Substrate Diversity of L-Threonic Acid Dehydrogenase Homologs
C. F. Zhang1#, Y. P. Liu1#, X. X. Wu1, X. S. Zhang2,a*, and H. Huang1,b*
1Guangdong Provincial Key Laboratory of Biotechnology for Plant Development, School of Life Sciences, South China Normal University, Guangzhou, 510631 Guangdong, China2Institute of Ecological Science, School of Life Sciences, South China Normal University, Guangzhou, 510631 Guangdong, China
# These authors contributed equally to this study.
* To whom correspondence should be addressed.
Received November 26, 2019; Revised January 7, 2020; Accepted January 21, 2020
Despite physiological importance of aldonic sugar acids for living organisms, little is known about metabolic pathways of these compounds. Here, we investigated the functional diversity of homologs of L-threonic acid dehydrogenase (ThrDH; UniProt ID: Q0KBC7), an enzyme composed of two NAD-binding domains (PF14833 and PF03446). Ten ThrDH homologs with different genomic context were studied; seven new enzymatic activities were identified, such as (R)-pantoate dehydrogenase, L-altronic acid dehydrogenase, 6-deoxy-L-talonate dehydrogenase, L-idonic acid dehydrogenase, D-xylonic acid dehydrogenase, D-gluconic acid dehydrogenase, and 2-hydroxy-3-oxopantoate reductase activities. Two associated metabolic pathways were identified: L-idonic acid dehydrogenase was found to be involved in the degradation of L-idonic acid through oxidation/decarboxylation in Agrobacterium radiobacter K84, while 2-hydroxy-3-oxopantoate reductase was found to participate in D-glucarate catabolism through dehydration/cleavage in Ralstonia metallidurans CH34.
KEY WORDS: (R)-pantoate dehydrogenase, L-idonic acid catabolism, D-glucarate catabolism, L-altronic acid dehydrogenase, D-xylonic acid dehydrogenaseDOI: 10.1134/S0006297920040069
Abbreviations: EryDH, D-erythronic acid dehydrogenase; PanDH, (R)-pantoate dehydrogenase; SSN, sequence similarity network; ThrDH, L-threonic acid dehydrogenase.
Sugar acids are saccharides with a carboxyl group; they are classified
into aldonic, ulosonic, uronic, and aldaric acids. Aldonic acids, in
which the aldehyde functional group of aldose is oxidized into carboxyl
group, have important physiological functions. For example, D-gluconic
acid is an acidity regulator and metal chelator [1]; ascorbic acid (vitamin C) is an essential body
chemical [2]; sialic acid is an important
constituent of the cell surface [3]. According to
the Human Metabolome Database (http://www.hmdb.ca/), aldonic acids are
detected in human biological fluid and excreta. Thus, D-glyceric,
D-gluconic, and L-xylonic acids have been found in saliva, while
L-threonic and D-xylonic acids are present in blood and urine [4]. Therefore, studying the metabolism of aldonic
acids can help clarify their biological functions at the molecular
level. Most aldonic acids are catabolized through the
dehydration/cleavage process, in which aldonic acids or their
phosphates are dehydrated into keto deoxy intermediates later cleaved
into two fragments by aldolase [5-9].
With the rapid increase in the number of entries to the protein databases, the diversity of aldonic acid catabolism has been proven largely underestimated. In an earlier report [10], Gerlt’s group characterized L-threonic acid dehydrogenase (ThrDH) and D-erythronic acid dehydrogenase (EryDH) which direct tetronic acids into different catabolic pathways. To explore new aldonic acid dehydrogenases and related metabolic pathways, we selected four homologs of EryDH (UniProt ID: Q0KBD2) and 10 homologs of ThrDH (UniProt ID: Q0KBC7) based on the sequence similarity, genomic context diversity [see Fig. S1 in Supplement to this paper on the journal website (http://protein.bio.msu.ru/biokhimiya) and Springer site (Link.springer.com)], and availability of genomic DNA. We were able to demonstrate broad substrate diversity of seven new aldonic acid dehydrogenases, as well as to identify two new catabolic pathways.
MATERIALS AND METHODS
Materials. All sugars were purchased from Carbosynth China (China) or synthesized by Huaxuejia (China). Other chemicals were from Sigma-Aldrich (USA); nucleotides were from General Biosystems (China); molecular biology enzymes were from New England Biolabs (USA). The plasmids were purchased from Novagen (Germany); genomic DNA was from ATCC (USA) and DSMZ (Germany). Varian 600 MHz NMR spectrometer, Nanodrop 2000, and UV/visible spectrophotometer (AOE instruments UV-1800) were used in the experiments.
