REVIEW: Engineering Artificial Biodiversity of Lantibiotics to Expand Chemical Space of DNA-Encoded Antibiotics
S. O. Pipiya1, S. S. Terekhov1,2, Yu. A. Mokrushina1,2, V. D. Knorre1, I. V. Smirnov1,2,a*, and A. G. Gabibov1,2
1Shemyakin and Ovchinnikov Institute of Bioorganic Chemistry, Russian Academy of Sciences, 117997 Moscow, Russia2Faculty of Chemistry, Lomonosov Moscow State University, 119991 Moscow, Russia
* To whom correspondence should be addressed.
Received August 11, 2020; Revised September 28, 2020; Accepted October 6, 2020
The discovery of antibiotics was one of the fundamental stages in the development of humanity, leading to a dramatic increase in the life expectancy of millions of people all over the world. The uncontrolled use of antibiotics resulted in the selection of resistant strains of bacteria, limiting the effectiveness of antimicrobial therapy nowadays. Antimicrobial peptides (AMPs) were considered promising candidates for next-generation antibiotics for a long time. However, the practical application of AMPs is restricted by their low therapeutic indices, impaired pharmacokinetics, and pharmacodynamics, which is predetermined by their peptide structure. Nevertheless, the DNA-encoded nature of AMPs enables creating broad repertoires of artificial biodiversity of antibiotics, making them versatile templates for the directed evolution of antibiotic activity. Lantibiotics are a unique class of AMPs with an expanded chemical space. A variety of post-translational modifications, mechanisms of action on bacterial membranes, and DNA-encoded nature make them a convenient molecular template for creating highly representative libraries of antimicrobial compounds. Isolation of new drug candidates from this synthetic biodiversity is extremely attractive but requires high-throughput screening of antibiotic activity. The combination of synthetic biology and ultrahigh-throughput microfluidics allows implementing the concept of directed evolution of lantibiotics for accelerated creation of new promising drug candidates.
KEY WORDS: directed evolution of antimicrobial activity, DNA-encoded antibiotics, ultrahigh-throughput screening, microfluidics, lantibiotic bioengineering, drug discovery, antibiotic resistanceDOI: 10.1134/S0006297920110048
Abbreviations: AMP, antimicrobial peptides; Dha, dehydroalanine; Dhb, dehydrobutyrine; PE, phosphatidylethanolamine; RiPPs, ribosomally synthesized and post-translationally modified peptides; UEV domain, ubiquitin E2 variant domain.
INTRODUCTION
Antibiotics have served humanity for decades, saving millions of lives from deadly infections. After the discovery of penicillin, the so-called “the Golden Age” of antibiotic discovery began. Over the period from 1928 to 1980, more than 10 types of antibiotics were discovered [1]. However, nowadays humanity faces a serious threat from the widespread emergence of microbial resistance to antimicrobial agents due to their uncontrolled use in agriculture and improper and excessive use in medicine. Humanity is entering the post-antibiotic era [2] according to the World Health Organization (WHO). This scenario is further complicated by the emergence of multiple drug-resistant strains that are no longer sensitive to several types of antibiotics. The most common in clinical practice are methicillin-resistant Staphylococcus aureus (MRSA), vancomycin-resistant Staphylococcus aureus (VRSA), and vancomycin-resistant Enterococci (VRE). They represent the leading cause of serious nosocomial infections. Clostridial infection is another example of common infectious cause of the hospital-acquired diarrhea. In recent years, the number of infected people and mortality rates are increasing, and new treatment options are needed [3]. Thus, the scientific and medical community emphasizes the essential role of studies devoted to design of alternative antimicrobial drugs.
Several approaches are currently being developed to find new antibiotics. Synthetic biology techniques and rational design methods are used to create compounds with improved antimicrobial and stability properties [4]. The development of bioinformatic methods and the expansion of genome databases made it possible to find new biosynthetic clusters of antimicrobial compounds using genome mining techniques [5]. High-throughput screening techniques are used to find natural antibiotics in exotic microbial communities [6]. The use of artificial intelligence in virtual screening of chemical compound libraries for the ability to exhibit significant antimicrobial activity also offers some promise in this field [7].
Safety and efficacy are crucial criteria for the development of new compounds useful in clinical practice. Peptides are promising candidates for the role of model entities for the creation of new drugs. They have high selectivity and efficiency of interaction with the targets and are easily metabolized in the body. Advances in peptide synthesis and biotechnology promote the reduction of production costs [8]. To date, more than 60 peptide drugs are on the market. They are used for the treatment of diabetes, cancer, and orphan diseases [9].
Peptides have high potential in the area of development of novel antimicrobials. However, antimicrobial peptides (AMP) have not been widely applied in practice, despite numerous attempts to create therapeutic drugs on their basis. Clinical applications of AMPs are limited due to the many complexities often associated with low selectivity [10]. The natural level of AMP production is relatively low and chemical synthesis is often associated with technical difficulties and large financial investments [11]. One of the most serious drawbacks of AMPs is their sensitivity to proteolysis, which dramatically impairs pharmacokinetic properties. Nonetheless, there are promising classes of AMPs that can serve as a template for the generation of peptide antibiotics by the methods of directed evolution and high-throughput screening.
Ribosomally synthesized and post-translationally modified peptides (RiPPs) attract particular interest in this case. Biosynthesis of RiPPs begins with the formation of a precursor peptide. It consists of a leader sequence that is recognized by the modifying enzymes and a structural sequence that undergoes subsequent modifications. At the last stage, the leader sequence is processed by proteases, and the mature RiPP is exported into the extracellular environment [12].
The genetically encoded sequence of these peptides makes it easy to introduce changes in their structure [13]. A large set of possible modifications potentially creates a wide range of structures and functional groups combined in a single molecule, which, in turn, can be produced by recombinant organisms. Thus, RiPPs have unique advantages for creating compounds with different biological activities, improving the stability of these molecules [14], and making them attractive for therapeutic use [15, 16].
Considerable interest in this area is supported by the development of the whole-genome sequencing technologies and metagenomic mining, intensifying the identification of gene clusters potentially responsible for RiPPs biosynthesis [17, 18]. Approximately 20 families of RiPPs are known that have characteristic structures, biosynthetic pathways, and biological activity [19]. They include lantipeptides, cyanobactins, botramycins, lasso peptides, microviridins, and others.
LANTIPEPTIDES AND LANTIBIOTICS
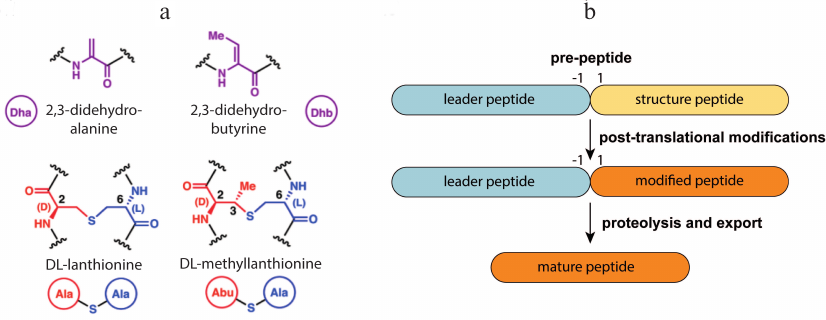
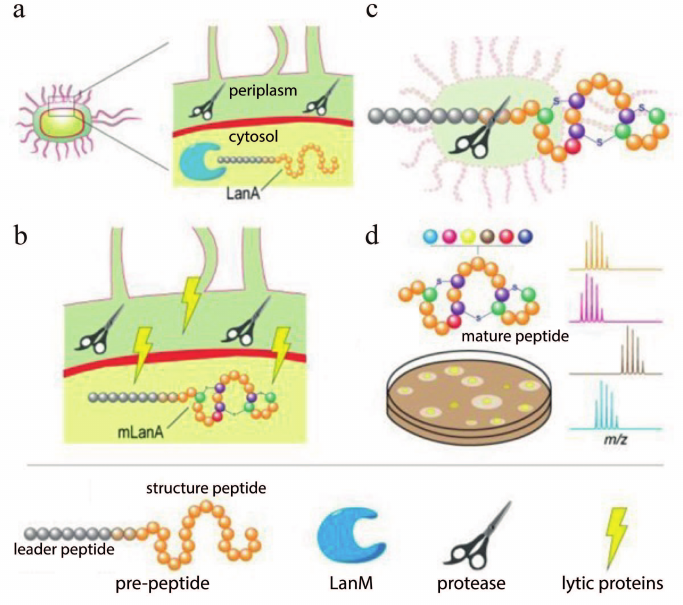
Lantipeptides are a group of RiPPs produced by Gram-positive bacteria [20]. They contain residues of non-canonical amino acids lanthionine and methyllanthionine, which form intramolecular thioether bonds (Fig. 1a), and several other modifications such as dehydration of Thr or Ser residues forming dehydrobutyrine (Dhb) and dehydroalanine (Dha), N-terminal acetylation, etc. Lantipeptides are encoded in the producer’ s genome in the form of a pre-peptide consisting of a leader sequence and a structural peptide. The leader sequence is recognized by the modifying enzyme and directs the maturation of the lantipeptide. Structural peptides undergo modification, cleavage of the leader sequence, and then become active molecules (Fig. 1b). It is known that lantipeptides can act as signaling or regulatory molecules [21, 22], but the most attractive group of them are the so-called lantibiotics (lantipeptides with antibacterial activity).
Fig. 1. a) Structure of dehydrated amino acid residues and thioether cross-links typical for lantipeptides. b) General lantipeptide biosynthetic pathway; adapted from [20]. Designations: Dha, dehydroalanine; Dhb, dehydrobutyrine; Abu, alpha-aminobutyric acid.
Genes encoding enzymes for the lantipeptide biosynthesis are often identified as clusters on chromosomes, mobile elements, or plasmids of Gram-positive bacteria. According to the commonly accepted designations [23], the gene coding for the structural peptide is called lanA. The enzymes that carry out basic modifications are lanB and lanC or lanM, lanL (for multifunctional enzymes modifying lantibiotics of classes 2 and 4), lanT for the transporter protein, lanP – specific protease that processes the leader sequence, additional modifying enzymes have their unique names. Lan in the name of a gene or its product is replaced with the relevant abbreviation for a specific lantibiotic.
