Inhibition of Chlamydial Infection by CRISPR/Cas9-SAM Mediated Enhancement of Human Peptidoglycan Recognition Proteins Gene Expression in HeLa Cells
P. A. Bobrovsky1,a*, V. D. Moroz1, V. N. Lavrenova1,2, V. A. Manuvera1, and V. N. Lazarev1
1Federal Research and Clinical Center of Physical-Chemical Medicine of Federal Medical Biological Agency, 119435 Moscow, Russia2Faculty of Biology, Lomonosov Moscow State University, 119991 Moscow, Russia
* To whom correspondence should be addressed.
Received July 7, 2020; Revised August 22, 2020; Accepted August 23, 2020
The global problem of emerging resistance of microorganisms to antibiotics makes the search for new natural substances with antibacterial properties relevant. Such substances include peptidoglycan recognition proteins (PGLYRP), which are the components of the innate immunity of many organisms, including humans. These proteins have a unique mechanism of action that allows them to evade the resistance of bacteria to them, as well as to be active against both Gram-positive and Gram-negative bacteria. However, the use of antimicrobial recombinant proteins is not always advisable due to the complexity of local delivery of the proteins and their stability; in this regard it seems appropriate to activate the components of the innate immunity. The aim of this study was to increase the expression level of native peptidoglycan recognition protein genes in HeLa cells using genome-editing technology with synergistic activation mediators (CRISPR/Cas9-SAM) and evaluate antichlamydial effect of PGLYRP. We demonstrated activation of the chlamydial two-component gene system (ctcB-ctcC), which played a key role in the mechanism of action of the peptidoglycan recognition proteins. We generated the HeLa cell line transduced with lentiviruses encoding CRISPR/Cas9-SAM activation system with increased PGLYRP gene expression. It was shown that activation of the own peptidoglycan recognition proteins gene expression in the cell line caused inhibition of the chlamydial infection development. The proposed approach makes it possible to use the capabilities of innate immunity to combat infectious diseases caused by Gram-positive and Gram-negative bacteria.
KEY WORDS: peptidoglycan, Chlamydia trachomatis, PGLYRP, CRISPR/Cas9DOI: 10.1134/S0006297920110036
Abbreviations: PGLYRP, peptidoglycan recognition proteins; DMEM, Dulbecco’s modified Eagle’s medium; SAM, synergistic activation mediator; HSF1, heat shock factor 1; p65, subunit of NF-κB transcription factor; CRISPR, clustered regularly interspaced short palindromic repeats; dCas9, nuclease-dead Cas9, PEI, polyethyleneimine.
INTRODUCTION
At present the increasing number of antibiotic-resistant strains represents a global problem. Emergence of the resistant strains results in the decreased efficiency of the therapeutic treatments of the socially significant infectious diseases. Diseases caused by the resistant strains are more difficult to treat and they are often more severe [1]. Serious side effects of some antibiotics have been described [2, 3]. In this regard, the search for new natural substances with bactericidal properties becomes relevant. Peptidoglycan recognition proteins (PGLYRP) found in many organisms from insects to mammals are promising candidates for this role. These proteins are components of the innate immune system, which plays the role of the first barrier of the body’s immune defense [4]. All PGLYRPs contain peptidoglycan-recognition domains that bind peptidoglycan that opens during cell division in the peptidoglycan-binding groove [5]. They also contain binding sites for lipopolysaccharides and lipoteichoic acids [6, 7]. Thus, PGLYRP can recognize peptidoglycan, lipopolysaccharides, and lipoteichoic acids of the cell walls of Gram-positive and Gram-negative bacteria [7]. In mammals, four PGLYRPs have been described: PGLYRP1, PGLYRP3, PGLYRP4, which have been identified as bactericidal proteins, and PGLYRP2, which additionally has an amidase activity. In 2011, the mechanism of action of these proteins was suggested. PGLYRP binds to peptidoglycan of the bacterial cell wall or lipopolysaccharides of the outer membrane and activates the protein-sensitive two-component stress response system [5]. Bacteria have many two-component systems that consist of sensory and regulatory components that allow bacteria to respond quickly to changing environmental conditions. The two-component systems CssR-CssS in Bacillus subtilis and CpxA-CpxR in Escherichia coli are functional homologues; moreover, similar two-component systems are typical for most bacteria [8, 9]. These two-component systems detect and eliminate misfolded secreted and membrane proteins, and they respond to both their own and foreign proteins, regardless of the cause of protein accumulation in the membrane compartment [10, 11]. In addition, they can also send a signal to the cell to activate production of hydroxyl radicals, depolarize the membrane, and stop synthesis of proteins, RNA and DNA, which eventually leads to the bacterial cell death [12]. Since these proteins are part of the human innate immune system, high bactericidal activity against pathogenic microorganisms and resistance of their own microflora to them [13] are indisputable advantages over antibiotics. Of particular interest is the activity of PGLYRP against intracellular parasites such as, for example, Chlamydia, which is a pathogenic Gram-negative bacterium with a unique life cycle. Chlamydia cause serious pathologies during pregnancy and childbirth, obstruction of the fallopian tubes and chlamydial conjunctivitis [14, 15]. Chlamydiae exist in the body in two forms: infectious, metabolically inactive elementary bodies, and non-infectious, metabolically active reticular bodies [16]. The unique features of the relationship of chlamydia with the human immune system often contribute to the development of a persistent form of infection [17]. Previously, we demonstrated antibacterial activity of the recombinant human PGLYRP against the intracellular pathogenic bacterium Chlamydia trachomatis and activation of gene expression of the two-component CtcB-CtcC system after contact of chlamydial elementary bodies with PGLYRP [18]. An alternative to the use of recombinant antimicrobial proteins obtained in heterologous expression systems is the use of various gene therapy approaches. One of the ways to fight infectious diseases using gene therapy is introduction of the genetic constructs encoding various components of innate immunity, for example, antimicrobial peptides [19-21], or activation of the expression of innate immunity proteins [22]. In this study, we modeled inhibition of the development of chlamydial infection by increasing expression level of peptidoglycan recognition proteins genes in HeLa cells using genome editing technology with synergistic activation mediators (SAM, Synergistic Activation Mediator). The obtained data demonstrate that this approach can be a promising tool for specific and targeted activation of the innate immunity components to cure severe infectious diseases.
