Bovine bta-microRNA-1271 Promotes Preadipocyte Differentiation by Targeting Activation Transcription Factor 3
H. Y. Xu1#, J. Shao1#, B. Z. Yin1, L. M. Zhang1, J. C. Fang2, J. S. Zhang1, and G. J. Xia1,a*
1Agriculture College, Yanbian University, 133002 Yanji, Jilin, China2Faculty of Agriculture and Life Science, Hirosaki University, 036-8560 Hirosaki, Japan
# These authors contributed equally to this work.
* To whom correspondence should be addressed.
Received March 4, 2020; Revised May 21, 2020; Accepted May 21, 2020
Yanbian yellow cattle are one of the top five largest breeds of cattle in China. We had previously found that bta-miR-1271 is differentially expressed in the longissimus dorsi muscles of Yanbian yellow bulls and steers. However, whether bta-miR-1271 affects bovine fat formation is unclear. In this study, we used target gene prediction, dual-luciferase reporter assay, and transfection-mediated overexpression and inhibition of bta-miR-1271 in a culture of Yanbian yellow cattle preadipocytes to investigate the role of bta-miR-1271 in adipogenesis. We showed that bta-miR-1271 directly targets the 3′-untranslated region (3′-UTR) of the activating transcription factor 3 (ATF3) mRNA and downregulates its expression. Overexpression of bta-miR-1271 enforced by the miRNA mimics promoted triglyceride accumulation and significantly upregulated expression of the adipogenic peroxisome proliferator-activated receptor γ (PPARγ) and CCAAT enhancer-binding protein α (C/EBPα) genes at both the protein and mRNA levels, as demonstrated by RT-qPCR and Western blot analyses. Conversely, inhibition of bta-miR-1271 expression produced the opposite effect. Our results show that bta-miR-1271 regulates differentiation of Yanbian yellow cattle preadipocytes by inhibiting ATF3 expression, which highlights the importance of microRNA-mediated regulation of adipogenesis. miR-1271 and its target gene(s) may provide a new research direction for investigating biological agents affecting intramuscular fat deposition in cattle.
KEY WORDS: ATF3, bta-miR-1271, preadipocyte differentiation, Yanbian yellow cattleDOI: 10.1134/S0006297920070032
Abbreviations: ATF3, activation transcription factor 3; C/EBPα, CCAAT/enhancer binding protein α; miRNA, microRNA; MUT, mutant type; NC, negative control; PPARγ, peroxisome proliferator-activated receptor γ; UTR, untranslated region; WT, wild type.
INTRODUCTION
Due to religious beliefs, consumer habits, and production of beef with higher protein and higher unsaturated fatty acid content, beef is gaining popularity amongst consumers [1]. Yanbian yellow cattle are one of the top five largest breeds of cattle in China. They are large, highly adaptable, cold-tolerant, robust animals that have stable genetic characteristics and produce beef with a unique flavor.
The intramuscular fat content affects meat quality and taste. In turn, it is affected by many factors, including breed, gender, and feed composition [2, 3]. The deposition of intramuscular fat is caused by an increase in the number and volume of adipocytes. Adipocytes are derived from mesenchymal stem cells (MSCs), which first differentiate into adipoblasts, then into preadipocytes, and finally, into mature adipocytes [4, 5]. The differentiation of precursor adipocytes occurs in four stages: proliferation, mitotic cloning, early differentiation, and terminal differentiation [6]. When preadipocytes grow to a sufficient density, it results in contact inhibition, when preadipocytes gradually stop proliferating and begin to differentiate [7]. The differentiation stage is regulated by many transcription factors. For example, C/EBPβ induces expression of the peroxisome proliferator-activated receptor γ (PPARγ), and PPARγ activates C/EBPα, which promotes preadipocyte differentiation. Eventually, precursor adipocytes differentiate into mature adipocytes containing large lipid droplets [8].
MicroRNAs (miRNAs), that were first discovered by R. Lee in Caenorhabditis elegans in 1993, are widely expressed in eukaryotic cells [9]. miRNAs are transcribed from the genomic DNA, but not translated into proteins (i.e., they are non-coding). Instead, they bind to the target mRNAs and regulate expression of target genes by inhibiting translation of the transcribed mRNAs or causing their degradation [10]. miRNAs exert their biological functions at the RNA level and regulate series of important physiological processes, including fat metabolism and cellular differentiation [11]. For example, miR-16-5p [12], miR-26b [13], and miR-181a [14] promote adipogenesis; whereas miR-124-3p [15], miR-375 [16], and miR-127 [17] inhibit this process. We have previously analyzed miRNA expression in the latissimus dorsi muscle of Yanbian yellow cattle bulls and steers using Affymetrix GeneChip miRNA 3.0 Array and discovered differential expression of bta-miR-1271 in this muscle [18]. We also used next generation sequencing methods (miRNA-seq and RNA-seq) to analyze the longissimus dorsi muscles of 30-month-old Yanbian yellow steers with high-fat group (three individuals each with a high marbling grade ) and low-fat group (three individuals each with a low marbling grade), and found both bta-miR-1271 and ATF3 to be differentially expressed between high- and low-fat groups (Fig. S1 in the Supplement). The ATF3 gene was predicted as a target gene of bta-miR-1271 by Targetscan. Therefore, we speculated that bta-miR-1271 and ATF3 could affect the intramuscular fat deposition in Yanbian yellow cattle, although, to our knowledge, no reports have been published that linked bta-miR-1271 and any of its target genes to adipogenesis.