CLASSIFICATION OF LANTIBIOTICS
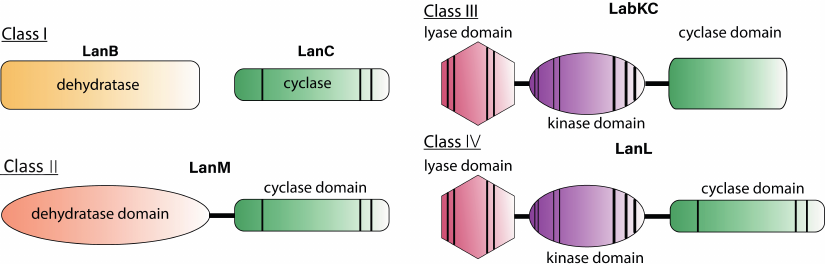
The modern classification system for lantibiotics is based on the difference in the structure of enzymes that dehydrate and cyclize the structural peptide and mechanisms by which these modifications are introduced [19]. Today there are 4 main classes of lantipeptides (Fig. 2).
Fig. 2. Schematic representation of enzymes involved in the biosynthesis of lantibiotics. Vertical bands indicate conserved regions that are required for catalytic activity, bandwidth schematically illustrates the relative size of the conserved region.
Class I. This class includes lantibiotics in which the main modifications are made by two separate enzymes: LanB, which performs dehydration, and LanC, which enables cyclization and theformation of lanthionine and methylanthionine residues. The biosynthetic cluster also includes specific proteases, ABC-transporters, enzymes that introduce additional modifications, regulatory elements, and proteins that provide self-immunity to lantibiotic. Genomic studies have shown that lantibiotic class I biosynthetic clusters are widespread among the bacteria from different phyla [24].
Lantipeptides of this class are divided into subgroups: nisin-like, epidermin-like, and Pep5-like lantipeptides, based on the similarity of the pre-peptide leader sequence recognized by the modification system.
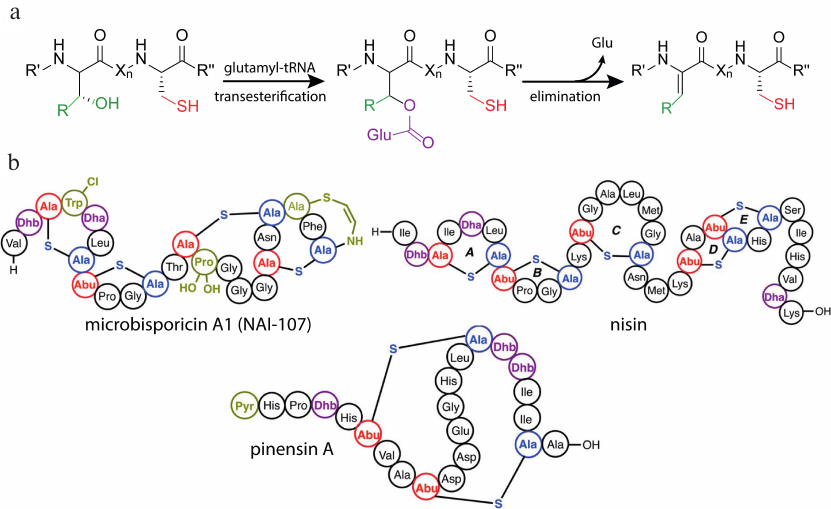
A distinctive feature of the class I lantibiotics is the mechanism of dehydration of the structural peptide. Mechanistic studies of biosynthesis were mainly focused on the nisin as the best-studied and most significant representative of the class. Despite the success in producing the heterologous system of nisin biosynthesis [25], the researchers were unable to reconstruct this process in vitro for a long time. The first positive results were reported by Garg et al. [26], in which the addition of ATP, MgCl2, and cell lysate from Escherichia coli to NisB in the reaction mixture resulted in complete dehydration of the NisA pre-peptide. Examination of the mutated forms of the enzyme showed the formation of a glutamylated intermediate product, that could be converted into the dehydrated peptide without additional components. The ultimate mechanism of dehydration of NisB was discovered after determining the role of the source of glutamate residues, glutamyl-tRNA [27]. LanB activates the hydroxyl group on the Ser/Thr side group by adding a glutamate residue from the glutamyl-tRNA. Then directed dissociation of the proton from the activated amino acid occurs, which leads to the elimination of glutamate with the formation of the dehydroamino acid (Fig. 3a).
Fig. 3. a) Scheme describing dehydration of the class I lantibiotics by LanB enzyme. b) Structures of some class I lantibiotics with various post-translational modifications. Designations: Lac, lactate; Dha, dehydroalanine: Dhb, dehydrobutyrine; Pyr, 2-oxopropionyl; Abu, alpha-aminobutyric acid; R′, R′′, lantipeptide polypeptide chain; Xn, fragment of the polypeptide chain consisting of n amino acid residues, where n is a number from 1 to 20; adapted from [20].
Cyclization and formation of the thioether ring are mediated by the enzyme LanC. It has structural similarity to the cyclase domain of multifunctional enzymes LanM [28] and LanL [29] of lantibiotics class II and IV, respectively. This similarity originates from the presence of the zinc-binding domain in these groups of enzymes, which may indicate similar cyclization mechanisms.
The mature peptide is transported by the specific LanT transporters. The membrane association, as well as the presence of two ATP-binding Walker motifs, indicates that they belong to the ABC-transporter family [30]. LanTs recognize the leader sequence during the transport process, while the structural peptide may differ from the natural one [31]. This flexibility makes it possible to adapt the machinery of lantibiotic biosynthesis for the creation of biologically active compound libraries in vivo. The leader peptide is processed by the subtilisin-like serine protease LanP. It can be anchored to the membrane or localized in the cytoplasm. However, the order of proteolytic activation is not completely established for all LanPs. Extracellular localization is shown for NisP, and proteolysis of the leader sequence occurs after the pre-peptide secretion. Effective recognition of the substrate and subsequent proteolysis requires the presence of at least one lanthionine in the structural part of the pre-peptide [32].
In addition to the main modifications, the presence of lactate at the N-terminus of the polypeptide chain [33] or acetylation [34], as well as the formation of amino vinyl cysteine (AviCys) at the C-terminus [35], is a common feature for this class of lantipeptides (Fig. 3b). The most famous representative of class I lantibiotics is nisin, which has been successfully used as a preservative in the food industry for more than 50 years [36]. The lantibiotic microbisporicin A1, also known as NAI-107, exhibits substantial activity against clinically significant pathogens such as S. aureus MRSA and Clostridium difficile. NAI-107 is undergoing pre-clinical trials for the treatment of infections caused by multidrug-resistant bacteria [37, 38]. The specific feature of this lantibiotic is the presence of a halogenated Trp residue and mono- or dihydroxylation of the Pro residue, which are necessary for its antimicrobial activity [39].
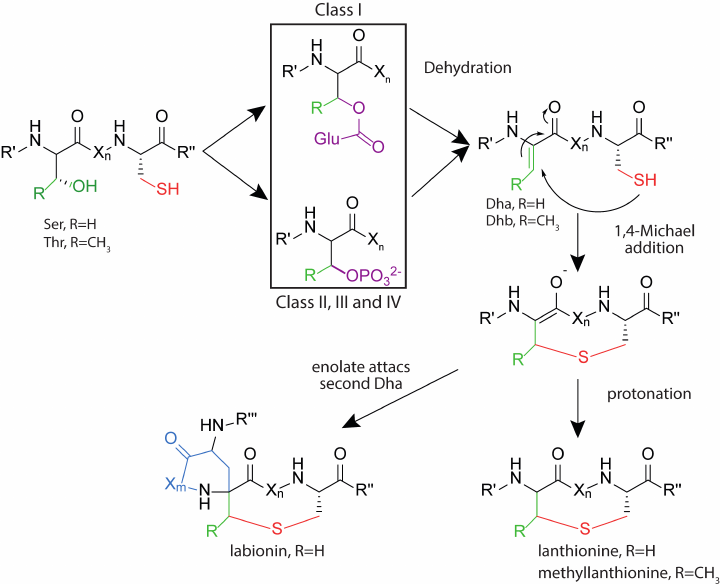
Class II. This class includes lantibiotics, which are dehydrated and cyclized by the multifunctional enzyme LanM. At the N-terminus of the enzyme, there is a domain responsible for dehydration of the pre-peptide. The C-terminal domain is homologous to the LanC cyclase of the class I lantibiotics and has a zinc-binding domain. Studies of the enzyme activity in vitro showed that the presence of ATP and Mg2+ ions is necessary for dehydration, and the use of analogs of the natural substrate led to the formation of an intermediate phosphorylated at Ser/Thr residues [40]. Thus, in class II lantibiotics, as well as in class III and IV described below, dehydration of Ser and Thr residues occurs through the phosphorylation stage, while for the class I lantibiotics, the process proceeds through the glutamylated derivatives (Fig. 4). The N-terminal domain of LanM has structural homology with eukaryotic lipid kinases, such as phosphoinositide 3-kinase (PI3K) [41], while these enzymes have low sequence identity.
Fig. 4. Formation of thioester amino acids in the biosynthesis of lantipeptides of different classes. Designations: Dha, dehydroalanine; Dhb, dehydrobutyrine; R′, R′′, parts of the polypeptide chain of lantipeptide; Xn,m, fragment of the polypeptide chain, consisting of n- or m-number of amino acid residues, where n and m are numbers from 1 to 20.