MATERIALS AND METHODS
Bacterial strains and cell lines. In this study E. coli TOP10 (Invitrogen, USA), genotype F- mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 nupG recA1 araD139 Δ(ara-leu)7697 galE15 galK16 rpsL(StrR) endA1 λ-, C. trachomatis D/UW-3/Cx (ATCC VR-885) elementary bodies, purified by ultracentrifugation in an urografin gradient as previously described [23], were used.
We used HeLa (ATCC CCL-2, USA) and HEK-293FT (Thermo Fisher Scientific, USA) cell lines. HeLa cells were cultured in a Dulbecco’s modified Eagle’s medium (DMEM, Thermo Fisher Scientific) supplemented with 10% inactivated fetal bovine serum (FBS) (Thermo Fisher Scientific) and 10 µg/ml of gentamicin (Thermo Fisher Scientific). When simulating chlamydial infection, HeLa cells were cultured until the monolayer reached 80-90% confluence using a medium without antibiotics. The cells were infected with elementary bodies of C. trachomatis at multiplicity of infection from 0.9 to 1 IFU/cell (inclusion forming units) using centrifugation for 1 h at 900g. After centrifugation, the cells were cultured at 37°C in the presence of 5% CO2. For PGLYRP production, the PGLYRP-producing HeLa cells obtained previously in our laboratory [18] were cultured in a DMEM medium with 10 µg/ml gentamicin and without FBS for 72 h.
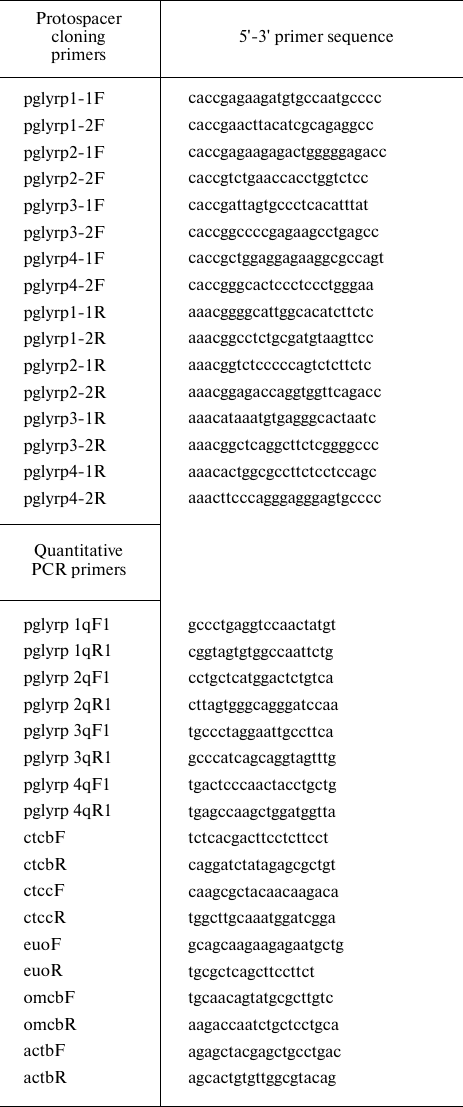
Quantitative PCR. RNA isolation from the samples was performed with Trizol reagent (Thermo Fisher Scientific). Total RNA was treated with 2 U of DNase I (Thermo Fisher Scientific) in the presence of 20 U of ribonuclease inhibitor (Thermo Fisher Scientific). First cDNA strand was synthesized using a RevertAid RT Reverse Transcription Kit (Thermo Fisher Scientific) using hexamer primers (table). Quantitative PCR was performed using a CFX96 Touch amplifier (BioRad, USA) with Taq polymerase (Lytech, Russia) and 0.1x SYBR-Green (Thermo Fisher Scientific). The amount of RNA corresponding to the target and reference genes was determined by the difference in the threshold reaction cycle (Ct) for each sample. Specific primers were used to determine the level of pglyrp1-4 expression (table). The data were normalized according to the level of the β-actin reference gene (actbF/actbR primers). The ctcbF/ctcbR and ctccF/ctccR primer pairs were used to determine the level of activation of the two-component system of chlamydia. The data were normalized to the level of the euo and omcB genes (using the euoF/euoR and omcbF/omcbR primer pairs) (table).
The list of primers used in this study
Construction of plasmid vectors for activation of expression of PGLYRP genes. The synergistic activation mediator (SAM) system includes three plasmid vectors: lenti sgRNA (MS2) puro (Addgene, USA), lenti MS2-P65-HSF1_Hygro (Addgene), and lenti dCAS-VP64_Blast (Addgene) [24]. To obtain a vector encoding a chimeric guide RNA, the lenti sgRNA (MS2) puro vector was treated with BsmBI restriction endonuclease (Thermo Fisher Scientific), after which oligonucleotide duplexes corresponding to protospacer sequences located in the promoter regions of peptidoglycan recognition proteins were cloned. The selection of protospacer sequences was performed using the crispor.org web tool based on the presence of specific PAM sequences in the promoter regions [25].