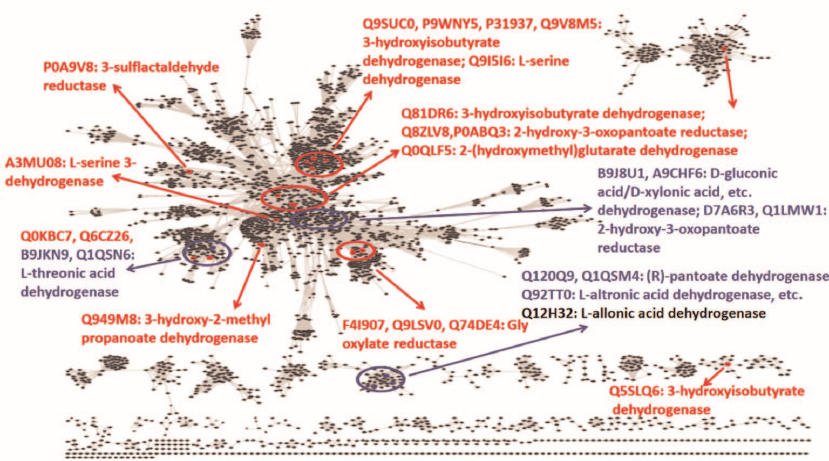
SSN generation. Sequence Similarity Network (SSN) enables analysis and visualization of the structure-function relationships for large protein families. The SSNs were constructed as described elsewhere [11]. In short, a protein sequence, Uniprot ID, or Pfam family number (for example, PF14833 in Fig. 1) were used as an input in the EFI-EST webtool (http://efi.igb.illinois.edu/efi-est/). In a representative node (rep-node) network, for example, 50% rep, means that sequences with more than 50% sequence identity will be collected in the same node. Generally, SSN is initially generated with the alignment score that corresponds to ~30% sequence identity; then, the stringency of the alignment score could be increased to distinguish between different protein functions (nodes with the same function segregate into one cluster).
Fig. 1. Overview of the sequence-function space for ThrDH homologs in the SSN (PF14833, 61,955 members, e–67, 40% identity, 50% rep). Known enzyme functions are shown in red; enzymes characterized in this work are shown in blue; Q12H32 is shown in gray (as a “slow” enzyme).
Enzyme purification. Gene fragments encoding ThrDH and EryDH homologs in bacterial species were amplified from the corresponding genomic DNAs by a standard PCR procedure with appropriate primers (Table S1 in the Supplement) and ligated into pET-28a vector (Fig. S2 in the Supplement). The resulting plasmids with the verified nucleotide sequences were used for the transformation of E. coli BL21(DE3) cells for protein expression. Transformed cells were inoculated into one liter of Luria–Bertani (LB) medium containing 50 µM kanamycin, and the culture was grown (37°C, 200 rpm) to OD600 = 0.5-0.8. Next, 0.4 mM IPTG was added to induce protein expression, and the culture was incubated overnight (20°C, 180 rpm). The cells were harvested, resuspended in 50 mM Tris-HCl buffer (pH 8.0), and disintegrated with a French press. The obtained cell lysate was clarified by centrifugation, and the supernatant was loaded on a Ni-NTA agarose column. The column was washed with three volumes of buffer A (25 mM Tris-HCl, pH 8.0) and two volumes of 25 mM imidazole in buffer A. The proteins were eluted with 250 mM imidazole in buffer A; the collected protein fractions were analyzed by SDS-PAGE (Fig. S3 in the Supplement). Fractions containing purified proteins were combined and dialyzed against three changes of buffer A. The resulted protein fraction was flash-frozen in liquid nitrogen and stored at –80°C until further use.
Enzyme assay. Analyzed enzymes (four EryDH homologs and 10 ThrDH homologs) were first assayed for the dehydrogenase activity against a library of aldonic sugar acids (34 monocarboxylic sugar acids; Table S2 in the Supplement) in a 96-well microplate. The dehydrogenase activity was evaluated by measuring NADH formation (ε = 6.2 mM–1·cm–1 at 340 nm). The reaction mixture (150 μl, 25°C) contained 1 µM dehydrogenase, 100 mM Tris-HCl buffer (pH 8.0), 1 mM Mg2+, 0.5 mM NAD+, 1 mM substrate; the absorbance of the reaction mixture was recorded every 1 s for 5 min. The most active substrates were selected for detail kinetic measurements by the same method (Table 1). For this, the reaction mixture (200 μl) contained 100 mM Tris-HCl buffer (pH 8.0), 1 mM Mg2+, 0.5 mM NAD+, and varying concentrations of aldonic acid substrate; the reaction was initiated by adding an appropriate concentration of the enzyme. Initial reaction rates at different substrate concentrations were fitted to the Michaelis–Menten equation (Table 2).