Dehydration and cyclization are carried out by the LanM enzymes independently, which is confirmed by experiments with an individual expression of each domain [42, 43]. An important property of lantipeptide synthetases is their strict stereoselectivity in the formation of thioether bridges and regioselectivity. An exceptional case is the enzyme CylM, which modifies a two-component class II lantibiotic cytolysin from Enterococcus faecalis. Cytolysin consists of two peptides: cytolysin L and cytolysin S, which contain two variants of methyllanthionine: one in the classic DL configuration, which is characteristic of most lantibiotics, and the other in the rarer LL configuration [44]. Thus, CylM can form thioether bridges of different configurations within one peptide. Lantipeptide synthetase ProcM has a broad substrate specificity. Analysis of the cluster of prochlorosin biosynthesis including ProcM, showed the presence of 30 peptides with different amino acid sequences. Despite a large number of substrate variants, ProcM modifies each pre-peptide into a lantipeptide with a strictly defined morphology [45]. The family of ProcM-like enzymes displays the widest range of substrate specificity known so far [46].
In most cases, transport is carried out by the specific ABC-transporter LanT that also has a protease domain [47]. During the transport of the pre-peptide across the membrane, the leader sequence is removed, and the active lantibiotic is secreted into the extracellular environment. Proteolysis occurs in the region of the Gly-Gly motif at the C-terminus of the leader. The leader peptide sequence is important for the LanT-driven transport, while the structural peptide may differ from the native one [48]. However, in the case of prochlorosins and lichnecidin, the transporter is also tolerant to the leader peptide, which is confirmed by the variability found in the leader sequence among the lantibiotics of the same cluster. Additional proteases are found for several lantibiotics of the class II, cleaving amino acids from the N-terminus of the structural peptide after the leader sequence has been processed [49].
Class II lantibiotics are distinguished by a more globular structure compared to the class I, furthermore, a unique group is also clustered from them – the two-peptide lantibiotics. Antibiotic activity of the two-peptide lantibiotics is based on a dual mechanism of binding to lipid II and pore formation. Among the two-peptide lantibiotics, lacticin 3147 has a remarkable activity. It is active against a wide range of Gram-positive bacteria, including methicillin-resistant S. aureus, vancomycin-resistant E. faecalis, Propionibacterium acne, and Streptococcus mutans [50]. Another two-peptide lantibiotic, haloduracin, is more active against vancomycin-resistant E. faecalis than nisin and is more stable under physiological conditions [51]. Traditionally, lactic acid bacteria have been considered as sources of new lantibiotics. However, with the development of whole-genome sequencing and genome mining, more and more active lantibiotics are found outside of this group of bacteria. The ticin A4 biosynthesis cluster was found in the bacterium Bacillus thuringiensis BMB3201 [52]. This lantibiotic exhibited activity similar to that of nisin against a wide range of Gram-positive bacteria. It was found that ticin A4 retained its antibiotic properties in the pH range 2-9 and was moderately resistant to the treatment with proteases such as trypsin, papain, and β-amylase.
Another example of the class II lantibiotic that has potential in medical application is NVB302. It is a semi-synthetic derivative of deoxyactagardine, lantibiotic from the bacterium Actinoplanes garbadinensis. In 2012, Novacta announced the successful completion of the first phase of clinical trials with NVB302 for the treatment of C. difficile infections. NVB302 has an efficacy similar to vancomycin against C. difficile; its advantage is a faster recovery of the Bacteroides fragilis population after the end of therapy [53]. B. fragilis is a part of the normal gut microbiota. It is assumed that the less severe antibiotic effect on this group of bacteria during clostridial infections leads to fewer fluctuations in the anaerobic intestinal microflora and reduces the risk of relapses [54].
Class III. This class of lantipeptides has been identified relatively recently. The main modifications are performed by the multifunctional enzyme LanKC [55]. Unlike LanM, LanKC consists of three domains: N-terminal lyase, central kinase, and C-terminal cyclase. The lyase and kinase domains perform dehydration, while the cyclase domain forms thioether rings. LanKC does not have a zinc-binding motif, but it exhibits homology with other cyclase domains [56]. Differences are also observed in the structure of the peptides since the formation of labionin is characteristic among some representatives of this class. Such peptides are classified into the labyrinthopeptin group. Formation of the additional carbon-carbon bond in labionin occurs upon the attack of dehydroalanine on enol intermediate, which is protonated in the case of lanthionin formation [57]. Reconstruction of the biosynthesis of labirynthopeptins in vitro showed that the dehydration process occurred sequentially from the C-terminus of the peptide to the N-terminus, and labionin was formed from the Ser(Xxx)2Ser(Xxx)3Cys motif. Dehydration occurs via the phosphorylated derivative as in the class II lantibiotics. LanKC enzymes utilize different nucleoside triphosphates as a substrate: LabKC uses only GTP when modifying labyrinthopeptin [58], while AviKC, which synthesizes catenulipeptin, can use ATP, TTP, GTP, and CTP [59].
For a long time, it was not known how exactly the leader sequence is processed since no specific protease or transporter with a protease domain was found within the biosynthesis cluster. The presence of several residues from the leader sequence at the N-terminus of the structural peptide was often observed. The main assumption was that endopeptidase, which processes the leader, and aminopeptidase, which removes the remaining amino acids, are present. The discovery of the NAI-122 lantipeptide and the study of its biosynthetic cluster showed the presence of a specific protease AplP with a dual function [60]. It is a zinc-dependent bifunctional protease with endo- and aminopeptidase activity. First, AplP, as an endopeptidase, cleaves the leader sequence at the conserved EL-Q motif, then removes the remaining amino acids, as aminopeptidase [61]. Further analysis showed the presence of AplP-like proteases in all genomes of the class III lantipeptide producers outside the lantipeptide biosynthetic cluster. The ability of these proteases to form mature lantipeptides was confirmed in vitro.
Currently, the known class III lantipeptides do not exhibit obvious antimicrobial properties, therefore the term lantibiotics is inapplicable for them. They are thought to have regulatory functions as signaling molecules. SapT has been shown to stimulate the formation of air hyphae in streptomycetes [62]. The potential use of lantipeptides is not limited to antibiotic activity. Studies in a mouse model of pain have shown an antinociceptive effect of the NAI-122 lantipeptide from Actinoplanes DSM 24059 [63]. Systemic administration of 1-10 µg/kg NAI-122 resulted in the dose-dependent decrease in the development of formalin-induced pain behavior in mice. Lantipeptide was also active in the experiments on the relief of chronic pain. Several lantipeptides have been shown to have antiviral properties. Labyrinthopeptin A1 exhibited significant activity against human immunodeficiency virus (HIV) and herpes virus in the cell models [64]. Synergistic effects with the clinically approved antiretrovirals and low toxicity to vaginal strains of Lactobacillus demonstrate its potential in the treatment of sexually transmitted diseases.
Class IV. It is one of the least studied groups of lantipeptides known today. The first characterized biosynthetic system of the class IV lantipeptides was found in Streptomyces venezuelae after genomic screening for proteins homologous to LanC. The peptide was named venezuelin, and its synthetase was named VenL [65]. Lantipeptide synthetases of the class IV LanL are multifunctional enzymes consisting of three domains: the lyase and cyclase domains are homologous to those of LanKC, and the cyclase domain is a homolog of LanM, since it has a characteristic zinc-binding motif. Despite the presence of all components of the venezuelin biosynthesis cluster, its production was not detected in the wild-type strain. Determination of the structure and properties of the LanL peptide and enzyme was carried out in vitro. Families of the lantipeptide-like genetic clusters were identified during the phylogenetic analysis of actinobacteria for the presence of LanC-like proteins [66]. In particular, with this analysis, several venezuelin-like peptides common among bacteria of the genus Streptomyces were found.
Streptocollin – another lantipeptide from this family produced by Streptomyces collinus Tu 365, was discovered similarly [67]. Dehydration and cyclization processed catalyzed by the LanL enzyme were studied using globisporicin biosynthesis as an example [68]. It was found that the SgbL enzyme recognized the α-helical region of the leader peptide located in its N-terminal part. The recognition occurred in the kinase part of SgbL, but not in the lyase one. The dehydration process occurred sequentially from the N-terminus to the C-terminus of the structural peptide through the phosphorylated derivative.
No specific protease processing the leader sequence has been found so far for the class IV lantipeptides. It is assumed that the maturation of a lantipeptide occurs with the participation of the producer’ s endopeptidases and aminopeptidases.
Currently, the biological function of the class IV lantipeptides remains unknown. For streptocollin, a mediocre inhibitory activity against the protein tyrosine phosphatase 1B (PTP1B) was found – 33% inhibition at the streptocollin concentration of 50 µM [67].
BIOLOGICAL ACTIVITY OF LANTIBIOTICS
The antimicrobial effect of lantibiotics was first discovered in 1927 in the study of the inhibitory effect of Lactococcus lactis on the growth of Lactobacillus bulgaricus [69]. In 1947 Mattick et al. [70] isolated the lantibiotic nisin from the cultivation medium of L. lactis. Now it is widely used in the food industry worldwide as a preservative (food additive E243). Nisin is the most researched representative of lantibiotics with an extensively studied spectrum of antimicrobial activity, mechanism of action, and biosynthesis.
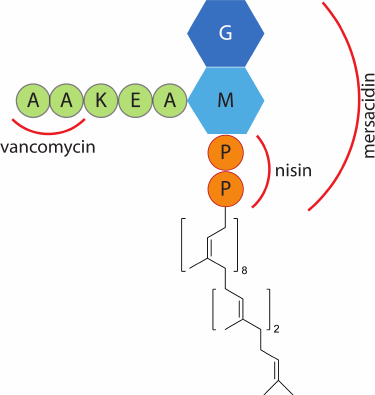
Lipid II is the main target on the surface of Gram-positive bacteria for many lantibiotics. This molecule plays a critical role in the formation of the bacterial cell wall. It acts as a carrier of peptidoglycans of the bacterial cell wall from the inner side of the membrane, where they are formed, to the outer side, where the cell wall matures. Since the number of lipid II molecules in the bacterial membrane is limited [71], lipid II binding to lantibiotic molecules blocks the cell wall synthesis, which subsequently leads to cell death. An important feature of lipid II is its conservatism among prokaryotes. Therefore, antibiotics that bind lipid II display a broad spectrum of activity and have the potential for the development of new drugs [72]. Lipid II is unique to prokaryotes, making these antimicrobial agents potentially safe for eukaryotic cells. The group of antibiotics that bind lipid II includes vancomycin, a tricyclic glycopeptide used in medicine for the treatment of serious infectious diseases, including those caused by multi-resistant bacteria, and is one of the “drugs of last resort”. However, the number of pathogenic bacterial strains resistant to vancomycin has been growing recently. In this regard, lantibiotics can take over, since they bind to another site on the lipid II molecule (Fig. 5), and can inhibit the growth of bacteria resistant to known antibiotics. In vitro experiments have shown that the lantibiotic microbisporicin was capable of inhibiting the growth of the E. faecalis strain resistant to vancomycin with a minimum inhibitory concentration of 0.5 µg/ml [39], nisin – 4 µg/ml [39], the two-peptide lantibiotic lacticin 3147 – 1.9 µg/ml [73].