In this study, we studied adipogenesis in preadipocytes from newborn Yanbian yellow cattle. Bovine preadipocytes provide significant advantages over more commonly used murine 3T3-L1 cell line for studying adipogenesis in Yanbian yellow cattle, as unique species-specific pathways of fat synthesis and metabolism in bovine cells cannot be accurately recapitulated in murine cells [19]. The purpose of this study was to elucidate the role of bta-miR-1271 in preadipocyte differentiation and to provide a basis for further investigations into the miRNA-mediated physiological regulation in bovine adipose tissue.
MATERIALS AND METHODS
Cell culture and adipogenic differentiation. Yanbian yellow cattle preadipocytes were obtained from the Jilin Gongzhuling Animal Husbandry Branch; human hepatocyte HepG2 cells were from the Yanbian University School of Pharmacy. The cells were cultured at 37°C in a 5% CO2 incubator in DMEM (Gibco, China) supplemented with 10% FBS (BI, Israel). When the preadipocytes grew to 70-80% confluence, they were passaged into 6-well plates. When the cell confluence reached 100%, the culture medium was replaced with DMEM containing 10 µg/ml insulin, 0.5 mM 3-isobutyl-1-methylxanthine, 1.0 µM dexamethasone (Sigma-Aldrich, China), and 10% FBS. After two days, the medium was replaced with DMEM containing 10 µg/ml insulin and 10% FBS, and then replaced again DMEM containing 10% FBS after two days. The cells were grown in a culture for 9 days.
Bioinformatics. The sequence of mature bta-miR-1271 was obtained from the miRBase (http://www.mirbase.org/). The target genes were predicted using the TargetScan (http://www.targetscan.org/vert_71/).
Transfection of bovine preadipocytes. Bovine preadipocytes were seeded into 6-well plates at a density of 1 × 106 cells/well in 2 ml of DMEM containing 10% FBS. For transfection, 5 µl of LipofectamineTM 2000 (11668019, Invitrogen, China) and 10 µl of 20 µM of bta-miR-1271 mimics or an equivalent amount of negative control (NC) mimics, miRNA inhibitor, or NC inhibitor were added to 100 µl of Opti-MEM medium (Gibco, USA) and incubated for 5 min at room temperature before adding to the cells in DMEM containing 10% FBS. All miRNAs were purchased from the Jiangsu GenePharma Company (Suzhou, China). The miRNA sequences are listed Table S1 in the Supplement.
Dual-luciferase reporter assay. Based on the 3′-untranslated region (3′-UTR) sequence of the bovine ATF3 gene provided by the National Center for Biotechnology Information (NCBI), we designed primers for amplification of a gene fragment that perfectly matched the bta-miR-1271 seed sequence (2nd to 8th nucleotides at the 5′-end of the miRNA). The sequences coding for the SacI and XbaI restriction endonuclease sites were added to the 5′-ends of the forward and reverse primers, respectively. The obtained PCR product was ligated into the pmirGLO vector (Promega, China). Mutations to the putative binding site (3 nucleotides) were introduced using mutation primers; the mutant sequences were amplified by PCR and cloned into the pmirGLO vector. The primer sequences are listed Table S2 in the Supplement.
HepG2 cells were seeded into 96-well plates and co-transfected with 0.1 µg of dual-luciferase reporter vector and 0.5 µl of 20 µM bta-miR-1271 or NC mimics using LipofectamineTM 3000 (Invitrogen). At 48 h post-transfection, the luciferase activity was detected using the Dual-Glo™ Luciferase Assay System (Promega) and quantified with a luminometer (GloMax 20/20, Promega). All experiments were performed in triplicate.
Oil Red O staining and triglyceride assay. The cells were washed twice with PBS and fixed in situ in 4% paraformaldehyde solution at room temperature for 30 min. The fixed cells were rinsed twice with PBS. Oil Red O (Sigma-Aldrich) stock solution (0.5 g of Oil Red O dissolved in 100 ml of isopropanol and filtered) was mixed with sterile water in at a 6 : 4 ratio and added to the cells in a 6-well plate for staining at room temperature for 30 min. Next, the Oil Red O solution was discarded; the cells were washed three times with water and observed under a microscope. The Triglyceride Assay Kit (Nanjing Jiancheng Bioengineering Institute, China) was used to quantify the triglyceride content in adipocytes.
RNA isolation and RT-qPCR. RNA was extracted from the cells using the Eastep® Super Total RNA Extraction Kit (Promega). miRNAs were extracted with the miRcute miRNA Isolation Kit (Tiangen, China). RNA was reverse transcribed into cDNA using the PrimeScript RT reagent Kit with genomic DNA Eraser (Takara, China). RT-qPCR was performed using the SYBR Premix Ex Taq II kit (Takara). For miRNA quantification, reverse transcription was performed using the miRcute Plus miRNA First-Strand cDNA Kit (Tiangen); qPCR was performed using miRcute Plus miRNA qPCR Kit (SYBR Green) (Tiangen). RT-qPCR reactions were performed with a PCRmax Eco 48 real-time PCR machine (PCRmax, UK). All experiments were performed in triplicate. The relative levels of mRNA and miRNA expression were calculated using the 2−ΔΔCt method. The β-actin gene was used as a housekeeping gene to normalize expression of protein-coding genes; let-7a miRNA was used to normalize miRNA expression. The sequences of primers used for qPCR are shown in Table S3 in the Supplement.