Table 1. Aldonic sugar acids used as
substrates for ThrDH and EryDH homologs tested in this paper
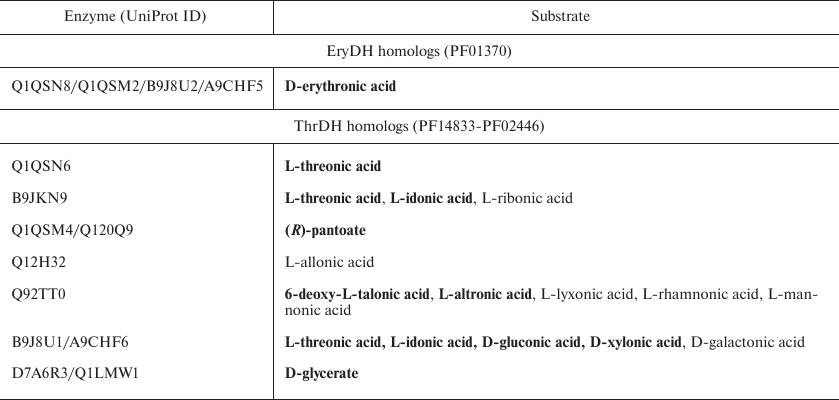
Notes: Substrates with a high activity are highlighted in bold.
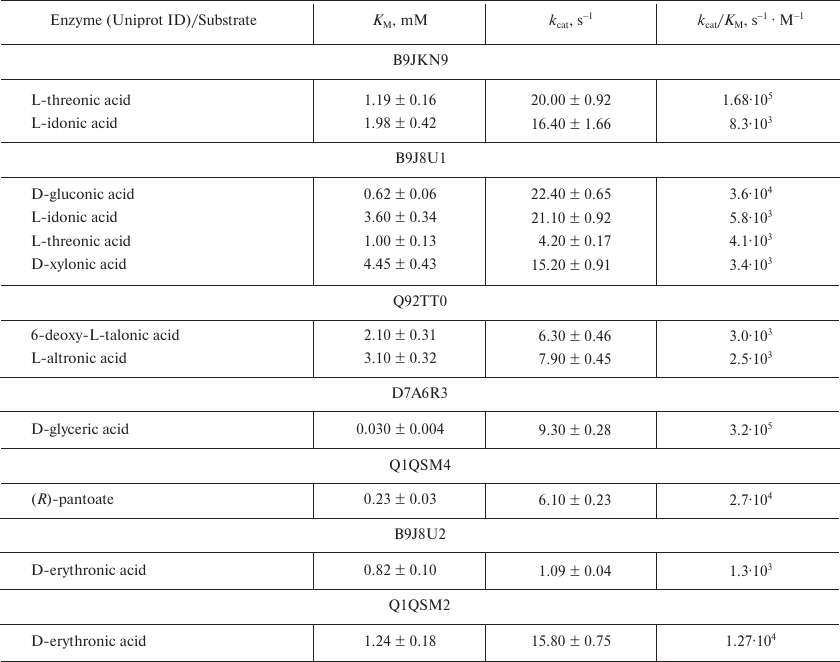
Table 2. Representative kinetic data for the
ThrDH homologs
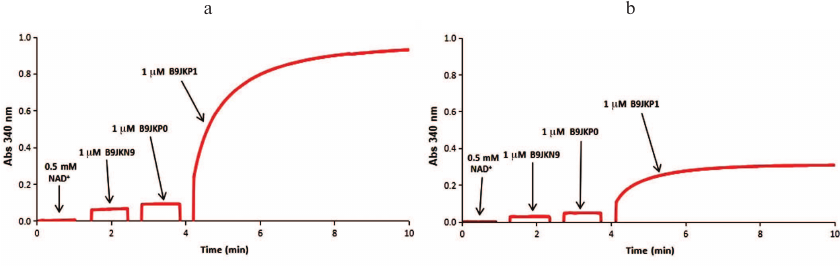
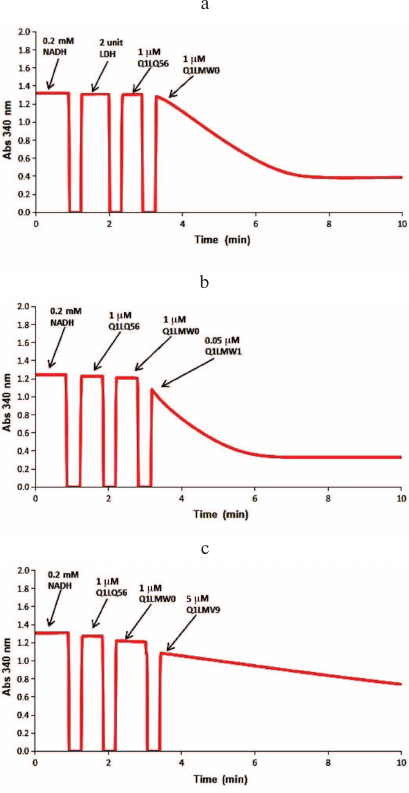
Identification of metabolic pathways. The sequence of reactions in the catabolic pathways for L-threonic and L-idonic acids was determined by sequentially adding the corresponding enzymes and recoding the changes in the absorbance at 340 nm. Thus, 1 μM dehydrogenase (B9JKN9) and 1 μM isomerase (B9JKP0) were added to 200 μl of the reaction mixture (100 mM Tris-HCl buffer, pH 8.0, 1 mM Mg2+, 0.5 mM NAD+, 0.5 mM ATP, 1 mM L-threonic acid or 1 mM L-idonic acid); after the reaction reached the equilibrium, 1 μM DUF1537 kinase (B9JKP1) was added (Fig. 2). The reactions catalyzed by D-glucarate dehydratase (Q1LQ56) and aldolase (Q1LMW0) in the D-glucarate catabolic pathway were confirmed by the production of pyruvate that was monitored by coupling with the reaction catalyzed by lactate dehydrogenase (LDH) in the presence of NADH. For this, the reaction mixture (200 μl, 25°C) containing 100 mM Tris-HCl buffer (pH 8.0), 1 mM Mg2+, 0.2 mM NADH, and 1 mM D-glucarate was supplemented with 2 units of LDH, 1 μM dehydratase (Q1LQ56), and 1 μM aldolase (Q1LMW0) to initiate the reaction, and changes in the absorbance at 340 nm were recorded. The dehydrogenase activity of Q1LMW1 was tested by following the same protocol except that 0.05 μM Q1LMW1 was added instead of LDH (Fig. 3).
Fig. 2. Spectrophotometric analysis of the L-threonic acid (a) and L-idonic acid (b) catabolic pathways. The reaction mixtures contained 100 mM Tris-HCl buffer (pH 8.0), 1 mM Mg2+, 0.5 mM NAD+, 0.5 mM ATP, and 1 mM L-threonic acid (a) or 1 mM L-idonic acid (b) as substrates. The sequence for the addition of enzymes ThrDH (B9JKN9), AP_endo (B9JKP0), and DUF1537 (B9JKP1) is shown in each panel.