Fig. 5. Schematic representation of lipid II structure. Designations: G, N-acetylglucosamine; M, N-acetylmuramic acid; P, phosphate group; red lines mark the binding areas of vancomycin, nisin, and mersacidin.
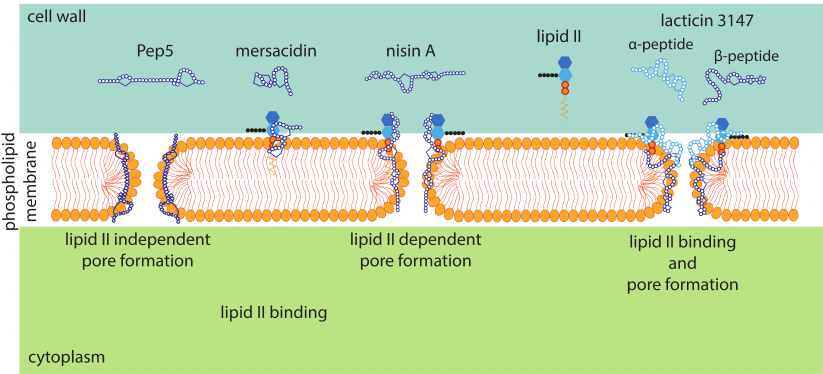
In addition to inhibiting the cell wall biosynthesis, some lantibiotics form pores in the cytoplasmic membrane (Fig. 6), which also leads to bacterial cell death [74]. Pore formation is lipid II-dependent in most cases since lantibiotics bind to the bacterial membrane through lipid II. In the case of nisin, two rings in the N-terminal part form a pyrophosphate-binding pocket, which binds the undecaprenyl pyrophosphate part of lipid II, and the C-terminus is involved in pore formation [75]. A study by Hasper et al. [76] showed that pore formation involved 8 nisin and 4 lipid II molecules, which ensured the binding of the lantibiotic to the membrane.
Fig. 6. Schematic representation of different mechanisms of the lantibiotic antimicrobial action on Gram-positive bacteria.
Cinnamycin-like lantibiotics, which also include duramycin and ancoverin, form a separate group of lantibiotics. Apart from the globular structure and the presence of a noncanonical amino acid residue, lysinoalanine, they differ in the target to which they bind on the cell membrane [77]. Phosphatidylethanolamine (PE) is an important representative of phospholipids. PE participates in the negative charge distribution on the membrane, it also plays the role of a precursor for the synthesis of other phospholipids, takes part in signaling cascades [78], and imparts special physicochemical properties to the plasma membrane necessary for the functioning of membrane proteins [79]. In eukaryotic cells, PE is mainly present on the inner surface of the plasma membrane. For most bacteria, PE is the main zwitterionic phospholipid. Moreover, in Gram-negative bacteria, the content of PE in the membrane is higher than in Gram-positive bacteria. [80]. High selectivity of cinnamycin-like lantibiotics to PE is achieved with the formation of a network of hydrogen bonds with lipid phosphates. In contrast to the bulkier choline group of phosphatidylcholine, the compact structure of the ammonium-binding domain of cinnamycin-like lantibiotics is complementary to a small group of PE [81]. The exact mechanism of antimicrobial action of this group of lantibiotics is still unknown. However, it is assumed that the binding of cinnamycin to phosphatidylethanolamine leads to an increase in the permeability of the cell membrane, which leads to cell death [82]. For duramycin, changes in morphology and disruption of cell integrity were shown in susceptible strains Bacillus BC15 and Bacillus subtilis 168 [83]. It was found that susceptible bacterial strains also developed resistance to higher concentrations following incubation with the low concentration of lantibiotic. Analysis of the composition of bacterial membranes before and after the emergence of resistance showed that bacteria with the acquired resistance had a reduced content of PE. Thus, the sensitivity of microorganisms to lantibiotics from the cinnamycin family correlates with the phospholipid composition of the bacterial membranes, which limits their use as broad-spectrum antibiotics. Nevertheless, selectivity to PE has potential applications in medicine. Duramycin is in the second phase of clinical trials as a drug for the treatment of cystic fibrosis [84]. Binding of duramycin to PE on the surface of lung epithelial cells stimulates the secretion of chloride ions, which, in turn, entails the secretion of water and stimulates cleansing of mucus from the lungs. Cinnamycin-like lantibiotics also find applications in bioimaging. PE becomes widespread on the cell surface during apoptosis [85], as well as during eukaryotic cell division [86], which enables the use of PE as a molecular marker in the study of the cellular processes. Biotinylated or radiolabeled duramycin and cinnamycin are used for detection of PE on the cell surface [87].
BIOENGINEERING OF LANTIBIOTICS AND DIRECTED EVOLUTION OF
ANTIBIOTIC ACTIVITY
Lantibiotics have great potential in the development of new antimicrobial drugs. The traditional strategy in antibiotic discovery is to isolate a natural producer strain from various sources and analyze the antimicrobial properties of its metabolites. This approach has limitations as it is only suitable for cultured bacteria. The development of whole-genome sequencing and genomic mining technologies enables the discovery of new lantibiotics that were not previously isolated from natural sources, such as unculturable microorganisms. Various approaches of synthetic biology are used to study biosynthetic clusters of lantibiotics, from cell-free systems to heterologous production.
Cell-free systems are practically unlimited in the size of the combinatorial library since there is no need for transformation of the producer cells – the stage that limits this parameter in cellular biosynthesis systems. An in vitro system was developed for the synthesis of a lantipeptide library based on mRNA display [88]. In vitro transcribed mRNA was used as a template for peptide synthesis. An oligonucleotide carrying puromycin at the 3′-end of the open reading frame was used to connect mRNA to the translated peptide. This oligonucleotide was attached to the 3′-end of the mRNA by photoinduced crosslinking due to the presence of psoralen at its 5′-end. The lysine residue was replaced by 4-selenolysine, which was transformed into dehydroalanine upon treatment with hydrogen peroxide. After immobilization of the pre-peptide on the column and corresponding change of buffers, lanthionines were formed. Thus, a completely artificial enzyme-free system was designed to create more than 1011 lantipeptide variants. Molecules with high specificity for sortase A, the enzyme responsible for the virulence of S. aureus, were isolated from this library.
Heterologous systems for expression of the lantibiotic clusters are used in the high-throughput screening of libraries of the mutant variants of lantibiotics for more active and stable compounds. The use of heterologous expression systems enables the production of lantibiotics in quantities sufficient for carrying out comprehensive studies of their structure and functions.
L. lactis is a lactic acid bacterium that is often used for heterologous protein expression. The NICE (NIsin-Controlled Expression system) expression system was created based on the L. lactis nisin biosynthetic cluster [89]. Nisin production in L. lactis and other nisin-producing bacteria is autoinduced. When nisin appears in the medium, the membrane-associated kinase NisK is autophosphorylated. After that, it phosphorylates the intracellular regulatory protein NisR, which binds to the nisA promoter and triggers the expression of the downstream genes. Thus, the controlled overexpression of the desired recombinant lantibiotic is realized. The biosynthetic cluster of the class II lantibiotic nukacin ISK-1 was investigated using the NICE system [90]. This approach allowed to reduce the level of proteolytic degradation of the lantibiotic, which was observed in the natural producer, and to determine the minimum set of genes required for production of the active lantibiotic.
Numerous structural and functional studies of lantibiotics have shown the presence of characteristic motifs and certain topology of the rings facilitating their inherent mechanism of action, e.g., a pyrophosphate-binding cavity in the nisin-like lantibiotics [75] or the lipid II binding motif [91]. The library of the artificial lantipeptides was created based on a similar modular system. It consisted of a combination of modules of 12 known lantibiotics. The corresponding genetic constructs were used to transform L. lactis with a nisin biosynthetic cluster in its genome carrying out post-translational modifications and transport of the mature lantipeptide into the extracellular environment. For the high-throughput testing of the biological activity of bacteria, lantibiotic producers were placed in the alginate beads together with the indicator bacterial strain and specific protease that processed the leader peptide to release the active lantibiotic. After incubation and staining with a fluorescent dye, the beads were sorted using flow cytometry. Chimeric molecules composed of gallidermin and nisin modules showed improved activity against Streptococcus pneumoniae compared to the natural lantibiotics. This technology offers promise for testing the activity of potential peptide drugs in vivo and screening for compounds with new spectra of biological activity.
E. coli is the most used microorganism for heterologous expression of proteins and peptides, including those with antimicrobial activity [92]. The advantages of E. coli are simplicity of cultivation, simplicity of genetic manipulations, and availability of various expression options: from adding tags to increase solubility and simplifying purification of the recombinant protein to directing production to the periplasmic space [93]. Several systems for the biosynthesis of lantibiotics have been successfully reconstructed using E. coli as a producer [25]. Prochlorosins synthesized in E. coli had a significantly higher yield compared to the natural producer Prochlorococcus MIT 9313, while being completely modified.
Screening of the improved variants of lantibiotics follows the development of the heterologous expression systems. The leader sequence inhibits the antimicrobial properties of the completely modified lantibiotic. Therefore, commercially available proteases that process polypeptides are used after the production and purification of pre-peptides. The need for purification, as well as subsequent treatment with proteases, complicates the process of analyzing the activity of lantibiotics, especially during the screening of a large number of variants. The leader peptide processing in E. coli colonies was used to solve this problem [94]. The lantipeptide was synthesized intracellularly, while the production of the specific lantipeptide protease LicP was directed to the periplasm (Fig. 7). After the temperature-induced autolysis of cells, protease and pre-peptide were released from the cells. Subsequently, the active lantibiotic was processed and detected with the indicator bacteria by the appearance of growth inhibition zones. The successful application of this system was demonstrated for the two-peptide lantibiotic haloduracin and lantibiotic lacticin 481.