Protein extraction and Western blotting. The cells were lyzed by incubation with RIPA Lysis Buffer (Beyotime, China) containing 1 mM PMSF (Beyotime) on ice. The total protein concentration the cell lysates was measured with the Enhanced BCA Protein Assay Kit (Beyotime) according to the manufacturer’s instructions. Proteins samples was subjected to electrophoresis in 12% SDS-PAG gels (20 µg protein per lane) and transferred to polyvinylidene difluoride (PVDF) membranes (Millipore, USA) according to the manufacturer’s instructions (Bio-Rad, USA). The PVDF membrane was rinsed with TBS containing 0.1% Tween 20 (TBST) and blocked with 5% ovalbumin (Solarbio, China) in TBST for 2 h at room temperature. The membrane was then incubated with the primary antibodies in the blocking buffer overnight at 4°C, washed five times with TBST, and incubated with horseradish peroxidase-conjugated secondary antibodies at 4°C for 2 h. After rinsing five times with TBST, the immunoblots were developed using a chemiluminescence solution and analyzed with an Alliance MINI HD9 AUTO Western Blot Imaging System (UVITEC, USA). The intensities of the target protein bands were normalized to the intensities of the β-actin band and computed with the ImageJ program. Rabbit anti-ATF3 (bs-0519R), rabbit anti-CEBPα (bs-1630R), and rabbit anti-PPARγ (bs-0530R) antibodies were from Bioss (China); mouse anti-β-actin (BS6007M) antibody was from Bioworld Technology (USA). All experiments were performed in triplicate.
Statistical analysis. All the results are presented as mean ± SD. The data were evaluated with the Student’s t-test (RT-qPCR, Western blotting, triglyceride assay) or the one-way analysis of variance (ANOVA) (dual-luciferase reporter assay). The differences between the groups were considered statistically significant at p < 0.05. All statistical analysis procedures were performed with the SPSS 20 software.
RESULTS
miR-1271 is highly conserved in mammals. The gene for bovine (bta) miR-1271 is located on chromosome 7 and encodes only one mature miRNA (bta-miR-1271), while the human (hsa) miR-1271 gene is located on chromosome 5 and generates two mature miRNA products (hsa-miR-1271-5p and hsa-miR-1271-3p). Interestingly, hsa-miR-1271-5p and bta-miR-1271 are homologous. The sequence of mature miRNA is also highly conserved across mammals, including gorillas, monkeys, pigs, goats, etc. Although there are some small differences in the miRNA sequences, the seed sequence is the same (Fig. 1a). The target site of bta-miR-1271 predicted in the ATF3 mRNA sequence is shown in Fig. 1b. The target genes predicted by TargetScan were compared with differentially expressed genes identified by RNA-seq analysis described previously (Fig. S1 in the Supplement). The ATF3 gene was found to be a candidate target gene of bta-miR-1271.
Fig. 1. Homologous miR-1271 sequences and bioinformatics prediction of the miR-1271 target gene. a) miR-1271 sequences in mammals with the seed sequences indicated in yellow. Asterisks show positions of perfectly conserved residues: bta, Bos taurus; hsa, Homo sapiens; ggo, Gorilla; ppy, Pongo pygmaeus; ptr, Pan troglodytes; mml, Macaca mulatta; ssc, Sus scrofa; eca, Equus caballus; cfa, Canis familiaris; chi, Capra hircus. b) The target site for bta-miR-1271 in the 3′-UTR of the ATF3 mRNA (red).
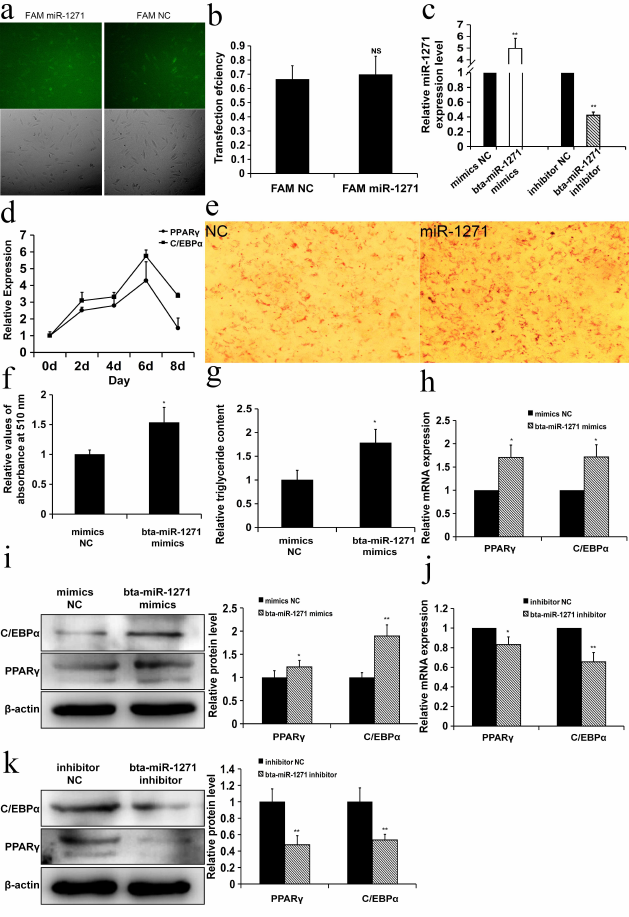