Fig. 3. Spectrophotometric analysis of the D-glucarate catabolic pathway. The initial reaction mixture contained 100 mM Tris-HCl buffer (pH 8.0), 1 mM Mg2+, 0.2 mM NADH, 1 mM D-glucarate. a) Two units of LDH, 1 μM dehydratase (Q1LQ56), and 1 μM aldolase (Q1LMW0) were added to the reaction mixture to confirm the activity of Q1LQ56 and Q1LMW0; b) 1 μM dehydratase (Q1LQ56), 1 μM aldolase (Q1LMW0), and 0.05 μM ThrDH (Q1LMW1) were added to confirm the 2-hydroxy-3-oxopantanoic acid reductase activity of Q1LMW1; c) 1 μM dehydratase (Q1LQ56), 1 μM aldolase (Q1LMW0), and 5 μM Q1LMV9 were added to test the activity of Q1LMV9 toward pyruvate and 2-hydroxy-3-oxopantanoic acid.
RESULTS
Substrate specificity of dehydrogenases. We found that four EryDH homologs (Uniprot ID: Q1QSN8, Q1QSM2, B9J8U2, and A9CHF5; 38-76% amino acid sequence identity) were specific for D-erythronic acid, which is in agreement with the previous report [10], while ThrDH homologs were able to catalyze reactions with a wide variety of substrates (Table 1).
As seen from the SSN for the ThrDH homologs (Fig. 1), Q1QSN6 specifically oxidizes L-threonic acid (localizes to the upper left blue circle); however, B9JKN9 (~49% sequence identity with Q1QSN6) can oxidize both L-threonic and L-idonic acids (Tables 1 and 2). D7A6R3 and Q1LMW1 (63% sequence identity, localize to the upper right blue circle) specifically oxidize D-glyceric acid into 2-hydroxy-3-oxopantoate, while B9J8U1 and A9CHF6 (67% sequence identity) exhibit substrate promiscuity toward four substrates (L-threonic, L-idonic, D-xylonic, and D-gluconic acids) (Table 1). In addition, there are novel (R)-pantoate dehydrogenase (PanDH, Q120Q9 and Q1QSM4; 61% sequence identity) and L-altronic acid dehydrogenase (localizes to the bottom blue circle). The kinetic values (kcat/KM) of the above enzymes with the aldonic acid sugar substrates ranged from 103 to 105 s–1·M–1 (Table 2).