Fig. 7. Schematic representation of a system for production of the mature class II lantibiotics in E. coli. a) Periplasmic compartmentalization of the protease. b) Temperature induction of expression of autolytic proteins. c) Processing of the leader peptide after autolysis. d) Structural and functional screening of the lantipeptide variants on colonies using the agar diffusion method and matrix assisted laser desorption ionization-time of flight mass spectrometry (MALDI-ToF MS) analysis; adapted from [94].
The genetically encoded nature of lantibiotics simplifies the process of modification of their structure. This allows the use of directed or random mutagenesis methods to create lantibiotics with improved properties, as well as to understand their structural and functional features. For instance, mutations R13A and K2A/R13A in the lantibiotic mutacin 1140 led to increased resistance to proteolysis by trypsin [95]. The substitution of the Met21 residue for valine in the nisin molecule led to the increase in activity against Listeria monocytogenes, while the N20P mutant was more active against S. aureus [96].
Lantibiotics mainly act on Gram-positive bacteria, since the Gram-negative bacteria are protected from their action by the outer membrane. To expand the spectrum of action of lantibiotics, various attempts are being made to create chimeric molecules capable of crossing the outer membrane and attacking Gram-negative bacteria. To overcome this limitation, it was suggested to use conjugates of the lantibiotic gallidermin with siderophore molecules [97]. It was assumed that the presence of specific receptors for siderophores on the outer membrane would help the conjugate to penetrate the outer membrane and make the inner membrane accessible to the lantibiotic. However, Gram-negative bacteria were found to be immune to the antimicrobial effect of the conjugate despite the activity against the indicator Gram-positive bacterium Lactococcus lactis subsp. cremoris HP. Alternatively, the short peptides with known activity against Gram-negative bacteria were attached to the C-terminus of the full-length or truncated nisin [98]. Several variants of such chimeric molecules inhibited the growth of indicator strains. The rational design was carried out for the most promising variants. As a result, a compound that was 4-12-fold more active against a number of important Gram-negative pathogens in comparison with nisin was obtained.
The presence of macrocycles in the structure of lantibiotics makes them convenient targets for the development of inhibitors of protein-protein interactions since they can act as structural analogs of the natural ligands. The genetically encoded nature of lantibiotics enables the creation of diverse libraries of variants and combine peptide synthesis with the screening inside the cell. There are many approaches to the selection of compounds with the required properties from the libraries. One of them is the reverse two-hybrid system (RTHS). This method was used to obtain inhibitors of the interaction between the p6 protein of HIV with the ubiquitin E2 variant (UEV) domain of the human TSG101 protein [99]. These inhibitors have a high therapeutic potential in antiviral therapy since they disrupt the process of HIV budding from the infected cell. The sequence of the lantipeptide ProcA2.8 was used as a template for creating a library of macrocycles. Formation of the thioether bonds was carried out by the lantipeptide synthetase ProcM, known for its high substrate tolerance. The p6 and UEV proteins are produced intracellularly as chimeric molecules with functional repressors. Their physical interaction leads to inhibition of the expression of the reporter genes that ensure bacterial growth on a selective medium. Thus, only peptide variants inhibiting the interaction between p6 and UEV allowed bacterial growth. The selected XY3-3 inhibitor showed potent antiviral activity detected using a cellular model of in vitro maturation of viral particles.
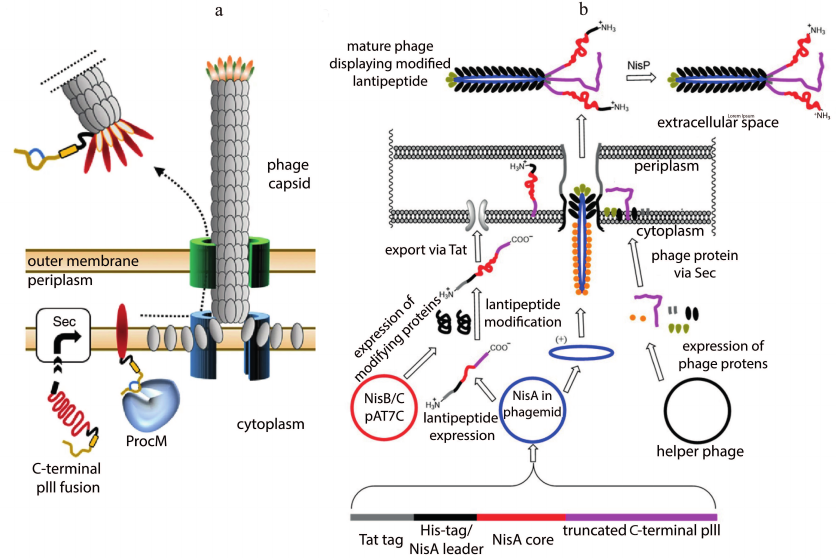
Phage display is a powerful in vitro selection tool, widely used for screening peptide ligands for a selected target [100]. This technology is also used to screen lantipeptide libraries. The prochlorosin biosynthetic cluster carrying ProcM synthetase was taken as a template for the creation of C-terminal peptide library [101]. The pre-peptide was fused with the C-terminus of pIII of the capsid protein of M13 bacteriophage. All modifications took place in the cytoplasm. Phage display of the modified peptides was provided by incorporation of the pIII conjugate into the phage envelope, followed by the release of a phage particle into the medium (Fig. 8a).
Fig. 8. Maturation of phage particles. a) C-terminal display of lantipeptides modified by ProcM; adapted from [101]; b) N-terminal display of a lantipeptide library based on the nisin biosynthesis system; adapted from [102].
Ligands to streptavidin and urokinase plasminogen activator were obtained based on the selection results, which showed the potential of using the lantipeptide biosynthesis system in tandem with the phage display technology.
The N-terminal phage display technology was developed to display a library of the lantibiotic nisin [102]. In contrast to the work mentioned above, secretion of the conjugate into the periplasm was carried out by the Tat pathway, which allowed translocation of the folded proteins (Fig. 8b). The selection was focused on the improved stability of the nisin derivatives at higher pH values. However, antimicrobial activity was lost despite the successful selection of several variants binding lipid II. A variant of the yeast display was also presented. The lantibiotic lacticin 481 was fused with a protein of the yeast cell wall. Modifications took place in the endoplasmic reticulum. After confirming the presence of all the necessary modifications by analyzing biological activity and mass spectrometry data of the purified lantibiotic, a library of 481 lacticin analogs was created. Peptides binding to the αvβ3 integrin were obtained after the selection. The results of the study illustrate the versatility of lantipeptide biosynthetic clusters for the discovery of compounds with new biological properties.
CONCLUSIONS AND FUTURE PROSPECTS
In the era of rapidly spreading antibiotic resistance, it is essential to develop systems facilitating the identification of new antimicrobial agents. Thus, it is crucial to find new sources of biological diversity and create approaches for efficient analysis of a large amount of data. The technologies of directed evolution of antibiotic activity allow us to create an artificial diversity of the antimicrobial agents and to select the most effective variants from this diversity, primarily taking into account their practical application. High-throughput screening of compounds generated by lantibiotic biochemical machinery is of particular interest. Combining biosynthesis and biological activity screening in a single step would reduce the number of stages of analysis of the candidate molecules, as well as time and costs of the process.
Currently, microfluidic technologies have been intensively developed. In particular, droplet microfluidics is successfully used in various biological research. Its main advantage is the ability to confine reactions within the individual droplets, which allows hundreds of millions of reactions to be carried out in parallel [103]. This feature was successfully used in the development of the full-cycle platforms for ultrahigh-throughput screening and directed evolution of polypeptides [104]. Encapsulation of single cells enables studying the effect of different factors on the physiological state of microorganisms in detail [105], as well as carrying out high-throughput screening of antibiotic activity among the representatives of different microbiological communities [106].
Lantibiotics, in turn, have great potential as a universal template for creating molecules with improved biological functions, from inhibitors of protein-protein interactions to therapeutic drugs and new generation antibiotics. Combining biosynthetic machinery that generates many variants of lantibiotics with microfluidic technologies allows high-throughput screening of compounds active against a given target. The use of heterologous systems for the production of lantibiotics and the adaptation of cell-free systems for the generation of biologically active lantibiotics in vitro are the cornerstones for further development of this technology. Integrating biosynthesis and biological activity screening inside individual microcompartments will allow achieving outstanding performance in the functional profiling of synthetic diversity of the recombinant lantibiotics. It opens up unique opportunities for analyzing the landscapes of antimicrobial activity of genetically encoded antibiotics and creating highly effective therapeutic agents.
Finding. The research was financially supported by the Russian Foundation for Basic Research (project no. 18-29-08054) and by the Russian Science Foundation (project no. 17-74-30019).
Ethics declarations. The authors declare no conflicts of interest in financial or any other sphere. This article does not contain any studies with human participants or animals performed by any of the authors.
REFERENCES
1.Davies, J., and Davies, D. (2010) Origins and
evolution of antibiotic resistance, Microbiol. Mol. Biol. Rev.,
74, 417-433, doi: 10.1128/mmbr.00016-10.
2.O’Neill, J. (2014) Antimicrobial
Resistance: Tackling A Crisis For The Health And Wealth Of Nations,
Review on Antimicrobial Resistance, London.
3.Czepiel, J., Drozdz, M., Pituch, H., Kuijper, E.
J., Perucki, W., et al. (2019) Clostridium difficile infection:
review, Eur. J. Clin. Microbiol. Infect. Dis., 38,
1211-1221, doi: 10.1007/s10096-019-03539-6.
4.Wang, J., Xiong, Z., Meng, H., Wang, Y., and Wang,
Y. (2012) Synthetic biology triggers new era of antibiotics
development, Subcell. Biochem., 64, 95-114, doi:
10.1007/978-94-007-5055-5_5.