Lentivirus production and transduction. To assemble lentiviruses, the HEK-293FT culture medium was replaced with DMEM with 25 µM chloroquine diphosphate (Sigma, USA) and incubated for 5 h. The cells cultivated in 225 cm2 culture flasks were simultaneously transfected with four plasmids using polyethyleneimine: PEIMAX 40K (1 mg/ml) (Polysciences, USA) (DNA : PEI ratio = 1 : 3) according to the previously described method [26]. After incubation for 20 min at room temperature, the DNA-PEI mixture was added dropwise to the cells followed by 6-h incubation. Next, the medium was changed to OptiMEM with 2 mM sodium butyrate (Sigma). After 48 h, the medium containing lentiviruses was concentrated on Amicon 30 kDa columns (Merk, USA) until the volume was reduced 50-fold, after which it was frozen and stored at –70°C.
Concentrated viral particles were added to HeLa cells in DMEM medium containing 50 µg/ml protamine sulfate (Ellara, Russia). The cells were incubated for 6 h. Then, the medium was changed to DMEM with an appropriate antibiotic, and the cells were cultured at 37°C and 5% CO2 until the death of all non-transduced cells.
Obtaining transduced lines with activated PGLYRP expression. To obtain transduced cell lines with enhanced expression of their own PGLYRP, sequential transduction was performed with three lentiviruses with intermediate selection. Blasticidin (3 µg/ml) was used to select HeLa cells transduced with the dCAS-VP64_Blast lentivirus. For the selection of cells transduced with MS2-P65-HSF1_Hygro, blasticidin and hygromycin (3 µg/ml and 550 µg/ml, respectively) were used. The resulting cell line was named HeLa-dCas9-MS2. For the selection of cells transduced with the sgRNA (MS2) _puro-PGLYRP1-4 lentivirus, blasticidin, hygromycin, and puromycin (3 µg/ml, 550 µg/ml and 0.5 µg/ml, respectively) were used. To analyze antichlamydial activity, cells were cultivated in DMEM without antibiotics.
Fluorescent microscopy. Cells infected with C. trachomatis were stained with RecombiSlide antibodies (Galart Diagnosticum, Russia) 48 h after infection as previously described [18]. The extent of infection was quantified by counting the number of cells and inclusions using fluorescence microscopy in at least 10 fields of view in triplicate. Images were acquired using a Nikon Eclipse E800 microscope (Nikon, Japan) with argon (488 nm) and helium-neon (594 nm) lasers. Images were generated using an EZ-C1 software (Nikon) and analyzed using ImageJ 1.48 software [27]. To determine antichlamydial activity of PGLYRP, the number of inclusions in cells synthesizing PGLYRP relative to unmodified HeLa cells was determined.
Statistical analysis. The Pfaffl method was used to quantitatively analyze the pglyrp genes expression [28]. The 2–ΔΔCT method was used for quantitative analysis of the two-component system gene expression [29]. Data analysis was performed using Student’s t-test and multiple comparisons test (two-way ANOVA) using Statistica 8.0 software (StatSoft, Russia). Data are presented as mean ± standard error. Differences between the mean values were considered significant at p < 0.05.
RESULTS
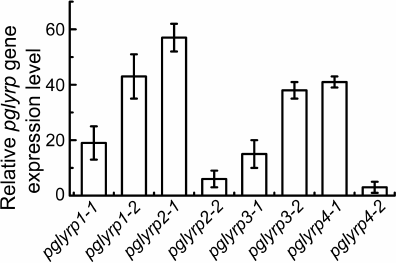
Modification of HeLa cells using CRISPR/Cas9-SAM technology for transcriptional activation of pglyrp genes. Two protospacer sequences for each pglyrp gene were selected in the promoter regions. As a result, eight plasmid vectors were constructed that encode guide RNAs for recognizing the promoter regions of the pglyrp genes. These vectors were used to generate lentiviruses and transduce HeLa-dCas9-MS2 cells. These cells synthesize defective Cas9 nuclease, incapable of making double stranded breaks in the DNA after binding with guide RNA, fused with VP16 tetramer to activate transcription. These cells also synthesize a chimeric protein of the MS2 phage fused with the activation domain of the heat shock factor 1 (HSF1) and the p65 subunit of the transcription factor NF-κB to attract transcription factors. To determine transcriptional activation of the pglyrp genes, total RNA was isolated; cDNA was obtained using hexamer primers, after which quantitative PCR was performed. It was shown that in the cells transduced with lentiviruses encoding guide RNAs with protospacer sequences pglyrp1-2, pglyrp2-1, pglyrp3-1, and pglyrp4-1, relative level of the expression of target genes was 43.2 ± 8.1, 57.0 ± 5.2, 38.5 ± 3.1, and 41.8 ± 2.6, respectively (Fig. 1). For further research, we used transduced cells with the highest expression level.
Fig. 1. Relative change in the expression level of pglyrp genes in the HeLa cell line transduced with lentiviruses encoding components of the CRISPR/Cas9-SAM activation system. For each pglyrp gene, two protospacers were selected to construct guide RNA. Bars on the histogram correspond to different protospacer sequences of guide RNAs.