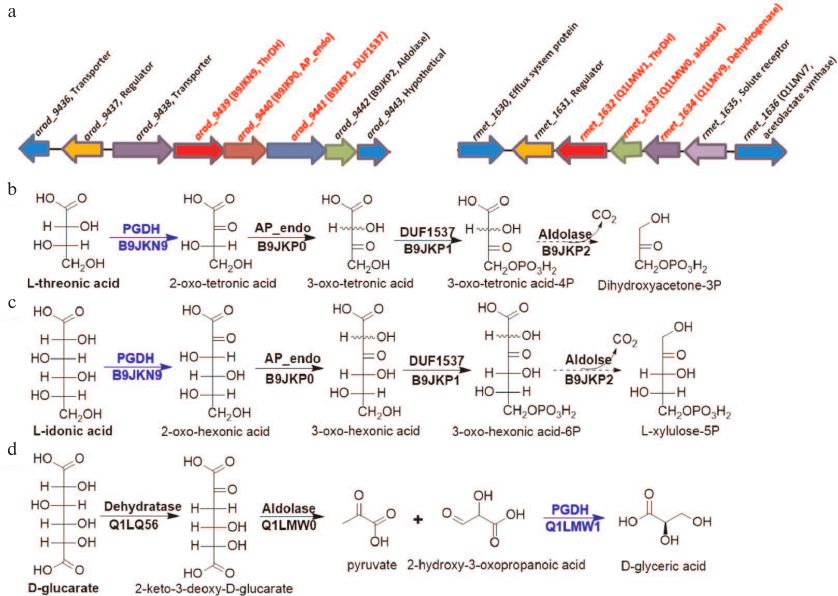
Identification of catabolic pathway. The genomic contexts for B9JKN9 and Q1LMW1 are shown in Fig. 4a. In vitro experiments have verified that L-idonic acid can be degraded in a pathway similar to the known pathway for L-threonic acid catabolism (Fig. 4b) [10]. In this pathway, L-idonic acid is oxidized by B9JKN9 into 2-oxo-hexonic acid, which is then isomerized by AP_endo isomerase (B9JKP0) and phosphorylated by DUF1537 kinase (B9JKP1). Finally, unstable 3-oxo-hexonic acid-6P may decompose into L-xylulose-5P either spontaneously or under the action of aldolase (B9JKP2) [10, 12] (Figs. 2 and 4c).
In Ralstonia metallidurans CH34, Q1LMW1 is involved in the typical D-glucarate catabolic pathway [13]. First, D-glucarate is dehydrated into 2-keto-3-deoxy-D-glucarate by D-glucarate dehydratase (Q1LQ56); this intermediate can be cleaved into pyruvate and 2-hydroxy-3-oxopropanoic acid (tartronate semialdehyde) by aldolase (Q1LMW0). Finally, Q1LMW1 transforms 2-hydroxy-3-oxopropanoic acid into D-glyceric acid (Figs. 3 and 4d).
Fig. 4. Gene contexts for L-idonic acid dehydrogenase (B9JKN9) and 2-hydroxy-3-oxopantanoic acid reductase (Q1LMW1) (a) and the corresponding catabolic pathways (b-d). a) Gene contexts for B9JKN9 (arad_9439) and Q1LMW1 (rmet_1632) are highlighted in red. The substrate for Q1LMV9 (rmet_1634) is unknown; however, this dehydrogenase exhibits reductase activity toward phenylpyruvic and α-ketoglutaric acids. b, c) L-Threonic acid and L-idonic acid catabolic pathways in Agrobacterium radiobacter K84; promiscuous enzymes in the pathway degrade both aldonic sugar acids into corresponding keto sugar phosphate. d) D-Glucarate catabolic pathway in R. metallidurans CH34; ThrDH homologs are highlighted in blue.
DISCUSSION
Aldonic sugar acids widely occur in natural environment and human biological fluids, where they display multiple important physiological functions [14, 15]. However, until now, the studies on the metabolism of aldonic sugar acids are either sporadic or missing [16, 17]. In this study, we aimed to elucidate these metabolic pathways through a detailed analysis of substrate diversity for the two known sugar acid dehydrogenase families – EryDHs (PF01370) and ThrDHs (PF14833-PF03446). First, we selected enzymes that may have diverse functions using combined bioinformatics analysis (homology assessment with the SSN and analysis of individual genomic contexts). Second, based on the structure of substrates (-COOH/-SO3H) for characterized ThrDH, selected ThrDH homologs were screened for substrate specificity against 34 sugar acids. Next, ThrDH homologs were tested individually with the appropriate substrates for detailed kinetic comparison. Finally, based on the identified activities of ThrDH toward the tested substrates, metabolic pathways involving these enzymes were explored.