5.Foulston, L. (2019) Genome mining and prospects for
antibiotic discovery, Curr. Opin. Microbiol., 51, 1-8,
doi: 10.1016/j.mib.2019.01.001.
6.Terekhov, S. S., Smirnov, I. V., Malakhova, M. V.,
Samoilov, A. E., Manolov, A. I., et al. (2018) Ultrahigh-throughput
functional profiling of microbiota communities, Proc. Natl. Acad.
Sci. USA, 115, 9551-9556, doi: 10.1073/pnas.1811250115.
7.Stokes, J. M., Yang, K., Swanson, K., Jin, W.,
Cubillos-Ruiz, A., et al. (2020) A deep learning approach to antibiotic
discovery, Cell, 180, 688-702, doi:
10.1016/j.cell.2020.01.021.
8.Lau, J. L., and Dunn, M. K. (2018) Therapeutic
peptides: historical perspectives, current development trends, and
future directions, Bioorg. Med. Chem., 26, 2700-2707,
doi: 10.1016/j.bmc.2017.06.052.
9.Fosgerau, K., and Hoffmann, T. (2015) Peptide
therapeutics: current status and future directions, Drug Discov.
Today, 20, 122-128, doi: 10.1016/j.drudis.2014.10.003.
10.Lei, J., Sun, L., Huang, S., Zhu, C., Li, P., et
al. (2019) The antimicrobial peptides and their potential clinical
applications, Am. J. Transl. Res., 11, 3919-3931.
11.Kang, S. J., Park, S. J., Mishig-Ochir, T., and
Lee, B. J. (2014) Antimicrobial peptides: therapeutic potentials,
Expert. Rev. Anti. Infect. Ther., 12, 1477-1486, doi:
10.1586/14787210.2014.976613.
12.Ortega, M. A., and van der Donk, W. A. (2016) New
insights into the biosynthetic logic of ribosomally synthesized and
post-translationally modified peptide natural products, Cell Chem.
Biol., 23, 31-44, doi: 10.1016/j.chembiol.2015.11.012.
13.McIntosh, J. A., Donia, M. S., and Schmidt, E. W.
(2009) Ribosomal peptide natural products: bridging the ribosomal and
nonribosomal worlds, Nat. Prod. Rep., 26, 537-559, doi:
10.1039/b714132g.
14.Muller, M. M. (2018) Post-translational
modifications of protein backbones: unique functions, mechanisms, and
challenges, Biochemistry, 57, 177-185, doi:
10.1021/acs.biochem.7b00861.
15.Hudson, G. A., and Mitchell, D. A. (2018) RiPP
antibiotics: biosynthesis and engineering potential, Curr. Opin.
Microbiol., 45, 61-69, doi: 10.1016/j.mib.2018.02.010.
16.Mullane, K., Lee, C., Bressler, A., Buitrago, M.,
Weiss, K., et al. (2015) Multicenter, randomized clinical trial to
compare the safety and efficacy of LFF571 and vancomycin for
Clostridium difficile infections, Antimicrob. Agents
Chemother., 59, 1435-1440, doi: 10.1128/aac.04251-14.
17.Poorinmohammad, N., Bagheban-Shemirani, R., and
Hamedi, J. (2019) Genome mining for ribosomally synthesised and
post-translationally modified peptides (RiPPs) reveals undiscovered
bioactive potentials of actinobacteria, Antonie Van Leeuwenhoek,
112, 1477-1499, doi: 10.1007/s10482-019-01276-6.
18.Velasquez, J. E., and van der Donk, W. A. (2011)
Genome mining for ribosomally synthesized natural products, Curr.
Opin. Chem. Biol., 15, 11-21, doi:
10.1016/j.cbpa.2010.10.027.
19.Arnison, P. G., Bibb, M. J., Bierbaum, G.,
Bowers, A. A., Bugni, T. S., et al. (2013) Ribosomally synthesized and
post-translationally modified peptide natural products: overview and
recommendations for a universal nomenclature, Nat. Prod. Rep.,
30, 108-160, doi: 10.1039/C2NP20085F.
20.Repka, L. M., Chekan, J. R., Nair, S. K., and van
der Donk, W. A. (2017) Mechanistic understanding of lanthipeptide
biosynthetic enzymes, Chem. Rev., 117, 5457-5520, doi:
10.1021/acs.chemrev.6b00591.
21.Kleerebezem, M. (2004) Quorum sensing control of
lantibiotic production; nisin and subtilin autoregulate their own
biosynthesis, Peptides, 25, 1405-1414, doi:
10.1016/j.peptides.2003.10.021.
22.Willey, J. M., Willems, A., Kodani, S., and
Nodwell, J. R. (2006) Morphogenetic surfactants and their role in the
formation of aerial hyphae in Streptomyces coelicolor, Mol.
Microbiol., 59, 731-742, doi:
10.1111/j.1365-2958.2005.05018.x.
23.Bierbaum, G., Gotz, F., Peschel, A., Kupke, T.,
van de Kamp, M., and Sahl, H. G. (1996) The biosynthesis of the
lantibiotics epidermin, gallidermin, Pep5 and epilancin K7, Antonie
Van Leeuwenhoek, 69, 119-127, doi: 10.1007/BF00399417.
24.Zhang, Q., Yu, Y., Velasquez, J. E., and van der
Donk, W. A. (2012) Evolution of lanthipeptide synthetases, Proc.
Natl. Acad. Sci. USA, 109, 18361-18366, doi:
10.1073/pnas.1210393109.
25.Shi, Y., Yang, X., Garg, N., and van der Donk, W.
A. (2011) Production of lantipeptides in Escherichia coli, J.
Am. Chem. Soc., 133, 2338-2341, doi: 10.1021/ja109044r.
26.Garg, N., Salazar-Ocampo, L. M., and van der
Donk, W. A. (2013) In vitro activity of the nisin dehydratase
NisB, Proc. Natl. Acad. Sci. USA, 110, 7258-7263, doi:
10.1073/pnas.1222488110.
27.Ortega, M. A., Hao, Y., Zhang, Q., Walker, M. C.,
van der Donk, W. A., and Nair, S. K. (2015) Structure and mechanism of
the tRNA-dependent lantibiotic dehydratase NisB, Nature,
517, 509-512, doi: 10.1038/nature13888.
28.Siezen, R. J., Kuipers, O. P., and de Vos, W. M.
(1996) Comparison of lantibiotic gene clusters and encoded proteins,
Antonie Van Leeuwenhoek, 69, 171-184, doi:
10.1007/BF00399422.
29.Goto, Y., Okesli, A., and van der Donk, W. A.
(2011) Mechanistic studies of Ser/Thr dehydration catalyzed by a member
of the LanL lanthionine synthetase family, Biochemistry,
50, 891-898, doi: 10.1021/bi101750r.
30.Hollenstein, K., Dawson, R. J., and Locher, K. P.
(2007) Structure and mechanism of ABC transporter proteins, Curr.
Opin. Struct. Biol., 17, 412-418, doi:
10.1016/j.sbi.2007.07.003.
31.Kuipers, A., de Boef, E., Rink, R., Fekken, S.,
Kluskens, L. D., et al. (2004) NisT, the transporter of the lantibiotic
nisin, can transport fully modified, dehydrated, and unmodified
prenisin and fusions of the leader peptide with non-lantibiotic
peptides, J. Biol. Chem., 279, 22176-22182, doi:
10.1074/jbc.M312789200.
32.Lagedroste, M., Smits, S. H. J., and Schmitt, L.
(2017) Substrate specificity of the secreted nisin leader peptidase
NisP, Biochemistry, 56, 4005-4014, doi:
10.1021/acs.biochem.7b00524.
33.Velasquez, J. E., Zhang, X., and van der Donk, W.
A. (2011) Biosynthesis of the antimicrobial peptide epilancin 15X and
its N-terminal lactate, Chem. Biol., 18, 857-867, doi:
10.1016/j.chembiol.2011.05.007.
34.Huang, E., and Yousef, A. E. (2015) Biosynthesis
of paenibacillin, a lantibiotic with N-terminal acetylation, by
Paenibacillus polymyxa, Microbiol. Res., 181,
15-21, doi: 10.1016/j.micres.2015.08.001.
35.Allgaier, H., Jung, G., Werner, R. G., Schneider,
U., and Zähner, H. (1985) Elucidation of the structure of
epidermin, a ribosomally synthesized, tetracyclic heterodetic
polypeptide antibiotic, Angew. Chem. Internat. Ed. Engl.,
24, 1051-1053, doi: 10.1002/anie.198510511.
36.De Arauz, L. J., Jozala, A. F., Mazzola, P. G.,
and Vessoni Penna, T. C. (2009) Nisin biotechnological production and
application: a review, Trends Food Sci. Technol., 20,
146-154, doi: 10.1016/j.tifs.2009.01.056.
37.Lepak, A. J., Marchillo, K., Craig, W. A., and
Andes, D. R. (2015) In vivo pharmacokinetics and
pharmacodynamics of the lantibiotic NAI-107 in a neutropenic murine
thigh infection model, Antimicrob. Agents Chemother., 59,
1258-1264, doi: 10.1128/AAC.04444-14.
38.Thomsen, T. T., Mojsoska, B., Cruz, J. C.,
Donadio, S., Jenssen, H., Lobner-Olesen, A., and Rewitz, K. (2016) The
Lantibiotic NAI-107 efficiently rescues Drosophila melanogaster
from infection with methicillin-resistant Staphylococcus aureus
USA300, Antimicrob. Agents Chemother., 60, 5427-5436,
doi: 10.1128/AAC.02965-15.
39.Castiglione, F., Lazzarini, A., Carrano, L.,
Corti, E., Ciciliato, I., et al. (2008) Determining the structure and
mode of action of microbisporicin, a potent lantibiotic active against
multiresistant pathogens, Chem. Biol., 15, 22-31, doi:
10.1016/j.chembiol.2007.11.009.
40.Chatterjee, C., Miller, L. M., Leung, Y. L., Xie,
L., Yi, M., Kelleher, N. L., and van der Donk, W. A. (2005) Lacticin
481 synthetase phosphorylates its substrate during lantibiotic
production, J. Am. Chem. Soc., 127, 15332-15333, doi:
10.1021/ja0543043.