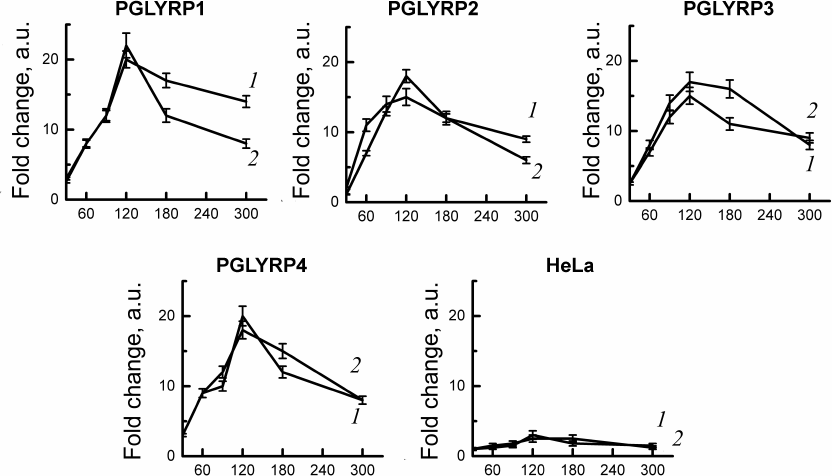
C. trachomatis two-component system activation assay. To study gene expression of the two-component system CtcB-CtcC of chlamydia activation in response to PGLYRP, we used stable cell lines producing recombinant PGLYRP, obtained by us earlier [18]. We isolated total RNA from the cells infected with C. trachomatis at 30, 60, 90, 120, 180, and 300 min after infection. Quantitative PCR has shown activation of the gene expression of the two-component system of chlamydia (ctcB and ctcC) after infection of cell lines producing recombinant PGLYRP. The mRNA levels of ctcB-ctcC were compared with the initial amount of mRNA of these genes in the intact elementary bodies. We have shown that incubation of the chlamydial elementary bodies in the culture medium with recombinant PGLYRP led to a significant increase in the mRNA level of the ctcB and ctcC genes (Fig. 2).
Fig. 2. Analysis of activation of the C. trachomatis two-component CtcB-CtcC system after interaction with recombinant PGLYRP. Total RNA was isolated from the cells 30, 60, 90, 120, 180, and 300 min after infection. Quantitative PCR data were normalized to the level of euo and omcB genes. Data are presented as change in expression level calculated by the 2–ΔΔCT method. Change in the expression level of the ctcB (curve 1) and ctcC (curve 2) genes was assessed relative to the RNA level in the intact elementary bodies not in contact with PGLYRP.
Maximum mRNA levels were observed 2 h after the contact of elementary bodies with PGLYRP. Expression of the ctcB gene increased 15-20-fold, and of ctcC – 15-22-fold relative to the initial amount of mRNA in the chlamydial elementary bodies. Addition of elementary bodies to the control HeLa cells did not produce any significant changes in the mRNA levels of ctcB-ctcC in the elementary bodies.
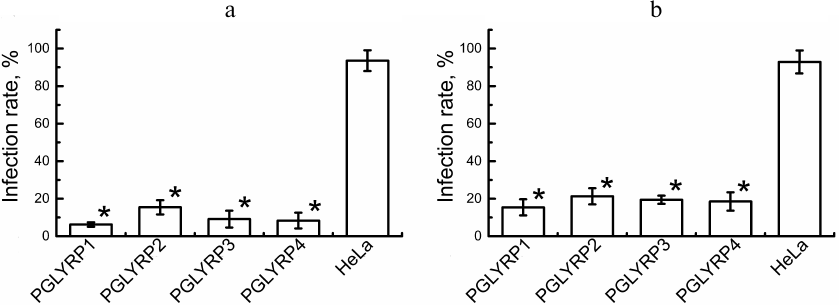
Antichlamydial activity of PGLYRP. To determine antichlamydial activity of PGLYRP, we counted the number of C. trachomatis inclusions formed in the stable HeLa cell lines producing recombinant PGLYRP and transduced HeLa cells with activated pglyrp gene expression 48 h after the addition of elementary bodies to the medium containing PGLYRP. The infection efficiency was determined ((number of inclusions/total number of cells) × 100%). The efficiency of infection of the recombinant cells’ producers of PGLYRP with C. trachomatis was 6.2 ± 1.0%, 15.4 ± 2.9%, 9.1 ± 3.3%, and 8.3 ± 4.0% for PGLYRP1, PGLYRP2, PGLYRP3, and PGLYRP4, respectively, while the infection efficiency for the control cells was 93.5 ± 6.2% (Fig. 3a). The infection efficiency of the pools of the transduced HeLa-PGLYRP cells with elementary bodies was 15.4 ± 3.2%, 21.3 ± 3.2%, 19.4 ± 1.8%, and 18.5 ± 3.7% for PGLYRP1, PGLYRP2, PGLYRP3, and PGLYRP4 producer cells, respectively, 48 h after infection. The infection efficiency of the control cells was 92.9 ± 6.9% (Fig. 3b). Data are expressed as mean ± standard error.
Fig. 3. Determination of PGLYRP antichlamydial activity. a) Histogram showing efficiency of infection of the recombinant PGLYRP producers HeLa cells. b) Histogram showing efficiency of infection of the transduced HeLa cells with enhanced pglyrp genes expression produced using CRISPR/Cas9-SAM genome editing technology. * p < 0.05.
Thus, we have shown that enhancement of expression of the human peptidoglycan recognition proteins genes in the cell line leads to inhibition of the development of chlamydial infection in vitro.