bta-miR-1271 promotes differentiation of Yanbian yellow cattle preadipocytes. To elucidate the role of bta-miR-1271 in the differentiation of preadipocytes, Yanbian yellow cattle preadipocytes were transfected with FAM-labeled bta-miR-1271 or NC mimics. The transfection efficiency was high, as determined by measuring the fluorescence of the FAM-labeled nucleic acids at 6 h post-transfection (Fig. 2a). The transfection efficiency was over 60% and did not differ significantly between the bta-miR-1271 and NC mimics (Fig. 2b). At 48 h post-transfection, the levels of miR-1271 were quantified by RT-qPCR. The expression of miR-1271 was significantly increased in cells transfected with the miRNA mimics vs. the cells transfected with the NC mimics (Fig. 2c). RNA was extracted on days 0, 2, 4, 6, and 8 of cell culture growth to examine all stages of preadipocyte differentiation, and the expression of adipogenic marker genes PPARγ and C/EBPα was evaluated by RT-qPCR. The expression of the PPARγ and C/EBPα mRNAs peaked on day 6 of induced preadipocyte differentiation (Fig. 2d). Therefore, the content of PPARγ and C/EBPα mRNAs and the amount of the corresponding proteins in the cells from different treatment groups were assessed by RT-qPCR and Western blotting, respectively, on day 6 of induced differentiation. Cells transfected with bta-miR-1271 demonstrated increased triglyceride accumulation in lipid droplets on day 9 of differentiation, as shown by Oil Red O staining (Fig. 2, e and f) and quantification of triglycerides (Fig. 2g). Compared to the NC mimics, overexpression of bta-miR-1271 enforced by transfection of bta-miR-1271 mimics significantly increased both mRNA and protein levels of PPARγ and C/EBPα. The mRNA and protein levels of PPARγ were 1.71- and 1.23-fold higher, respectively, and the mRNA and protein levels of C/EBPα were 1.72- and 1.89-fold higher, respectively, than those in the cells transformed with the NC mimics (p < 0.05) (Fig. 2, h and i). The bta-miR-1271 inhibitor produced the opposite effect. The mRNA and protein levels of PPARγ were lower (0.83- and 0.48-fold, respectively) and the mRNA and protein levels of C/EBPα were also lower (0.66- and 0.54-fold, respectively) than those in the NC inhibitor group (p < 0.05) (Fig. 2, j and k). Collectively, these data suggest that bta-miR-1271 promotes adipogenesis in the culture of Yanbian yellow cattle preadipocytes.
Fig. 2. bta-miR-1271 promotes differentiation of Yanbian yellow cattle preadipocytes. a) Preadipocytes were transiently transfected with FAM-labeled miR-1271 and NC mimics. The transfection efficiency was estimated 6 h later by the bright-light imaging and fluorescence microscopy. Green color, NC or miR-1271 mimics transfected into the cells (100×). b) Transfection efficiency of FAM-labeled miRNA (n = 3; NS, not significant). c) bta-miR-1271 levels detected by RT-qPCR 48 h after transfection (n = 3; ** p < 0.01). d) Changes in the levels of PPARγ and C/EBPα mRNAs during preadipocyte differentiation (n = 3; * p < 0.05). e) Lipid droplet formation in the cells transfected with NC and bta-miR-1271 mimics on day 9 after preadipocyte differentiation induction; Oil Red O staining (200×). f) The lipid content of differentiated cells was quantified by extracting Oil Red O with isopropanol and measuring its absorption at 510 nm (n = 3). g) Triglyceride content in the cells transfected with NC and bta-miR-1271 mimics (n = 3). h and i) The effect of bta-miR-1271 overexpression induced by transfection with the mimics on the levels of PPARγ and C/EBPα mRNAs (h) and PPARγ and C/EBPα protein content (i) quantified by RT-qPCR and Western blotting, respectively (n = 3). j and k) The effect of bta-miR-1271 knockdown after transfection with the bta-miR-1271 inhibitor on the levels of PPARγ and C/EBPα mRNAs (j) and proteins (k) vs. the cells transfected with the NC inhibitor (n = 3); * p < 0.05, ** p < 0.01.