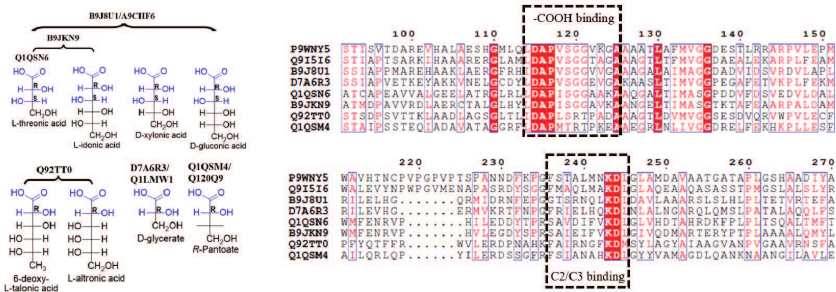
The experimental results clearly show that EryDH homologs are specific for D-erythronic acid, while the functions of ThrDH homologs are quite diverse. Some ThrDH homologs are promiscuous and exhibit high activity with multiple structurally similar sugar acids. For example, the substrates for Q92TT0 (6-deoxy-L-talonic acid, L-altronic acid) have the (2R)-configuration; while the substrates for B9JKN9/B9J8U1/A9CHF6 (L-threonic, L-idonic, D-xylonic, and D-gluconic acids) have the conserved (2R,3S)-configuration (Fig. 5). Some ThrDH homologs (Q1QSN6, Q1QSM4, Q120Q9, D7A6R3, and Q1LMW1) catalyzed reaction with substrates typical for L-threonic acid dehydrogenase, D-glyceric acid dehydrogenase, and a novel PanDH. The sequence alignment of ThrDHs and β-hydroxyacid dehydrogenases revealed the presence of conserved sequences for binding carboxyl groups (DAPVSGG motif) and adjacent carbons (FXXXXXXKDL motif) [18, 19]. However, due to the shortage of structures for close homology evaluation, detailed correlation between the substrate specificity and enzyme sequence is difficult.
Fig. 5. Substrate structure and sequence comparison for ThrDH. Conserved structures in substrates are highlighted in blue (left panel); substrate-binding motifs are shown within dashed-line boxes; conserved residues are shown in blue boxes (right panel). Structures of 3-hydroxyisobutyric acid dehydrogenase (P9WNY5, PDB: 5Y8N) from Mycobacterium tuberculosis and L-serine dehydrogenase (Q9I5I6, PDB: 3OBB) from Pseudomonas aeruginosa were used for the structure-based sequence alignment, and they have 22.4-29.5% sequence identity with the ThrDHs analyzed in this study.
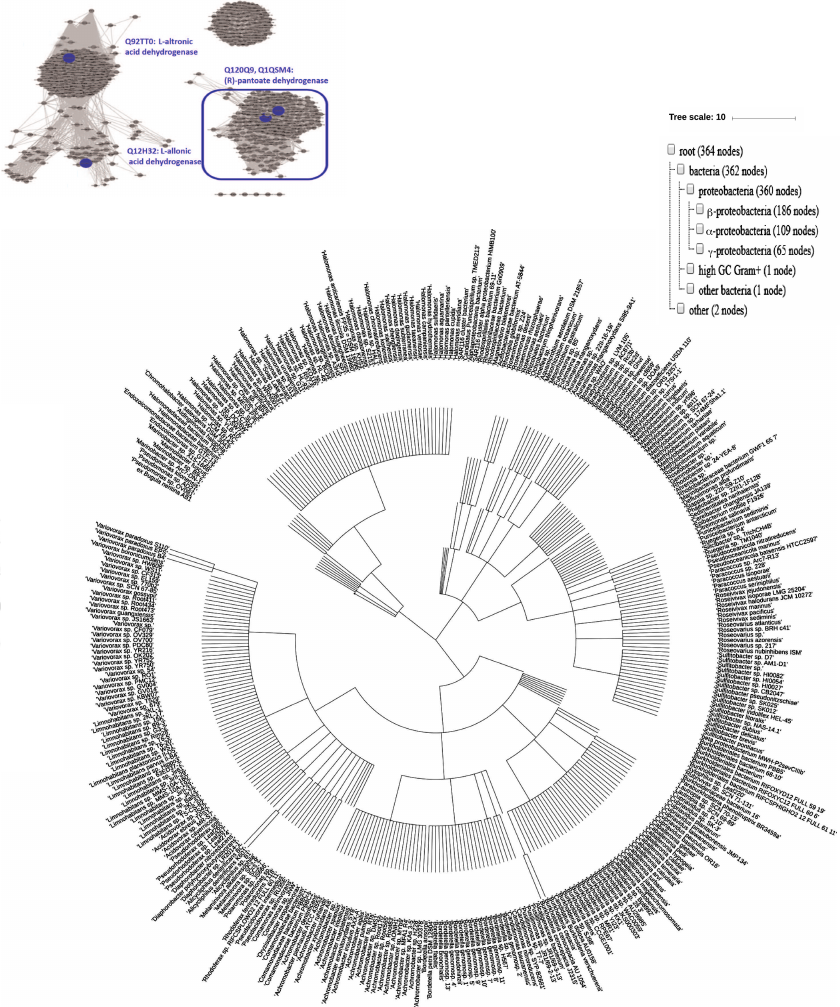
Unlike widely occurring PanDH (PF02558-PF08546), which is an essential enzyme of coenzyme A biosynthesis [20], (R)-pantoate dehydrogenase (Q1QSM4, PF14833-PF03446) has been found solely in proteobacteria (Fig. 6). Although Q12H32 (35.7% sequence identity with Q1QSM4), Q92TT0, and Q1QSM4, exhibiting different substrate specificities, localize to the same cluster in the ThrDH homology network (Fig. 1), increasing the stringency of the alignment score (e–72) allowed to segregate these enzymes into different clusters according to their functions (Fig. 6).