DISCUSSION
Skin and mucous membranes protect body from pathogenic microflora. These tissues form not only mechanical barrier but also synthesize and secrete various antimicrobial peptides and proteins [30, 31]. Antimicrobial proteins and peptides are considered the most promising anti-infective agents because they cause fewer side effects as compared to antibiotics [32]. Moreover, it is difficult to develop resistance to them due to the specific mechanism of action [33]. The study of the innate defense systems of the body may be an alternative way to search for new antimicrobial drugs. Peptidoglycan-recognition proteins are a class of bactericidal proteins of the innate immune system. These proteins are active against pathogenic Gram-negative and Gram-positive bacteria in vivo and in vitro [12, 34] and have a unique mechanism of action. PGLYRP binds to peptidoglycan of the bacterial cell wall during division and activates the bacterial two-component stress response system, resulting in the induction of membrane depolarization and production of hydroxyl radicals in the cytoplasm [12]. This process is accompanied by the termination of all main intracellular biosynthetic reactions, probably due to the lack of energy, depending on the membrane potential [35]. Two-component protein-sensitive systems that play a role in this mechanism of action are designed to detect and utilize misfolded bacterial proteins that could not enter periplasm or be excreted from the cell and are stuck in the membrane [8, 9]. PGLYRP binding to peptidoglycan causes a change in PGLYRP, which blocks peptidoglycan in the peptidoglycan-binding groove and makes binding irreversible [36]. PGLYRP also have a hydrophobic part on the opposite side of the peptidoglycan-binding groove, as a result of which these proteins are recognized as misfolded proteins, which usually have hydrophobic regions exposed or aggregated proteins since misfolded proteins often aggregate and bind to peptidoglycan [37]. Peculiarity of the mechanism of action of PGLYRP is that PGLYRP kills bacteria while subjecting them to oxidative, thiol-disulfide, and metal stress. Oxidative stress induced by PGLYRP occurs due to the increased production of hydrogen peroxide and hydroxyl radicals [5, 38, 39]. Thiol-disulfide stress occurs due to depletion of the pool of intracellular thiols, especially glutathione, by more than 90%, and metal stress arises in connection with the increase in the intracellular concentration of Zn2+ and Cu2+ ions [38, 39]. Induction of all three stress responses is required for initiation of bactericidal effect. Each stress response separately is only bacteriostatic, and only the combined induction of all three events is bactericidal [39]. Resistance to PGLYRP could arise if all three stress responses (i.e., oxidative, thiol, and metallic stress) were induced by PGLYRP via a single signaling pathway. However, PGLYRP induces oxidative, thiol, and metal stress via three independent pathways [38]. Moreover, each PGLYRP has a different spectrum of bactericidal activity [34]. Apparently, bacteria of normal flora have developed resistance to bactericidal proteins in organs and tissues where PGLYRP is constantly secreted (for example, PGLYRP-3 and PGLYRP-4 are secreted in the skin, eyes and mucous membranes) and can colonize these areas. Bacteriostatic effect of PGLYRP on bacteria of normal flora has physiological significance for the host – bacteria of normal flora do not die, but their growth is limited [13].
Peptidoglycan is a major component of bacterial cell walls and is a target for some antibacterial drugs. Previously, it was thought that chlamydiae lack peptidoglycan, although its genome contains all the components for its biosynthesis [40, 41]; therefore, the effect of PGLYRP on chlamydia has not been studied. In 2014, the presence of peptidoglycan in chlamydia was shown [42]. Chlamydia also has a two-component system (CtcB-CtcC) [43], which may take part in the mechanism of action of PGLYRP. Chlamydiae are obligate intracellular Gram-negative bacteria that can bypass the immune defense mechanisms and remain viable in the cells for a long period of time [44]. Previously, we demonstrated for the first time antichlamydial activity of isolated and purified recombinant peptidoglycan recognition proteins produced in the human Expi293 cells, determined minimum inhibitory concentrations [18], and estimated the level of protein accumulation in the cell lines [45]. In this study, we evaluated antichlamydial effect of the stable HeLa cell lines that secrete recombinant human PGLYRP1, PGLYRP2, PGLYRP3, and PGLYRP4. We have shown that upon infection of proteins producer cells, the number of identifiable chlamydial inclusions inside the cells is lower than upon infection of control cells (Fig. 3a).
We demonstrated that the contact of PGLYRP with C. trachomatis elementary bodies increased the level of gene expression of the two-component system, which could lead to inhibition of the chlamydial infection (Fig. 2). Moreover, the maximum amount of mRNA of genes of the two-component system was observed 2 h after contact with a solution of the recombinant protein. For some bacterial two-component systems, a mechanism of autoactivation was described, which could explain the excessive induction of the stress response [46]. Thus, the mechanism of action of PGLYRP on chlamydia is likely to be similar to the action of these proteins on other bacteria.
There are limitations in the use of recombinant proteins for treatment of infectious diseases. These include low stability of some isolated recombinant proteins [47], as well as limited possibility of local application [48]. An alternative option for using recombinant proteins can be gene therapy approaches by activating the innate immune system’s genes to fight infection [49]. Gene therapy for infectious diseases has significant potential for treating infections that do not respond to standard clinical treatment [50-54]. At the same time, one of the methods of therapy is the induction of innate immunity, for example, through activation of the signaling pathways of Toll-like receptors [55, 56].
To determine antichlamydial activity of the human PGLYRP, we increased expression level of the pglyrp genes in HeLa cells using the CRISPR/Cas9-SAM system. The system of synergistic activation mediators consists of three components: a specific chimeric guide RNA, which has two MS2 aptamers for binding to dimerized coat proteins of the bacteriophage MS2. Then, MS2 proteins with various activators, such as the p65 activation domain of the NF-κB transcription factor, and the activation domain of the heat shock protein HSF1, which recruit transcription factors around the promoter of the target gene. Finally, it has the defective dCas9-VP64 nuclease, which is fused to the VP16 tetramer, which is a transcription activator. Such system provides a significant increase in the expression of target genes [57].
After transduction with lentiviruses encoding the CRISPR/Cas9-SAM activation system, HeLa cells demonstrated an increased level of gene expression of peptidoglycan-recognizing proteins (Fig. 1), as well as the presence of proteins in the culture medium (data not shown). We showed that the efficiency of infection of the cells with activated pglyrp genes expression with chlamydia is reduced by 79.5-85% in comparison to the control (Fig. 3b).
The use of antibiotics promotes the emergence of resistant strains. One of the ways to solve the problem of bacterial resistance to the drugs can be the use of innate immunity proteins. This is because the development of resistance in this case is difficult due to the specific mechanisms of action of these proteins. Even though the use of recombinant proteins of innate immunity obtained in hetero- and homologous expression systems is beneficial due to the absence of toxic effects, their use is limited due to the difficulty of local application. This work describes an approach that allows us to solve this disadvantage of the therapeutic use of innate immunity proteins by activating expression of the peptidoglycan recognition proteins genes in eukaryotic cells. The proposed approach makes it possible to use the resources of innate immunity to fight infections, including those caused by antibiotic-resistant strains.