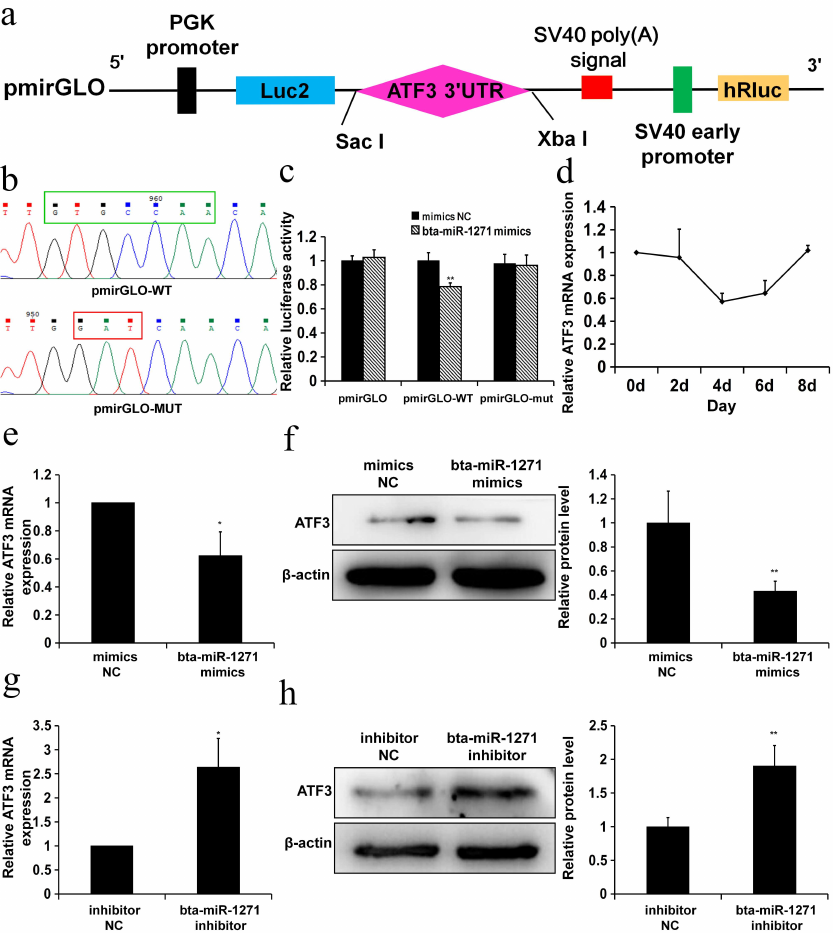
bta-miR-1271 directly targets the 3′-UTR of the ATF3 mRNA. To determine whether bta-miR-1271 directly targets the ATF3 mRNA, we created the pmirGLO-wild type (WT) and pmirGLO-mutant type (MUT) vectors (Fig. 3, a and b). HepG2 cells were co-transfected with the bta-miR-1271 or NC mimics together with either pmirGLO, pmirGLO-WT, or pmirGLO-MUT vectors. At 24 h post-transfection, the luciferase activity in the cells transfected with the bta-miR-1271 mimics was lower than in the cells transfected with the NC mimics (Fig. 3c).
The ATF3 expression decreased during preadipocyte differentiation, reaching its minimum on day 4 of the cell culture (Fig. 3d). To determine if bta-miR-1271 promotes adipogenesis via ATF3 suppression, we transiently transfected bovine preadipocytes with the bta-miR-1271 mimics and bta-miR-1271 inhibitor. The levels of the ATF3 mRNA and protein during the mid-stage of preadipocyte differentiation (4 days after induction of differentiation) were significantly suppressed as a consequence enforced bta-miR-1271 overexpression (Fig. 3, e and f). Conversely, suppression of bta-miR-1271 expression by the bta-miR-1271 inhibitor resulted in the upregulation of the ATF3 expression (Fig. 3, g and h). Taken together, these data demonstrate that ATF3 is a direct target of bta-miR-1271.
Fig. 3. ATF3 is a target gene of bta-miR-1271. a) Cloning of the ATF3 target sequence downstream of the firefly luciferase gene in the pmirGLO vector. b) The binding region for bta-miR-1271 (green box) in the ATF3 3′-UTR that was cloned into the pmirGLO vector to create pmirGLO-WT. The three nucleotides in the bta-miR-1271-binding region (red box) were mutated to create pmirGLO-MUT. c) Luciferase activity in HepG2 cells transfected with the pmirGLO dual-luciferase reporter vectors. Relative luciferase activity was calculated by the ratio of Firefly luminescence to Renilla luminescence (n = 3; * p < 0.05). d) ATF3 expression during differentiation of Yanbian yellow cattle preadipocytes (n = 3). e and f) The effect of bta-miR-1271 overexpression (enforced by the bta-miR-1271 mimics) in bovine adipocytes on the amounts of the ATF3 mRNA (e) (n = 4) and protein (f) (n = 3). g and h) The effect of bta-miR-1271 knockdown (by the bta-miR-1271 inhibitor) in bovine adipocytes on the amounts of the ATF3 mRNA (g) (n = 4) and protein (h) (n = 3); * p < 0.05; ** p < 0.01.
DISCUSSION
The Yanbian yellow cattle are high-yield producers of meat with a unique flavor, that occupy a superior market position in the beef industry. This is a world-renowned beef cattle breed, although there are disadvantages of low slaughter rates and insufficient production of high-grade beef. Understanding the molecular mechanisms that regulate fat production in Yanbian yellow cattle has become an important part of its molecular breeding program. An in vitro culture of preadipocytes has attracted much attention in recent years, as it is makes possible to directly monitor adipogenesis regulation in response to a variety of regulatory factors, and thereby to gain insights into the adipose tissue formation in vivo [20]. The number of fat cells, their volume, and accumulation of lipid droplets in them directly affect fat deposition. In turn, the type and the content of bovine adipose tissue affect meat tenderness and flavor [21]. miRNAs are important regulators of adipogenesis and lipid metabolism. Some of them are pro-adipogenic (miR-214-3p, miR-20a-5p, and miR-144-3p) [22-24], while others negatively regulate adipogenesis (miR-18b-3p and miR-27) [25, 26].