Fig. 6. Left panel: SSN for 3000 homologs of Q1QSM4 (e–72, 50% ID, 90% rep). Four ThrDH homologs characterized in this work are labeled in blue. The cluster for (R)-pantoate dehydrogenase is selected for phylogenetic analysis. Right: occurrence of (R)-pantoate dehydrogenase (blue box in left panel) in organisms. Taxonomy IDs extracted from the SSN were applied to generate the phylogenetic tree using the NCBI Taxonomy common tree: https://www.ncbi.nlm.nih.gov/Taxonomy/CommonTree/wwwcmt.cgi; then the downloaded .phy file was viewed with iToL (https://itol.embl.de/upload.cgi).
The diversity of the substrate specificity of the above ThrDH homologs is consistent with the diversity of their genome context (Fig. S1). Similar to L-threonic acid dehydrogenase (Q1QSN6) and L-idonic acid dehydrogenase (B9JKN9), DUF1537 and neighboring aldolase have been proven to participate in the degradation of these sugars (Fig. 4) [10]. However, B9J8U1 and A9CHF6, which demonstrate similar activity for L-threonic and L-idonic acids, have the dehydratase (threonine dehydratase, dihydroxyacid dehydratase) gene context, which might indicate their involvement in different metabolic pathway. Unlike PanDH, which clusters with other pantothenate biosynthesis enzymes [21], (R)-pantoate dehydrogenase (Q120Q9, Q1QSM4) characterized in this work tends to pair with aldehyde dehydrogenase. In addition, two D-glyceric acid dehydrogenases (Q1LMW1, D7A6R3) have different genomic contexts: Q1LMW1 clusters with aldolase (Q1LMW0) and participates as a downstream enzyme in D-glucarate catabolism in R. metallidurans CH34 (Fig. 4), while D7A6R3 might be involved in glycolate metabolism because of close location to glyoxylate carboligase [22]. L-altronic acid dehydrogenase (Q92TT0), together with neighboring methyltransferase and glycosylase, imply an existence of unknown pathway that still has to be identified.
In conclusion, identification of substrates for ThrDH homologs not only clarifies the metabolic pathways of these compounds, but also paves the way for further elucidation of their physiological functions.
Funding. This work was supported by the National Natural Science Foundation of China (project No. 31970087) and the start-up grant from the South China Normal University.
Conflict of interest. The authors declare no conflict of interest.
Compliance with ethical standards. This article does not contain description of studies with human participants or animals performed by any of the authors.
REFERENCES
1.Ramachandran, S., Fontanille, P., Pandey, A., and
Larroche, C. (2006) Gluconic acid: properties, applications and
microbial production, Food Technol. Biotechnol., 44,
185-195.
2.Smirnoff, N. (2000) Ascorbic acid: metabolism and
functions of a multi-facetted molecule, Curr. Opin. Plant Biol.,
3, 229-235.
3.Vimr, E. R., Kalivoda, K. A., Deszo, E. L., and
Steenbergen, S. M. (2004) Diversity of microbial sialic acid
metabolism, Microbiol. Mol. Biol. R., 68,
132-153.
4.Wishart, D. S., Knox, C., Guo, A., Eisner, R.,
Young, N., et al. (2009) HMDB: a knowledgebase for the human
metabolome, Nucleic Acids Res., 37, D603-D610.
5.Holzer, H., and Holldorf, A. (1957) Isolation of
D-glycerate dehydrogenase, some properties of the enzyme and its
application to the enzymic – optic determination of
hydroxypyruvate in presence of pyruvate, Biochem. Z.,
329, 292-312.
6.Dreyer, J. L. (1987) The role of iron in the
activation of mannonic and altronic acid hydratases, two Fe-requiring
hydrolyases, Eur. J. Biochem., 166, 623-630.
7.Dahms, A. S., and Donald, A. (1982) D-xylo-aldonate
dehydratase, Methods Enzymol., 90, E302-E305.