Funding. This research was financially supported by the Russian Science Foundation (project no. 20-15-00270).
Ethics declarations. The authors declare no conflicts of interest in financial or any other sphere. This article does not contain any studies with human participants or animals performed by any of the authors.
REFERENCES
1.Prestinaci, F., Pezzotti, P., and Pantosti, A.
(2015) Antimicrobial resistance: a global multifaceted phenomenon,
Pathog. Glob. Health, 109, 309-318, doi:
10.1179/2047773215y.0000000030.
2.Grill, M. F., and Maganti, R. K. (2011) Neurotoxic
effects associated with antibiotic use: management considerations,
Br. J. Clin. Pharmacol., 72, 381-393, doi:
10.1111/j.1365-2125.2011.03991.x.
3.Roberts, M. C. (2002) Antibiotic toxicity,
interactions and resistance development, Periodontol. 2000,
28, 280-297, doi: 10.1034/j.1600-0757.2002.280112.x.
4.Liu, C., Xu, Z., Gupta, D., and Dziarski, R. (2001)
Peptidoglycan recognition proteins: a novel family of four human innate
immunity pattern recognition molecules, J. Biol. Chem.,
276, 34686-34694, doi: 10.1074/jbc.M105566200.
5.Kashyap, D. R., Wang, M., Liu, L. H., Boons, G. J.,
Gupta, D., and Dziarski, R. (2011) Peptidoglycan recognition proteins
kill bacteria by activating protein-sensing two-component systems,
Nat. Med., 17, 676-683, doi: 10.1038/nm.2357.
6.Sharma, P., Dube, D., Singh, A., Mishra, B., Singh,
N., et al. (2011) Structural basis of recognition of
pathogen-associated molecular patterns and inhibition of
proinflammatory cytokines by camel peptidoglycan recognition protein,
J. Biol. Chem., 286, 16208-16217, doi:
10.1074/jbc.M111.228163.
7.Tydell, C. C., Yuan, J., Tran, P., and Selsted, M.
E. (2006) Bovine peptidoglycan recognition protein-S: antimicrobial
activity, localization, secretion, and binding properties., J.
Immunol., 176, 1154-1162, doi:
10.4049/jimmunol.176.2.1154.
8.Hyyryläinen, H. L., Bolhuis, A., Darmon, E.,
Muukkonen, L., Koski, P., et al. (2001) A novel two-component
regulatory system in Bacillus subtilis for the survival of
severe secretion stress, Mol. Microbiol., 41, 1159-1172,
doi: 10.1046/j.1365-2958.2001.02576.x.
9.Kohanski, M. A., Dwyer, D. J., Wierzbowski, J.,
Cottarel, G., and Collins, J. J. (2008) Mistranslation of membrane
proteins and two-component system activation trigger
antibiotic-mediated cell death, Cell, 135, 679-690, doi:
10.1016/j.cell.2008.09.038.
10.Darmon, E., Noone, D., Masson, A., Bron, S.,
Kuipers, O. P., Devine, K. M., and van Dijl, J. M. (2002) A novel class
of heat and secretion stress-responsive genes is controlled by the
autoregulated CssRS two-component system of Bacillus subtilis,
J. Bacteriol., 184, 5661-5671, doi:
10.1128/jb.184.20.5661-5671.2002.
11.Westers, H., Westers, L., Darmon, E., van Dijl,
J. M., Quax, W. J., and Zanen, G. (2006) The CssRS two-component
regulatory system controls a general secretion stress response in
Bacillus subtilis, FEBS J., 273, 3816-3827, doi:
10.1111/j.1742-4658.2006.05389.x.
12.Dziarski, R., Kashyap, D. R., and Gupta, D.
(2012) Mammalian peptidoglycan recognition proteins kill bacteria by
activating two-component systems and modulate microbiome and
inflammation., Microb. Drug Res., 18, 280-285, doi:
10.1089/mdr.2012.0002.
13.Dziarski, R., and Gupta, D. (2006) Mammalian
PGRPs: novel antibacterial proteins, Cell. Microbiol., 8,
1059-1069, doi: 10.1111/j.1462-5822.2006.00726.x.
14.O’Connell, C. M., and Ferone, M. E. (2016)
Chlamydia trachomatis genital infections, Microb. Cell,
3, 390-403, doi: 10.15698/mic2016.09.525.
15.Kalayoglu, M. V. (2002) Ocular chlamydial
infections: pathogenesis and emerging treatment strategies, Curr.
Drug Targets Infect. Disord., 2, 85-91, doi:
10.2174/1568005024605918.
16.Moulder, J. W. (1991) Interaction of chlamydiae
and host cells in vitro, Microb. Rev., 55,
143-190.
17.Panzetta, M. E., Valdivia, R. H., and Saka, H. A.
(2018) Chlamydia persistence: a survival strategy to evade
antimicrobial effects in vitro and in vivo, Front.
Microbiol., 9, 3101, doi: 10.3389/fmicb.2018.03101.
18.Bobrovsky, P., Manuvera, V., Polina, N.,
Podgorny, O., Prusakov, K., Govorun, V., and Lazarev, V. (2016)
Recombinant human peptidoglycan recognition proteins reveal
antichlamydial activity, Infect. Immun., 84, doi:
10.1128/IAI.01495-15.
19.Zhu, M., Miao, B., Zhu, J., Wang, H., and Zhou,
Z. (2017) Expression and antimicrobial character of cells transfected
with human β-defensin-3 against periodontitis-associated
microbiota in vitro, Mol. Med. Rep., 16,
2455-2460, doi: 10.3892/mmr.2017.6913.