41.Dong, S. H., Tang, W., Lukk, T., Yu, Y., Nair, S.
K., and van der Donk, W. A. (2015) The enterococcal cytolysin
synthetase has an unanticipated lipid kinase fold, eLife,
4, e07607, doi: 10.7554/eLife.07607.
42.Ma, H., Gao, Y., Zhao, F., and Zhong, J. (2015)
Individual catalytic activity of two functional domains of bovicin HJ50
synthase BovM, Wei Sheng Wu Xue Bao, 55, 50-58.
43.Shimafuji, C., Noguchi, M., Nishie, M., Nagao,
J., Shioya, K., Zendo, T., Nakayama, J., and Sonomoto, K. (2015) In
vitro catalytic activity of N-terminal and C-terminal domains in
NukM, the post-translational modification enzyme of nukacin ISK-1,
J. Biosci. Bioeng., 120, 624-629, doi:
10.1016/j.jbiosc.2015.03.020.
44.Tang, W., Jimenez-Oses, G., Houk, K. N., and van
der Donk, W. A. (2015) Substrate control in stereoselective lanthionine
biosynthesis, Nat. Chem., 7, 57-64, doi:
10.1038/nchem.2113.
45.Thibodeaux, C. J., Ha, T., and van der Donk, W.
A. (2014) A price to pay for relaxed substrate specificity: a
comparative kinetic analysis of the class II lanthipeptide synthetases
ProcM and HalM2, J. Am. Chem. Soc., 136, 17513-17529,
doi: 10.1021/ja5089452.
46.Cubillos-Ruiz, A., Berta-Thompson, J. W., Becker,
J. W., van der Donk, W. A., and Chisholm, S. W. (2017) Evolutionary
radiation of lanthipeptides in marine cyanobacteria, Proc. Natl.
Acad. Sci. USA, 114, E5424-E5433, doi:
10.1073/pnas.1700990114.
47.Nishie, M., Sasaki, M., Nagao, J., Zendo, T.,
Nakayama, J., and Sonomoto, K. (2011) Lantibiotic transporter requires
cooperative functioning of the peptidase domain and the ATP binding
domain, J. Biol. Chem., 286, 11163-11169, doi:
10.1074/jbc.M110.212704.
48.Kuipers, A., Meijer-Wierenga, J., Rink, R.,
Kluskens, L. D., and Moll, G. N. (2008) Mechanistic dissection of the
enzyme complexes involved in biosynthesis of lacticin 3147 and nisin,
Appl. Environ. Microbiol., 74, 6591-6597, doi:
10.1128/AEM.01334-08.
49.Caetano, T., Barbosa, J., Moesker, E., Sussmuth,
R. D., and Mendo, S. (2014) Bioengineering of lanthipeptides in
Escherichia coli: assessing the specificity of
lichenicidin and haloduracin biosynthetic machinery, Res.
Microbiol., 165, 600-604, doi:
10.1016/j.resmic.2014.07.006.
50.Galvin, M., Hill, C., and Ross, R. P. (1999)
Lacticin 3147 displays activity in buffer against gram-positive
bacterial pathogens which appear insensitive in standard plate assays,
Lett. Appl. Microbiol., 28, 355-358, doi:
10.1046/j.1365-2672.1999.00550.x.
51.Oman, T. J., and van der Donk, W. A. (2009)
Insights into the mode of action of the two-peptide lantibiotic
haloduracin, ACS Chem. Biol., 4, 865-874, doi:
10.1021/cb900194x.
52.Xin, B., Zheng, J., Xu, Z., Li, C., Ruan, L.,
Peng, D., and Sun, M. (2015) Three novel lantibiotics, ticins A1, A3,
and A4, have extremely stable properties and are promising food
biopreservatives, Appl. Environ. Microbiol., 81,
6964-6972, doi: 10.1128/aem.01851-15.
53.Crowther, G. S., Baines, S. D., Todhunter, S. L.,
Freeman, J., Chilton, C. H., and Wilcox, M. H. (2013) Evaluation of
NVB302 versus vancomycin activity in an in vitro human gut model of
Clostridium difficile infection, J. Antimicrob.
Chemother., 68, 168-176, doi: 10.1093/jac/dks359.
54.Louie, T. J., Emery, J., Krulicki, W., Byrne, B.,
and Mah, M. (2009) OPT-80 eliminates Clostridium difficile and
is sparing of bacteroides species during treatment of C.
difficile Infection, Antimicrob. Agents Chemother.,
53, 261-263, doi: 10.1128/aac.01443-07.
55.Knerr, P. J., and van der Donk, W. A. (2012)
Discovery, biosynthesis, and engineering of lantipeptides, Annu.
Rev. Biochem., 81, 479-505, doi:
10.1146/annurev-biochem-060110-113521.
56.Kodani, S., Hudson, M. E., Durrant, M. C.,
Buttner, M. J., Nodwell, J. R., and Willey, J. M. (2004) The SapB
morphogen is a lantibiotic-like peptide derived from the product of the
developmental gene ramS in Streptomyces coelicolor, Proc.
Natl. Acad. Sci. USA, 101, 11448-11453, doi:
10.1073/pnas.0404220101.
57.Meindl, K., Schmiederer, T., Schneider, K.,
Reicke, A., Butz, D., et al. (2010) Labyrinthopeptins: a new class of
carbacyclic lantibiotics, Angew. Chem. Int. Ed. Engl.,
49, 1151-1154, doi: 10.1002/anie.200905773.
58.Krawczyk, B., Ensle, P., Müller, W. M., and
Süssmuth, R. D. (2012) Deuterium labeled peptides give insights
into the directionality of class III lantibiotic synthetase LabKC,
J. Am. Chem. Soc., 134, 9922-9925, doi:
10.1021/ja3040224.
59.Müller, W. M., Schmiederer, T., Ensle, P.,
and Süssmuth, R. D. (2010) In vitro biosynthesis of the prepeptide
of type-III lantibiotic labyrinthopeptin A2 including formation of a
C-C bond as a post-translational modification, Angew. Chem. Int. Ed.
Engl., 49, 2436-2440, doi: 10.1002/anie.200905909.
60.Völler, G. H., Krawczyk, J. M., Pesic, A.,
Krawczyk, B., Nachtigall, J., and Süssmuth, R. D. (2012)
Characterization of new class III lantibiotics–erythreapeptin,
avermipeptin and griseopeptin from Saccharopolyspora erythraea,
Streptomyces avermitilis and Streptomyces griseus
demonstrates stepwise N-terminal leader processing, Chembiochem,
13, 1174-1183, doi: 10.1002/cbic.201200118.
61.Iorio, M., Sasso, O., Maffioli, S. I.,
Bertorelli, R., Monciardini, P., et al. (2014) A glycosylated,
labionin-containing lanthipeptide with marked antinociceptive activity,
ACS Chem. Biol., 9, 398-404, doi: 10.1021/cb400692w.
62.Chen, S., Xu, B., Chen, E., Wang, J., Lu, J.,
Donadio, S., Ge, H., and Wang, H. (2019) Zn-dependent bifunctional
proteases are responsible for leader peptide processing of class III
lanthipeptides, Proc. Natl. Acad. Sci. USA, 116,
2533-2538, doi: 10.1073/pnas.1815594116.
63.Kodani, S., Lodato, M. A., Durrant, M. C.,
Picart, F., and Willey, J. M. (2005) SapT, a lanthionine-containing
peptide involved in aerial hyphae formation in the streptomycetes,
Mol. Microbiol., 58, 1368-1380, doi:
10.1111/j.1365-2958.2005.04921.x.
64.Ferir, G., Petrova, M. I., Andrei, G., Huskens,
D., Hoorelbeke, B., et al. (2013) The lantibiotic peptide
labyrinthopeptin A1 demonstrates broad anti-HIV and anti-HSV activity
with potential for microbicidal applications, PLoS One,
8, e64010, doi: 10.1371/journal.pone.0064010.
65.Goto, Y., Li, B., Claesen, J., Shi, Y., Bibb, M.
J., and van der Donk, W. A. (2010) Discovery of unique lanthionine
synthetases reveals new mechanistic and evolutionary insights, PLoS
Biol., 8, e1000339, doi: 10.1371/journal.pbio.1000339.
66.Zhang, Q., Doroghazi, J. R., Zhao, X., Walker, M.
C., and van der Donk, W. A. (2015) Expanded natural product diversity
revealed by analysis of lanthipeptide-like gene clusters in
actinobacteria, Appl. Environ. Microbiol., 81, 4339-4350,
doi: 10.1128/AEM.00635-15.
67.Iftime, D., Jasyk, M., Kulik, A., Imhoff, J. F.,
Stegmann, E., Wohlleben, W., Süssmuth, R. D., and Weber, T. (2015)
Streptocollin, a type IV lanthipeptide produced by Streptomyces
collinus Tü 365, Chembiochem., 16, 2615-2623,
doi: 10.1002/cbic.201500377.
68.Hegemann, J. D., and van der Donk, W. A. (2018)
Investigation of substrate recognition and biosynthesis in class IV
lanthipeptide systems, J. Am. Chem. Soc., 140, 5743-5754,
doi: 10.1021/jacs.8b01323.
69.Rogers, L. A. (1928) The inhibiting effect of
Streptococcus Lactis on Lactobacillus Bulgaricus, J.
Bacteriol., 16, 321-325, doi:
10.1128/JB.16.5.321-325.1928.
70.Mattick, A. T. R., Hirsch, A., and Berridge, N.
J. (1947) Further observations on an inhibitory substance (nisin) from
lactic streptococci, Lancet, 250, 5-8, doi:
10.1016/S0140-6736(47)90004-4.
71.Van Heijenoort, Y., Gomez, M., Derrien, M.,
Ayala, J., and van Heijenoort, J. (1992) Membrane intermediates in the
peptidoglycan metabolism of Escherichia coli: possible roles of
PBP 1b and PBP 3, J. Bacteriol., 174, 3549-3557, doi:
10.1128/jb.174.11.3549-3557.1992.