Although many studies have shown that miR-1271 regulates several key biological functions, including cancer cell proliferation [27, 28] and neuro-modulation [29], there have been no reports so far that implicate miR-1271 in adipogenesis. In our previous works on, we demonstrated using miRNA-seq and RNA-seq analysis that miR-1271 was differentially expressed in the longissimus dorsi in the high- and low-fat variants of Yanbian yellow cattle. Among the target genes of miR-1271 predicted by the bioinformatics methods, the ATF3 gene was also differentially expressed in these two groups of cattle. Therefore, we speculated that miR-1271 regulates lipid metabolism by targeting the ATF3 mRNA. In the present study, we explored whether bta-miR-1271 regulates adipogenesis in cultured preadipocytes from the Yanbian yellow cattle. It was found that overexpression of bta-miR-1271 enforced by transfection of the cells with the bta-miR-1271 mimics promoted adipocyte differentiation and lipid droplet formation. Furthermore, during preadipocyte differentiation, overexpression of bta-miR-1271 increased expression of adipogenic PPARγ and C/EBPα genes. We used miRNA-seq analysis to compare miRNA expression in the longissimus dorsi muscles from 30-month-old Yanbian yellow steers with different intramuscular fat content and found that the level of miR-1271 was in the upper-middle range among differentially expressed miRNAs. This proved that the endogenous expression of miR-1271 was in the upper-middle range. Therefore, we transfected the cells with the bta-miR-1271 inhibitor to inhibit endogenous bta-miR-1271 expression. Inhibition of endogenous miR-1271 downregulated expression of the PPARγ and C/EBPα genes and increased expression of the ATF3 gene in adipocytes. Together, these results suggest that bta-miR-1271 promotes fat differentiation in Yanbian cattle.
ATF3 is an early-response stress gene that belongs to the ATF/CREB transcription factor family [30]. ATF3 has been shown to inhibit adipogenic differentiation of murine 3T3-L1 cells by downregulating PPARγ and C/EBPα expression [31, 32]. ATF3 also inhibits expression of the adiponectin gene in 3T3-L1 cells [33]. ATF3 was reported to activate the Wnt/β-catenin pathway [34], which is considered to be a major negative regulator of preadipocyte differentiation. Signaling via this pathway inhibits adipogenesis [35, 36]. Jang et al. [37] found that the ATF3 expression was higher in white adipose tissue from high-fat diet-fed obese mice vs. control animals. Therefore, ATF3 expression might be involved in lipid droplet formation, which is consistent with the observed increase in the ATF3 expression after day 5. We also examined relative ATF3 expression during preadipocyte differentiation. The results showed that the ATF3 expression decreased during the initial stage of preadipocyte differentiation, reaching the minimal level on day 4 of differentiation, and then gradually increased until mature adipocytes were formed. In summary, before the production of lipid droplets, ATF3 was expressed at a low level; after day 4, adipocytes formed lipid droplets, and the ATF3 expression increased. In this study, we used the dual-luciferase reporter assay to show that bta-miR-1271 inhibits ATF3 expression by binding to the 3′-UTR of ATF3 mRNA. We further provided evidence that bta-miR-1271 overexpression suppresses expression of ATF3 mRNAs and inhibits ATF3 protein synthesis, while bta-miR-1271 knockdown promotes ATF3 expression, thus confirming ATF3 as a target gene of bta-miR-1271.
Although we failed to find published research on the role of miR-1271 in preadipocyte differentiation, there are reports that miR-1271 indirectly inhibits the Wnt/β-catenin pathway and suppresses TGFβ signaling by targeting other genes [38, 39]. Takin these data into consideration, we propose that bta-miR-1271 upregulates expression of the PPARγ and C/EBPα genes by inhibiting the ATF3 expression, which promotes adipocyte differentiation. Understanding the molecular pathways that regulate adipogenesis in cattle can inform molecular breeding programs. In addition, miR-1271 and its target gene(s) may play roles in the obesity development, opening a new research direction for investigating into biological treatments for obesity-related diseases.
Funding. This study was supported by the Science and Technology Development Plan of Jilin Province of China (project 20160204017NY).
Ethics declarations. The authors declare no conflict of interests. All applicable international, national, and/or institutional guidelines for the care and use of laboratory animals were followed in this study.
Electronic supplementary material. Supplementary materials are available in the electronic version on the journal website (http://protein.bio.msu.ru/biokhimiya) and Springer site (Link.springer.com).
REFERENCES
1.Yun, J., Jin, H., Cao, Y., Zhang, L., Zhao, Y.,
Jin, X., and Yu, Y. (2018) RNA-Seq analysis reveals a positive role of
HTR2A in adipogenesis in Yan yellow cattle, Int. J. Mol. Sci.,
19, 1760, doi: 10.3390/ijms19061760.
2.Han, J., Lee, J. E., Jin, J., Lim, J. S., Oh, N.,
Kim, K., Chang, S., Shibuya, M., Kim, H., and Koh, G. Y. (2011) The
spatiotemporal development of adipose tissue, Development,
138, 5027-5037, doi: 10.1242/dev.067686.
3.Tang, W., Zeve, D., Suh, J. M., Bosnakovski, D.,
Kyba, M., Hammer, R. E., Tallquist, M. D., and Graff, J. M. (2008)
White fat progenitor cells reside in the adipose vasculature,
Science, 322, 583-586,
doi: 10.1126/science.1156232.
4.Dani, C., Smith, A., Dessolin, S., Leroy, P.,
Staccini, L., Villageois, P., Darimont, C., and Ailhaud, G. (1997)
Differentiation of embryonic stem cells into adipocytes in
vitro, J. Cell Sci., 110, 1279-1285,
doi: 10.1159/000244147.
