Biochemistry (Moscow), 84, 346-357;
doi: 10.1134/S0006297919040023.
21.Herskowitz, I., and Jensen, R. E. (1991) [8]
Putting the HO gene to work: practical uses for mating-type switching,
in Methods in Enzymology, Academic Press, pp. 132-146; doi:
10.1016/0076-6879(91)94011-z.
22.Strauber, H., and Muller, S. (2010) Viability
states of bacteria – specific mechanisms of selected probes,
Cytometry A, 77, 623-634; doi: 10.1002/cyto.a.20920.
23.Moser, M. J., Geiser, J. R., and Davis, T. N.
(1996) Ca2+-calmodulin promotes survival of pheromone-induced growth
arrest by activation of calcineurin and Ca2+-calmodulin-dependent
protein kinase, Mol. Cell. Biol., 16, 4824-4831; doi:
10.1128/mcb.16.9.4824.
24.Crowley, J. H., Leak, F. W., Jr., Shianna, K. V.,
Tove, S., and Parks, L. W. (1998) A mutation in a purported regulatory
gene affects control of sterol uptake in Saccharomyces cerevisiae,
J. Bacteriol., 180, 4177-4183.
25.Joshua, I. M., and Hofken, T. (2017) From lipid
homeostasis to differentiation: old and new functions of the zinc
cluster proteins Ecm22, Upc2, Sut1 and Sut2, Int. J. Mol. Sci.,
18; doi: 10.3390/ijms18040772.
26.Daum, G., Lees, N. D., Bard, M., and Dickson, R.
(1998) Biochemistry, cell biology and molecular biology of lipids of
Saccharomyces cerevisiae, Yeast, 14, 1471-1510; doi:
10.1002/(SICI)1097-0061(199812)14:16<1471::AID-YEA353>3.0.CO;2-Y.
27.Knorre, D. A., Ojovan, S. M., Saprunova, V. B.,
Sokolov, S. S., Bakeeva, L. E., and Severin, F. F. (2008) Mitochondrial
matrix fragmentation as a protection mechanism of yeast
Saccharomyces cerevisiae, Biochemistry (Moscow), 73, 1254-1259;
doi: 10.1134/s0006297908110126.
28.De Godoy, L. M. F., Olsen, J. V., Cox, J.,
Nielsen, M. L., Hubner, N. C., Frohlich, F., Walther, T. C., and Mann,
M. (2008) Comprehensive mass-spectrometry-based proteome quantification
of haploid versus diploid yeast, Nature, 455, 1251-1254; doi:
10.1134/s0006297908110126.
29.Peng, M., Taouatas, N., Cappadona, S., van
Breukelen, B., Mohammed, S., Scholten, A., and Heck, A. J. R. (2012)
Protease bias in absolute protein quantitation, Nat. Methods, 9,
524-525; doi: 10.1038/nmeth.2031.
30.Kulak, N. A., Pichler, G., Paron, I., Nagaraj,
N., and Mann, M. (2014) Minimal, encapsulated proteomic-sample
processing applied to copy-number estimation in eukaryotic cells, Nat.
Methods, 11, 319-324; doi: 10.1038/nmeth.2834.
31.Maresova, L., Muend, S., Zhang, Y.-Q., Sychrova,
H., and Rao, R. (2009) Membrane hyperpolarization drives cation influx
and fungicidal activity of amiodarone, J. Biol. Chem., 284,
2795-2802; doi: 10.1074/jbc.M806693200.
32.Zweytick, D., Hrastnik, C., Kohlwein, S. D., and
Daum, G. (2000) Biochemical characterization and subcellular
localization of the sterol C-24(28) reductase, erg4p, from the yeast
Saccharomyces cerevisiae, FEBS Lett., 470, 83-87; doi:
10.1016/s0014-5793(00)01290-4.
33.Liu, G., Chen, Y., Faergeman, N. J., and Nielsen,
J. (2017) Elimination of the last reactions in ergosterol biosynthesis
alters the resistance of Saccharomyces cerevisiaeto multiple stresses,
FEMS Yeast Res., 17; doi: 10.1093/femsyr/fox063.
34.Parks, L. W., and Casey, W. M. (1995)
Physiological implications of sterol biosynthesis in yeast, Annu.
Rev. Microbiol., 49, 95-116; doi:
10.1146/annurev.mi.49.100195.000523.
35.Jimenez-Munguia, I., Volynsky, P. E., Batishchev,
O. V., Akimov, S. A., Korshunova, G. A., Smirnova, E. A., Knorre, D.
A., Sokolov, S. S., and Severin, F. F. (2019) Effects of sterols on the
interaction of SDS, benzalkonium chloride, and a novel compound,
Kor105, with membranes, Biomolecules, 9; doi: 10.3390/biom9100627.
36.Hofken, T. (2017) Ecm22 and Upc2 regulate yeast
mating through control of expression of the mating genes PRM1 and PRM4,
Biochem. Biophys. Res. Commun., 493, 1485-1490; doi:
10.1016/j.bbrc.2017.10.005.
37.Kodedova, M., and Sychrova, H. (2015) Changes in
the sterol composition of the plasma membrane affect membrane
potential, salt tolerance and the activity of multidrug resistance
pumps in Saccharomyces cerevisiae, PLoS One, 10, e0139306; doi:
10.1371/journal.pone.0139306.
38.Hongay, C., Jia, N., Bard, M., and Winston, F.
(2002) Mot3 is a transcriptional repressor of ergosterol biosynthetic
genes and is required for normal vacuolar function in Saccharomyces
cerevisiae, EMBO J., 21, 4114-4124; doi:
10.1093/emboj/cdf415.
39.Zhang, Y.-Q., Gamarra, S., Garcia-Effron, G.,
Park, S., Perlin, D. S., and Rao, R. (2010) Requirement for ergosterol
in V-ATPase function underlies antifungal activity of azole drugs,
PLoS Pathog., 6, e1000939; doi:
10.1371/journal.ppat.1000939.