72.Breukink, E., and de Kruijff, B. (2006) Lipid II
as a target for antibiotics, Nat. Rev. Drug Discov., 5,
321-323, doi: 10.1038/nrd2004.
73.Piper, C., Draper, L. A., Cotter, P. D., Ross, R.
P., and Hill, C. (2009) A comparison of the activities of lacticin 3147
and nisin against drug-resistant Staphylococcus aureus and
Enterococcus species, J. Antimicrob. Chemother.,
64, 546-551, doi: 10.1093/jac/dkp221.
74.Dischinger, J., Basi Chipalu, S., and Bierbaum,
G. (2014) Lantibiotics: promising candidates for future applications in
health care, Int. J. Med. Microbiol., 304, 51-62, doi:
10.1016/j.ijmm.2013.09.003.
75.Hsu, S.-T. D., Breukink, E., Tischenko, E.,
Lutters, M. A. G., de Kruijff, B., et al. (2004) The nisin–lipid
II complex reveals a pyrophosphate cage that provides a blueprint for
novel antibiotics, Nat. Struct. Mol. Biol., 11, 963-967,
doi: 10.1038/nsmb830.
76.Hasper, H. E., de Kruijff, B., and Breukink, E.
(2004) Assembly and stability of nisin-lipid II pores,
Biochemistry, 43, 11567-11575, doi:
10.1021/bi049476b.
77.Märki, F., Hänni, E., Fredenhagen, A.,
and van Oostrum, J. (1991) Mode of action of the lanthionine-containing
peptide antibiotics duramycin, duramycin B and C, and cinnamycin as
indirect inhibitors of phospholipase A2, Biochem. Pharmacol.,
42, 2027-2035, doi: 10.1016/0006-2952(91)90604-4.
78.Vance, J. E., and Tasseva, G. (2013) Formation
and function of phosphatidylserine and phosphatidylethanolamine in
mammalian cells, Biochim. Biophys. Acta, 1831, 543-554,
doi: 10.1016/j.bbalip.2012.08.016.
79.Gbaguidi, B., Hakizimana, P., Vandenbussche, G.,
and Ruysschaert, J. M. (2007) Conformational changes in a bacterial
multidrug transporter are phosphatidylethanolamine-dependent, Cell.
Mol. Life Sci., 64, 1571-1582, doi:
10.1007/s00018-007-7031-0.
80.Epand, R. M., and Epand, R. F. (2009) Lipid
domains in bacterial membranes and the action of antimicrobial agents,
Biochim. Biophys. Acta, 1788, 289-294, doi:
10.1016/j.bbamem.2008.08.023.
81.Vestergaard, M., Berglund, N. A., Hsu, P. C.,
Song, C., Koldso, H., Schiott, B., and Sansom, M. S. P. (2019)
Structure and dynamics of cinnamycin-lipid complexes: mechanisms of
selectivity for phosphatidylethanolamine lipids, ACS Omega,
4, 18889-18899, doi: 10.1021/acsomega.9b02949.
82.Choung, S. Y., Kobayashi, T., Takemoto, K.,
Ishitsuka, H., and Inoue, K. (1988) Interaction of a cyclic peptide,
Ro09-0198, with phosphatidylethanolamine in liposomal membranes,
Biochim. Biophys. Acta, 940, 180-187, doi:
10.1016/0005-2736(88)90193-9.
83.Hasim, S., Allison, D. P., Mendez, B., Farmer, A.
T., Pelletier, D. A., Retterer, S. T., Campagna, S. R., Reynolds, T.
B., and Doktycz, M. J. (2018) Elucidating duramycin' s bacterial
selectivity and mode of action on the bacterial cell envelope,
Front. Microbiol., 9, 219, doi:
10.3389/fmicb.2018.00219.
84.Jones, A. M., and Helm, J. M. (2009) Emerging
treatments in cystic fibrosis, Drugs, 69, 1903-1910, doi:
10.2165/11318500-000000000-00000.
85.Elvas, F., Stroobants, S., and Wyffels, L. (2017)
Phosphatidylethanolamine targeting for cell death imaging in early
treatment response evaluation and disease diagnosis, Apoptosis,
22, 971-987, doi: 10.1007/s10495-017-1384-0.
86.Emoto, K., Kobayashi, T., Yamaji, A., Aizawa, H.,
Yahara, I., Inoue, K., and Umeda, M. (1996) Redistribution of
phosphatidylethanolamine at the cleavage furrow of dividing cells
during cytokinesis, Proc. Natl. Acad. Sci. USA, 93,
12867-12872, doi: 10.1073/pnas.93.23.12867.
87.Zhao, M. (2011) Lantibiotics as probes for
phosphatidylethanolamine, Amino Acids, 41, 1071-1079,
doi: 10.1007/s00726-009-0386-9.
88.Hofmann, F. T., Szostak, J. W., and Seebeck, F.
P. (2012) In vitro selection of functional lantipeptides, J.
Am. Chem. Soc., 134, 8038-8041, doi: 10.1021/ja302082d.
89.Zhou, X. X., Li, W. F., Ma, G. X., and Pan, Y. J.
(2006) The nisin-controlled gene expression system: construction,
application and improvements, Biotechnol. Adv., 24,
285-295, doi: 10.1016/j.biotechadv.2005.11.001.
90.Aso, Y., Nagao, J.-I., Koga, H., Okuda, K.-I.,
Kanemasa, Y., Sashihara, T., Nakayama, J., and Sonomoto, K. (2004)
Heterologous expression and functional analysis of the gene cluster for
the biosynthesis of and immunity to the lantibiotic, nukacin ISK-1,
J. Biosci. Bioeng., 98, 429-436, doi:
10.1016/S1389-1723(05)00308-7.
91.Böttiger, T., Schneider, T.,
Martínez, B., Sahl, H.-G., and Wiedemann, I. (2009) Influence of
Ca2+ ions on the activity of lantibiotics containing a
mersacidin-like lipid II binding motif, Appl. Environ.
Microbiol., 75, 4427-4434, doi: 10.1128/aem.00262-09.
92.Li, Y. (2011) Recombinant production of
antimicrobial peptides in Escherichia coli: a review, Protein
Expr. Purif., 80, 260-267, doi:
10.1016/j.pep.2011.08.001.
93.Mesa-Pereira, B., Rea, M. C., Cotter, P. D.,
Hill, C., and Ross, R. P. (2018) Heterologous expression of
biopreservative bacteriocins with a view to low cost production,
Front. Microbiol., 9, 1654, doi:
10.3389/fmicb.2018.01654.
94.Si, T., Tian, Q., Min, Y., Zhang, L., Sweedler,
J. V., van der Donk, W. A., and Zhao, H. (2018) Rapid screening of
lanthipeptide analogs via in-colony removal of leader peptides in
Escherichia coli, J. Am. Chem. Soc., 140,
11884-11888, doi: 10.1021/jacs.8b05544.
95.Geng, M., and Smith, L. (2018) Modifying the
lantibiotic mutacin 1140 for increased yield, activity, and stability,
Appl. Environ. Microbiol., 84, doi:
10.1128/AEM.00830-18.
96.Field, D., Connor, P. M., Cotter, P. D., Hill,
C., and Ross, R. P. (2008) The generation of nisin variants with
enhanced activity against specific gram-positive pathogens, Mol.
Microbiol., 69, 218-230, doi:
10.1111/j.1365-2958.2008.06279.x.
97.Yoganathan, S., Sit, C. S., and Vederas, J. C.
(2011) Chemical synthesis and biological evaluation of
gallidermin-siderophore conjugates, Org. Biomol. Chem.,
9, 2133-2141, doi: 10.1039/c0ob00846j.
98.Li, Q., Montalban-Lopez, M., and Kuipers, O. P.
(2018) Increasing the antimicrobial activity of nisin-based
lantibiotics against Gram-negative pathogens, Appl. Environ.
Microbiol., 84, doi: 10.1128/AEM.00052-18.
99.Yang, X., Lennard, K. R., He, C., Walker, M. C.,
Ball, A. T., Doigneaux, C., Tavassoli, A., and van der Donk, W. A.
(2018) A lanthipeptide library used to identify a protein–protein
interaction inhibitor, Nat. Chem. Biol., 14, 375-380,
doi: 10.1038/s41589-018-0008-5.
100.Nixon, A. E., Sexton, D. J., and Ladner, R. C.
(2014) Drugs derived from phage display: from candidate identification
to clinical practice, MAbs, 6, 73-85, doi:
10.4161/mabs.27240.
101.Urban, J. H., Moosmeier, M. A., Aumuller, T.,
Thein, M., Bosma, T., et al. (2017) Phage display and selection of
lanthipeptides on the carboxy-terminus of the gene-3 minor coat
protein, Nat. Commun., 8, 1500, doi:
10.1038/s41467-017-01413-7.
102.Hetrick, K. J., Walker, M. C., and van der
Donk, W. A. (2018) Development and application of yeast and phage
display of diverse lanthipeptides, ACS Cent. Sci., 4,
458-467, doi: 10.1021/acscentsci.7b00581.
103.Convery, N., and Gadegaard, N. (2019) 30 years
of microfluidics, Micro Nano Eng., 2, 76-91, doi:
10.1016/j.mne.2019.01.003.
104.Fallah-Araghi, A., Baret, J. C., Ryckelynck,
M., and Griffiths, A. D. (2012) A completely in vitro
ultrahigh-throughput droplet-based microfluidic screening system for
protein engineering and directed evolution, Lab. Chip.,
12, 882-891, doi: 10.1039/c2lc21035e.
105.Mahler, L., Wink, K., Beulig, R. J., Scherlach,
K., Tovar, M., et al. (2018) Detection of antibiotics synthetized in
microfluidic picolitre-droplets by various actinobacteria, Sci.
Rep., 8, 13087, doi: 10.1038/s41598-018-31263-2.
106.Terekhov, S. S., Smirnov, I. V., Stepanova, A.
V., Bobik, T. V., Mokrushina, Y. A., et al. (2017) Microfluidic droplet
platform for ultrahigh-throughput single-cell screening of
biodiversity, Proc. Natl. Acad. Sci. USA, 114, 2550-2555,
doi: 10.1073/pnas.1621226114.