5.Pittenger, M. F., Mackay, A. M., Beck, S. C.,
Jaiswal, R. K., Douglas, R., Mosca, J. D., Moorman, M. A., Simonetti,
D. W. J. W. R., Craig, S., and Marshak, D. R. (1999) Multilineage
potential of adult human mesenchymal stem cells, Science,
284, 143-147, doi: 10.1126/science.284.5411.143.
6.Gregoire, F. M., Smas, C. M., and Sul, H. S. (1998)
Understanding adipocyte differentiation, Physiol. Rev.,
78, 783-809, doi: 10.1152/physrev.1998.78.3.783.
7.Otto, T. C., and Lane, M. D. (2005) Adipose
development: from stem cell to adipocyte, Crit. Rev. Biochem. Mol.
Biol., 40, 229-242, doi: 10.1080/10409230591008189.
8.Gregoire, F. M. (2001) Adipocyte differentiation:
from fibroblast to endocrine cell, Exp. Biol. Med., 226,
997-1002, doi: 10.1177/153537020122601106.
9.Bartel, D. P. (2004) MicroRNAs: genomics,
biogenesis, mechanism, and function, Cell, 116, 281-297,
doi: 10.1016/S0092-8674(04)00045-5.
10.Bartel, D. P. (2009) MicroRNAs: target
recognition and regulatory functions, Cell, 136, 215-233,
doi: 10.1016/j.cell.2009.01.002.
11.Vishnoi, A., and Rani, S. (2017) MiRNA biogenesis
and regulation of diseases: an overview, Methods Mol. Biol.,
1509, 1-10, doi: 10.1007/978-1-4939-6524-3_1.
12.Xu, J., Zhang, L., Shu, G., and Wang, B. (2019)
MicroRNA-16-5p promotes 3T3-L1 adipocyte differentiation through
regulating EPT1, Biochem. Biophys. Res. Commun., 514,
1251-1256, doi: 10.1016/j.bbrc.2019.04.179.
13.Li, G., Ning, C., Ma, Y., Jin, L., Tang, Q., Li,
X., Li, M., and Liu, H. (2017) miR-26b promotes 3T3-L1 adipocyte
differentiation through targeting PTEN, DNA Cell Biol.,
36, 672-681, doi: 10.1089/dna.2017.3712.
14.Zhang, Z., Gao, Y., Xu, M., Wang, C., Fu, X.,
Liu, J., Han, D., Jiang, H., Yuan, B., and Zhang, J. (2019) miR-181a
regulate porcine preadipocyte differentiation by targeting TGFBR1,
Gene, 681, 45-51,
doi: 10.1016/j.gene.2018.09.046.
15.Pan, Y., Jing, J., Qiao, L., Liu, J., An, L., Li,
B., Ren, D., and Liu, W. (2018) MiRNA-seq reveals that miR-124-3p
inhibits adipogenic differentiation of the stromal vascular fraction in
sheep via targeting C/EBPα, Domest. Anim. Endocrinol.,
65, 17-23, doi: 10.1016/j.domaniend.2018.05.002.
16.Liu, S., Sun, G., Yuan, B., Zhang, L., Gao, Y.,
Jiang, H., Dai, L., and Zhang, J. (2016) miR-375 negatively regulates
porcine preadipocyte differentiation by targeting BMPR2, FEBS
Lett., 590, 1417-1427,
doi: 10.1002/1873-3468.12169.
17.Gao, Y., Wang, Y., Chen, X., Peng, Y., Chen, F.,
He, Y., Pang, W., Yang, G., and Yu, T. (2019) MiR-127 attenuates
adipogenesis by targeting MAPK4 and HOXC6 in porcine adipocytes, J.
Cell. Physiol., 234, 21838-21850,
doi: 10.1002/jcp.28660.
18.Xia, G. (2014) Screening of Candidate Genes
Associated to Meat Quality Traits of Yanbian Yellow Cattle by a
Combination of miRNA and Functional Genes Transcriptome, PhD
Thesis, Yanji, Yanbian University [in Chinese].
19.Tran, K., Gealekman, O., Frontini, A.,
Zingaretti, M. C., Morroni, M., Giordano, A., Smorlesi, A., Perugini,
J., De Matteis, R., and Sbarbati, A. (2012) The vascular endothelium of
the adipose tissue gives rise to both white and brown fat cells,
Cell Metab., 15, 222-229,
doi: 10.1016/j.cmet.2012.01.008.
20.Fernyhough, M. E., Vierck, J. L., Hausman, G. J.,
Mir, P. S., Okine, E. K., and Dodson, M. V. (2004) Primary adipocyte
culture: adipocyte purification methods may lead to a new understanding
of adipose tissue growth and development, Cytotechnology,
46, 163-172, doi: 10.1007/s10616-005-2602-0.
21.Carnevalli, L. S., Masuda, K., Frigerio, F.,
Bacquer, O. L., Um, S. H., Gandin, V., Topisirovic, I., Sonenberg, N.,
Thomas, G., and Kozma, S. C. (2010) S6K1 plays a critical role in early
adipocyte differentiation, Dev. Cell, 18, 763-774,
doi: 10.1016/j.devcel.2010.02.018.
22.Xi, F., Wei, C., Xu, Y., Ma, L., He, Y., Shi, X.,
Yang, G., and Yu, T. (2019) MicroRNA-214-3p targeting Ctnnb1 promotes
3T3-L1 preadipocyte differentiation by interfering with the
Wnt/β-catenin signaling pathway, Int. J. Mol. Sci.,
20, 1816, doi: 10.3390/ijms20081816.
