REVIEW: Riboswitch Mechanisms: New Tricks for an Old Dog
Ascensión Ariza-Mateos1, Ashok Nuthanakanti1, and Alexander Serganov1,a*
1Department of Biochemistry and Molecular Pharmacology, New York University Grossman School of Medicine, New York, New York 10016, USA* To whom correspondence should be addressed.
Received May 1, 2021; Revised May 1, 2021; Accepted May 19, 2021
Discovered almost twenty years ago, riboswitches turned out to be one of the most common regulatory systems in bacteria, with representatives found in eukaryotes and archaea. Unlike many other regulatory elements, riboswitches are entirely composed of RNA and capable of modulating expression of genes by direct binding of small cellular molecules. While bacterial riboswitches had been initially thought to control production of enzymes and transporters associated with small organic molecules via feedback regulatory circuits, later findings identified riboswitches directing expression of a wide range of genes and responding to various classes of molecules, including ions, signaling molecules, and others. The 5′-untranslated mRNA regions host a vast majority of riboswitches, which modulate transcription or translation of downstream genes through conformational rearrangements in the ligand-sensing domains and adjacent expression-controlling platforms. Over years, the repertoire of regulatory mechanisms employed by riboswitches has greatly expanded; most recent studies have highlighted the importance of alternative mechanisms, such as RNA degradation, for the riboswitch-mediated genetic circuits. This review discusses the plethora of bacterial riboswitch mechanisms and illustrates how riboswitches utilize different features and approaches to elicit various regulatory responses.
KEY WORDS: riboswitch, transcription, translation, mRNA, metaboliteDOI: 10.1134/S0006297921080071
INTRODUCTION
Most bacteria live in ever-changing environmental conditions and must quickly adjust their metabolism to match nutrient availability and other environmental factors. Bacteria activate expression of genes to produce proteins and RNAs needed to maximize adaptation to surroundings. To save resources, bacteria also shut down genes whose products are not required at the moment. Gene control is often achieved through the feedback regulation by an end-product: when the molecule is synthesized at the over-threshold concentration, the gene can be turned off through sensing of the gene product. This type of regulation, when a molecule controls its own production, was termed autoregulation. Autoregulation does not necessarily require changes in the environment. Homeostatic maintenance of certain levels of a gene product and prevention of accumulation of not needed molecules could be equally important for cellular fitness. Autoregulation can be achieved by the mechanisms acting at different levels, including direct repression of transcription or translation and many posttranscriptional and posttranslational mechanisms, such as covalent modifications, inhibition of enzymatic activity, and others.
In addition to proteins being end-products, numerous examples of feedback regulation include small metabolites produced by proteins as end-products. In this case, small molecules are sensed and bound to proteins, and the resulting complexes modulate, for example, transcription of genes through binding to transcriptional operators [1]. In order to control expression of metabolite-associated genes, cells would have to make dozens of protein repressors and activators, one for each gene or operon, to respond to a plethora of cellular metabolites. While such approach is feasible, it would result in wasting cellular resources. Can regulation by a small molecule happen without an intermediary (sensing protein)?
The small molecule ppGpp can directly bind to RNA polymerase, mRNA, and protein production machinery and modulate their activity during adaptation to starvation for amino acids (reviewed in [2]). However, (p)ppGpp is a second messenger synthesized by special enzymes and is not an end-product of amino acid-producing enzymes. While direct binding to the polymerase and ribosome represents a convenient and efficient way for gene expression control, these cellular machineries cannot sense dozens of different cellular metabolites. Can a small molecule bind to DNA or mRNA to modulate transcription or translation? While the double-stranded genomic DNA does not have sufficient complexity for specific recognition of small molecules, RNA can fold into intricate three-dimensional structures capable of specific binding of small compounds [3]. Therefore, modulation of gene expression by a small molecule seems feasible, if the molecule binds to mRNA and allosterically changes its structure to hide or expose gene regulatory signals embedded into the mRNA sequence. This idea led to the discovery of riboswitches almost 20 years ago.
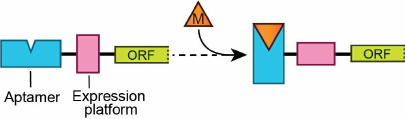
The term “riboswitches” was coined for regulatory RNA regions capable of modulating gene expression through direct sensing of cellular metabolites [4-6]. Riboswitches typically reside in the 5′-untranslated regions (UTRs) of mRNAs upstream of the genes involved in the metabolism or transport of riboswitch ligands or related cellular metabolites [7]. The vast majority of riboswitches provide feedback regulation by controlling the levels of transcription or translation. Riboswitches usually consist of two parts – evolutionarily conserved metabolite-sensing “aptamer” domain and variable adjacent “expression platform”, which carries gene expression signals (Fig. 1). Various expression platforms can accompany the same type of sensing domain but with different outcomes of gene expression.
Fig. 1. Domains of a typical riboswitch. Metabolite-sensing aptamer domain, expression platform, and open reading frame (ORF) of the gene are shown with rectangles. Metabolite (M) binding stabilizes an alternative conformation of the riboswitch domains. Two riboswitch conformations elicit two opposite regulatory responses (not shown).
The ability of riboswitches to adopt two mutually exclusive RNA conformations defines the basis for the genetic response. Specific metabolite binding induces folding of the metabolite sensor and expression platform that prompts one genetic response, while adaptation of the alternative conformation produces the opposite effect on the gene expression (Fig. 1). Riboswitches comprise over 40 classes that recognize ~30 different small molecules, ranging from ions to vitamins [8]. The molecular basis for the sensor specificity is formation of intricate three-dimensional structures tuned for exquisite recognition of a ligand or ligands and rejection of similar metabolites and precursors [9]. Since aptamers range from ~30 to ~250 nucleotides in size, their structures and metabolite recognition features vary dramatically. On the contrary, expression platforms most often fold into hairpins, which function as transcription terminators, antiterminators, or ribosome sequestering structures.