20.Zhang, J., Xie, L., Xu, D., Yue, S., Li, Y., Guo,
X., and Lai, X. (2017) Targeting expression of antimicrobial peptide
CAMA-Syn by adenovirus vector in macrophages inhibits the growth of
intracellular bacteria, Gene, 630, 59-67, doi:
10.1016/j.gene.2017.07.079.
21.Ramos-Espinosa, O.,
Hernández-Bazán, S., Francisco-Cruz, A., Mata-Espinosa,
D., et al. (2016) Gene therapy based in antimicrobial peptides and
proinflammatory cytokine prevents reactivation of experimental latent
tuberculosis, Pathog. Dis., 74, doi:
10.1093/femspd/ftw075.
22.Dolgachev, V., Panicker, S., Balijepalli, S.,
McCandless, L. K., Yin, Y., et al. (2018) Electroporation-mediated
delivery of FER gene enhances innate immune response and improves
survival in a murine model of pneumonia, Gene Ther., 25,
359-375, doi: 10.1038/s41434-018-0022-y.
23.Scidmore, M. A. (2005) Cultivation and laboratory
maintenance of Chlamydia trachomatis, Curr. Protoc.
Microbiol., 11, 11A.1, doi:
10.1002/9780471729259.mc11a01s00.
24.Joung, J., Konermann, S., Gootenberg, J. S.,
Abudayyeh, O. O., Platt, R. J., et al. (2017) Genome-scale CRISPR-Cas9
knockout and transcriptional activation screening, Nat. Protoc.,
12, 828-863, doi: 10.1038/nprot.2017.016.
25.Haeussler, M., Schönig, K., Eckert, H.,
Eschstruth, A., Mianné, J., et al. (2016) Evaluation of
off-target and on-target scoring algorithms and integration into the
guide RNA selection tool CRISPOR, Genome Biol., 17, 148,
doi: 10.1186/s13059-016-1012-2.
26.Tiscornia, G., Singer, O., and Verma, I. M.
(2006) Production and purification of lentiviral vectors, Nat.
Protoc., 1, 241-245, doi: 10.1038/nprot.2006.37.
27.Schneider, C. A., Rasband, W. S., and Eliceiri,
K. W. (2012) NIH Image to ImageJ: 25 years of image analysis, Nat.
Methods, 9, 671-675, doi: 10.1038/nmeth.2089.
28.Pfaffl, M. W. (2001) A new mathematical model for
relative quantification in real-time RT-PCR, Nucleic Acids Res.,
29, e45, doi: 10.1093/nar/29.9.e45.
29.Rao, X., Huang, X., Zhou, Z., and Lin, X. (2013)
An improvement of the 2ˆ(-delta delta CT) method for quantitative
real-time polymerase chain reaction data analysis, Biostat.
Bioinform. Biomath., 3, 71-85.
30.Schittek, B., Paulmann, M., Senyürek, I.,
and Steffen, H. (2008) The role of antimicrobial peptides in human skin
and in skin infectious diseases, Infect. Dis. Drug Targets,
8, 135-143, doi: 10.2174/1871526510808030135.
31.Schauber, J., and Gallo, R. L. (2008)
Antimicrobial peptides and the skin immune defense system, J.
Allergy Clin. Immunol., 122, 261-266, doi:
10.1016/j.jaci.2008.03.027.
32.Huang, G. T., Zhang, H. B., Kim, D., Liu, L., and
Ganz, T. (2002) A model for antimicrobial gene therapy: demonstration
of human beta-defensin 2 antimicrobial activities in vivo,
Hum. Gene Ther., 13, 2017-2025, doi:
10.1089/10430340260395875.
33.Joo, H. S., Fu, C. I., and Otto, M. (2016)
Bacterial strategies of resistance to antimicrobial peptides,
Philos. Trans. R. Soc. Lond. B Biol. Sci., 371, 20150292,
doi: 10.1098/rstb.2015.0292.
34.Lu, X., Wang, M., Qi, J., Wang, H., Li, X.,
Gupta, D., and Dziarski, R. (2006) Peptidoglycan recognition proteins
are a new class of human bactericidal proteins, J. Biol. Chem.,
281, 5895-5907, doi: 10.1074/jbc.M511631200.
35.Dimroth, P., Kaim, G., and Matthey, U. (2000)
Crucial role of the membrane potential for ATP synthesis by F(1)F(0)
ATP synthases, J. Exp. Biol., 203, 51-59.
36.Wang, Z. M., Li, X., Cocklin, R. R., Wang, M.,
Wang, M., et al. (2003) Human peptidoglycan recognition protein-L is an
N-acetylmuramoyl-L-alanine amidase, J. Biol. Chem., 278,
49044-49052, doi: 10.1074/jbc. M307758200.
37.Lim, J. H., Kim, M. S., Kim, H. E., Yano, T.,
Oshima, Y., et al. (2006) Structural basis for preferential recognition
of diaminopimelic acid-type peptidoglycan by a subset of peptidoglycan
recognition proteins, J. Biol. Chem., 281, 8286-8295,
doi: 10.1074/jbc.M513030200.
38.Kashyap, D. R., Kuzma, M., Kowalczyk, D. A.,
Gupta, D., and Dziarski, R. (2017) Bactericidal peptidoglycan
recognition protein induces oxidative stress in Escherichia coli
through a block in respiratory chain and increase in central carbon
catabolism, Mol. Microbiol., 105, 755-776, doi:
10.1111/mmi.13733.
39.Kashyap, D. R., Rompca, A., Gaballa, A., Helmann,
J. D., Chan, J., Chang, C. J., Hozo, I., Gupta, D., and Dziarski, R.
(2014) Peptidoglycan recognition proteins kill bacteria by inducing
oxidative, thiol, and metal stress, PLoS Pathog., 10,
e1004280, doi: 10.1371/journal.ppat.1004280.