8.Shimizu, T., Takaya, N., and Nakamura, A. (2012) An
L-glucose catabolic pathway in Paracoccus species 43P, J.
Biol. Chem., 287, 40448-40456.
9.Wichelecki, D. J., Balthazor, B. M., Chau, A. C.,
Vetting, M. W., Fedorov, A. A., Fedorov, E. V., Luk, T., Patskovsky, T.
Y., Stead, M. B., Hillerich, B. S., Seidel, R. D., Almo, S. C., and
Gerlt, J. A. (2014) Discovery of function in the enolase superfamily:
d-mannonate and d-gluconate dehydratases in the d-mannonate dehydratase
subgroup, Biochemistry, 53, 2722-2731.
10.Zhang, X., Carter, M. S., Vetting, M. W.,
Francisco, B. S., Zhao, S., Al-Obaidi, N. F., Solbiatia, J. O.,
Thiavilled, J. J., Crecy-Lagard, V., Jacobson, M. P., Almo, S. C., and
Gerlt, J. A. (2016) Assignment of function to a domain of unknown
function: DUF1537 is a new kinase family in catabolic pathways for acid
sugars, Proc. Natl. Acad. Sci. USA, 113, E4161-E4169.
11.Gerlt, J. A., Bouvier, J. T., Davidson, D. B.,
Imker, H. J., Sadkhin, B., Slater, D. R., and Whalen, K. L. (2015)
Enzyme Function Initiative-Enzyme Similarity Tool (EFI-EST): a web tool
for generating protein sequence similarity networks, Biochim.
Biophys. Acta, 1854, 1019-1037.
12.Luo, S., and Huang, H. (2018) Discovering a new
catabolic pathway of D-ribonate in Mycobacterium smegmatis,
Biochem. Biophys. Res. Commun., 505, 1107-1111.
13.Aghaie, A., Lechaplais, C., Sirven, P., Tricot,
S., Besnard-Gonnet, M., Muselet, D., Berardinis, V., Kreimeyer, A.,
Gyapay, G., Salanoubat, M., and Perret, A. (2008) New insights into the
alternative D-glucarate degradation pathway, J. Biol. Chem.,
283, 15638-15646.
14.Severi, E., Hood, D. W., and Thomas, G. H. (2007)
Sialic acid utilization by bacterial pathogens, Microbiology,
153, 2817-2822.
15.Adachi, O., Hours, R. A., Shinagawa, E., Akakabe,
Y., Ykushi, T., and Matsushita, K. (2011) Enzymatic synthesis of
4-pentulosonate (4-keto-D-pentonate) from D-aldopentose and D-pentonate
by two different pathways using membrane enzymes of acetic acid
bacteria, Biosci. Biotechnol. Biochem., 75,
2418-2420.
16.Arrigoni, O., and De Tulio, M. C. (2002) Ascorbic
acid: much more than just an antioxidant, Biochim. Biophys.
Acta, 1569, 1-9.
17.Yew, W. S., Fedorov, A. A., Fedorov, E. V.,
Rakus, J. F., Pierce, R. W., Almo, S. C., and Gerlt, J. A. (2006)
Evolution of enzymatic activities in the enolase superfamily:
L-fuconate dehydratase from Xanthomonas campestris,
Biochemistry, 45, 14582-14597.
18.Tchigvintsev, A., Singer, A., Brown, G., Flick,
R., Evdokimova, E., Tan, K., Gonzalez, C. F., Savchenko, A., and
Yakunin, A. F. (2012) Biochemical and structural studies of
uncharacterized protein PA0743 from Pseudomonas aeruginosa
revealed NAD-dependent L-serine dehydrogenase, J. Biol. Chem.,
287, 1874-1883.
19.Srikalaivani, R., Singh, A., Vijayan, M., and
Surolia, A. (2018) Structure, interactions and action of
Mycobacterium tuberculosis 3-hydroxyisobutyric acid
dehydrogenase, Biochem. J., 475, 2457-2471.
20.Zheng, R., and Blanchard, J. S. (2000) Kinetic
and mechanistic analysis of the E. coli panE-encoded
ketopantoate reductase, Biochemistry, 39, 3708-3717.
21.Webb, M. E., Smith, A. G., and Abell, C. (2004)
Biosynthesis of pantothenate, Nat. Prod. Rep., 21,
695-721.
22.Grostern, A., Sales, C. M., Zhuang, W. Q.,
Erbilgin, O., and Alvarez-Cohen, L. (2012) Glyoxylate metabolism is a
key feature of the metabolic degradation of 1,4-dioxane by
Pseudonocardia dioxanivorans strain CB1190, Appl. Environ.
Microbiol., 78, 3298-3308.
Supplementary Tables S1-S2 & Figures S1-S3 (PDF)