23.Zhu, E., Zhang, J., Zhou, J., Yuan, H., Zhao, W.,
and Wang, B. (2018) miR-20a-5p promotes adipogenic differentiation of
murine bone marrow stromal cells via targeting Kruppel-like factor 3,
J. Mol. Endocrinol., 60, 225-237,
doi: 10.1530/JME-17-0183.
24.Shen, L., Li, Q., Wang, J., Zhao, Y., Niu, L.,
Bai, L., Shuai, S., Li, X., Zhang, S., and Zhu, L. (2018) miR-144-3p
promotes adipogenesis through releasing C/EBPα from Klf3 and
CtBP2, Front. Genet., 9,
doi: 10.3389/fgene.2018.00677.
25.Sun, G., Li, F., Ma, X., Sun, J., Jiang, R.,
Tian, Y., Han, R., Li, G., Wang, Y., and Li, Z. (2019) gga-miRNA-18b-3p
inhibits intramuscular adipocytes differentiation in chicken by
targeting the ACOT13 gene, Cells, 8, 556,
doi: 10.3390/cells8060556.
26.Jang, S. Y., Chae, M. K., Lee, J. H., Lee, E. J.,
and Yoon, J. S. (2019) MicroRNA-27 inhibits adipogenic differentiation
in orbital fibroblasts from patients with Graves’ orbitopathy,
PLoS One, 14, e0221077,
doi: 10.1371/journal.pone.0221077.
27.Lin, M., Yang, Y., Peng, Z., Zhang, M., Liang,
J., Chen, W., Liu, X., and Zheng, Y. (2017) FOXK2, regulted by
miR-1271-5p, promotes cell growth and indicates unfavorable prognosis
in hepatocellular carcinoma, Int. J. Biochem. Cell Biol.,
88, 155-161, doi: 10.1016/j.biocel.2017.05.019.
28.Liu, X., Ma, L., Rao, Q., Mao, Y., Xin, Y., Xu,
H., Li, C., and Wang, X. (2015) MiR-1271 inhibits ovarian cancer growth
by targeting cyclin G1, Med. Sci. Monit., 21, 3152-3158,
doi: 10.12659/MSM.895562.
29.Jensen, K. P., and Covault, J. (2011) Human
miR-1271 is a miR-96 paralog with distinct non-conserved brain
expression pattern, Nucleic Acids Res., 39,
701-711, doi: 10.1093/nar/gkq798.
30.Hai, T., Wolfgang, C. D., Marsee, D. K., Allen,
A. E., and Sivaprasad, U. (1999) ATF3 and stress responses, Gene
Express., 7, 321-335, doi: 10.1248/bpb.29.2502.
31.Jang, M., and Jung, M. H. (2014) ATF3 represses
PPARγ expression and inhibits adipocyte differentiation,
Biochem. Biophys. Res. Commun., 454, 58-64,
doi: 10.1016/j.bbrc.2014.10.028.
32.Jang, M. K., Kim, C. H., Seong, J. K., and Jung,
M. H. (2012) ATF3 inhibits adipocyte differentiation of 3T3-L1 cells,
Biochem. Biophys. Res. Commun., 421, 38-43,
doi: 10.1016/j.bbrc.2012.03.104.
33.Kim, H. B., Kong, M., Kim, T. M., Suh, Y. H.,
Kim, W. H., Lim, J. H., Song, J. H., and Jung, M. H. (2006) NFATc4 and
ATF3 negatively regulate adiponectin gene expression in 3T3-L1
adipocytes, Diabetes, 55, 1342-1352,
doi: 10.2337/db05-1507.
34.Yan, L., Coletta, L. D., Powell, K. L., Shen, J.,
Thames, H. D., Aldaz, C. M., and Macleod, M. C. (2011) Activation of
the canonical Wnt/β-catenin pathway in ATF3-induced mammary
tumors, PLoS One, 6, e0016515,
doi: 10.1371/journal.pone.0016515.
35.Laudes, M. (2011) Role of WNT signalling in the
determination of human mesenchymal stem cells into preadipocytes, J.
Mol. Endocrinol., 46, R65-72,
doi: 10.1530/JME-10-0169.
36.Ross, S. E., Hemati, N., Longo, K. A., Bennett,
C. N., Lucas, P. C., Erickson, R. L., and Macdougald, O. A. (2000)
Inhibition of adipogenesis by Wnt signaling, Science,
289, 950-953, doi: 10.1126/science.289.5481.950.
37.Jang, M., Son, Y., and Jung, M. H. (2013). ATF3
plays a role in adipocyte hypoxia-mediated mitochondria dysfunction in
obesity, Biochem. Biophys. Res. Commun., 431,
421-427, doi: 10.1016/j.bbrc.2012.12.154.
38.Li, J., Xu, J., Yan, X., Jin, K., Li, W., and
Zhang, R. (2018) Suppression of Capn4 by microRNA-1271 impedes the
proliferation and invasion of colorectal cancer cells, Biomed.
Pharmacother., 99, 162-168,
doi: 10.1016/j.biopha.2017.12.107.
39.Xiang, X., Deng, J., Liu, Y., Wan, L., Feng, M.,
Chen, J., and Xiong, J. (2015) MiR-1271 inhibits cell proliferation,
invasion and EMT in gastric cancer by targeting FOXQ1, Cell.
Physiol. Biochem., 36, 1382-1394,
doi: 10.1159/000430304.
Supplementary Tables S1-S3, Fig S1 (PDF)