40.Chopra, I., Storey, C., Falla, T. J., and Pearce,
J. H. (1998) Antibiotics, peptidoglycan synthesis and genomics: the
chlamydial anomaly revisited, Microbiology (Reading),
144, 2673-2678, doi: 10.1099/00221287-144-10-2673.
41.Stephens, R. S., Kalman, S., Lammel, C., Fan, J.,
Marathe, R., et al. (1998) Genome sequence of an obligate intracellular
pathogen of humans: Chlamydia trachomatis, Science,
282, 754-759, doi: 10.1126/science.282.5389.754.
42.Liechti, G. W., Kuru, E., Hall, E., Kalinda, A.,
Brun, Y. V., VanNieuwenhze, M., and Maurelli, A. T. (2014) A new
metabolic cell-wall labelling method reveals peptidoglycan in
Chlamydia trachomatis, Nature, 506, 507-510, doi:
10.1038/nature12892.
43.Koo, I. C., and Stephens, R. S. (2003) A
developmentally regulated two-component signal transduction system in
Chlamydia, J. Biol. Chem., 278, 17314-17319, doi:
10.1074/jbc.M212170200.
44.Witkin, S. S., Minis, E., Athanasiou, A., Leizer,
J., and Linhares, I. M. (2017) Chlamydia trachomatis: the
persistent pathogen, Clin. Vaccine Immunol., 24, doi:
10.1128/cvi.00203-17.
45.Bobrovsky, P. A., Larin, A. K., Polina, N. F.,
and Lazarev, V. N. (2019) Transcriptional analysis of HELA cells
– producers of the recombinant peptidoglycan recognition protein
PGLYRP1 at different stages of the Chlamydia trachomatis
infection development, Biomed. Chem. Res. Methods, 2,
e00113, doi: 10.18097/BMCRM00113.
46.De Wulf, P., Kwon, O., and Lin, E. C. (1999) The
CpxRA signal transduction system of Escherichia coli:
growth-related autoactivation and control of unanticipated target
operons, J. Bacteriol., 181, 6772-6778.
47.Deller, M. C., Kong, L., and Rupp, B. (2016)
Protein stability: a crystallographer’s perspective, Acta
Crystallogr. F Struct. Biol. Commun., 72, 72-95, doi:
10.1107/s2053230x15024619.
48.Muheem, A., Shakeel, F., Jahangir, M. A., Anwar,
M., Mallick, N., Jain, G. K., Warsi, M. H., and Ahmad, F. J. (2016) A
review on the strategies for oral delivery of proteins and peptides and
their clinical perspectives, Saudi Pharm. J., 24,
413-428, doi: 10.1016/j.jsps.2014.06.004.
49.O’Neill, L. A., Bryant, C. E., and Doyle,
S. L. (2009) Therapeutic targeting of toll-like receptors for
infectious and inflammatory diseases and cancer, Pharmacol.
Rev., 61, 177-197, doi: 10.1124/pr.109.001073.
50.Sierra-Delgado, J. A., Bautista-Nino, P. K.,
Vargas-Castellanos, C. I., Serrano Diaz, N. C., and Rincon, M. Y.
(2019) Immune response and gene therapy with adenoassociated viral
vectors, Medicina, 79, 493-501.
51.Bergmann, B., Fei, Y., Jirholt, P., Hu, Z.,
Bergquist, M., et al. (2020) Pre-treatment with IL2 gene therapy
alleviates Staphylococcus aureus arthritis in mice, BMC
Infect. Dis., 20, 185, doi: 10.1186/s12879-020-4880-8.
52.Chandler, L. C., Yusuf, I. H., McClements, M. E.,
Barnard, A. R., MacLaren, R. E., and Xue, K. (2020) Immunomodulatory
effects of hydroxychloroquine and chloroquine in viral infections and
their potential application in retinal gene therapy, Int. J. Mol.
Sci., 21, doi: 10.3390/ijms21144972.
53.Verma, R., Sahu, R., Singh, D. D., and Egbo, T.
E. (2019) A CRISPR/Cas9 based polymeric nanoparticles to treat/inhibit
microbial infections, Semin. Cell Dev. Biol., 96, 44-52,
doi: 10.1016/j.semcdb.2019.04.007.
54.Lazarev, V. N., Polina, N. F., Shkarupeta, M. M.,
Kostrjukova, E. S., Vassilevski, A. A., Kozlov, S. A., Grishin, E. V.,
and Govorun, V. M. (2011) Spider venom peptides for gene therapy of
Chlamydia infection, Antimicrob. Agents Chemother.,
55, 5367-5369, doi: 10.1128/aac.00449-11.
55.Abe, T., Kaname, Y., Wen, X., Tani, H., Moriishi,
K., et al. (2009) Baculovirus induces type I interferon production
through toll-like receptor-dependent and -independent pathways in a
cell-type-specific manner, J. Virol., 83, 7629-7640, doi:
10.1128/jvi.00679-09.
56.Abe, T., Hemmi, H., Miyamoto, H., Moriishi, K.,
Tamura, S., Takaku, H., Akira, S., and Matsuura, Y. (2005) Involvement
of the toll-like receptor 9 signaling pathway in the induction of
innate immunity by baculovirus, J. Virol., 79, 2847-2858,
doi: 10.1128/jvi.79.5.2847-2858.2005.
57.Konermann, S., Brigham, M. D., Trevino, A. E.,
Joung, J., Abudayyeh, O. O., et al. (2015) Genome-scale transcriptional
activation by an engineered CRISPR-Cas9 complex, Nature,
517, 583-588, doi: 10.1038/nature14136.