Recent studies have identified (or suggested) conceptually new riboswitch mechanisms and found many variations of the common mechanisms, greatly expanding the repertoire of riboswitch functioning. In this review, we describe typical riboswitch mechanisms and discuss some new findings and hypotheses to illustrate the versatility of riboswitches in genetic control.
THE ROAD TO RIBOSWITCHES
Although identification of riboswitches has come to many researchers as a surprise, a number of studies have set the scene for their discovery. One of the influential factors was the discovery of the feedback control by protein autorepressors, such as ribosomal proteins [10]. To balance the biosynthesis of rRNA and ribosomal proteins for the ribosome assembly, several ribosomal proteins, synthesized in excess over rRNA, can bind to the 5′-UTR of their own mRNA and prevent translation initiation through sequestering the ribosome-binding site and the initiation codon. The mRNA and rRNA targets of these proteins bear some resemblance at either the sequence or structural levels, but the rRNA binding affinity always exceeds the mRNA binding affinity to ensure proper control [11]. The feedback control is not limited to proteins. The so-called “T-box” mRNAs specifically bind tRNA and, depending on the presence of a covalently attached amino acid on the tRNA, facilitate the folding of the downstream regions into transcription terminators or antiterminators, reminiscent of the controlling elements of transcriptional riboswitches [12]. Specific binding of small molecules to RNA was not an entirely new observation as well. Recruitment of guanosine cofactor by the self-splicing group I introns was described almost two decades prior to the discovery of riboswitches [12]. In vitro selection of RNA aptamers specific to various small molecules has reinforced the view that RNA can selectively recognize small molecules with different characteristics [13-15]. The binding of amino acids to mRNA and their involvement in regulation was also observed in attenuation, when proteins and small molecules collectively re-shape the mRNA structure and cause transcription termination [16].
Identification of first riboswitches was primarily driven by observations that thiamine, riboflavin, and cobalamin inhibit vitamin B1, B2, and B12 biosynthetic genes, respectively. mRNAs of these genes contained conserved regulatory mRNA sequences, or “boxes”; however, no protein repressor specific for these boxes was found [17, 18], suggesting direct binding of the vitamins to mRNAs [17, 19-21]. After unsuccessful attempts [22, 23], three vitamin derivatives, adenosylcobalamin (AdoCbl) [4], thiamine pyrophosphate (TPP) [5, 6], and flavin mononucleotide (FMN) [6], were convincingly shown to bind their respective mRNAs, modulate RNA conformation, and regulate gene expression. Examples from other domains of life have followed the initial finding of bacterial riboswitches although active eukaryotic riboswitches scarcely distributed and only represented by TPP binders [24-26]. In contrast, many bacterial species possess riboswitches, many bacteria contain different classes of riboswitches, and some species have a few riboswitches of the same class involved in controlling different genes and operons [8].
CANONICAL RIBOSWITCH MECHANISMS
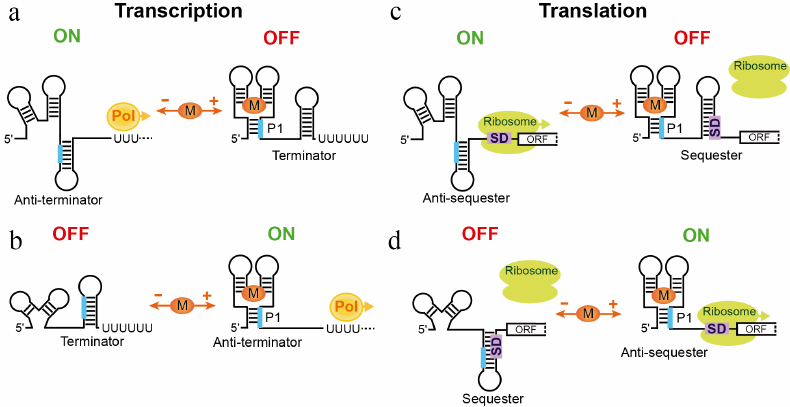
The first reports on the identification of riboswitches have immediately suggested four major mechanisms of gene expression control (Fig. 2) [7] for riboswitches that contain a defined metabolite sensor and an expression platform. Metabolite binding stabilizes the overall structure of the sensing domain and facilitates formation and stabilization of a helix that usually closes the sensing domain. This regulatory helix, designated as “Pairing 1” (P1), involves a region, typically on the 3′-end, capable of alternative base-pairing with a complementary downstream sequence when the metabolite is not bound. The alternative base-pairing of the P1 segment is a quintessence of the regulation for most riboswitches; however, the cellular processes under control and machineries involved differ dramatically. In bacteria, canonical riboswitch mechanisms rely on the transcription termination and translation initiation.
Fig. 2. Canonical mechanisms of riboswitches. a) Inhibition of gene expression through transcription termination mechanism. In the absence of metabolite, the aptamer domain is not folded and the riboswitch forms an antiterminator hairpin. RNA polymerase (Pol) transcribes the entire gene, turning expression of the gene on. Metabolite (M) binding stabilizes the ligand-bound form, closed by helix P1, and facilitates formation of the intrinsic transcription terminator, thereby turning gene expression off. A fragment of the riboswitch (thick line) engages in two alternative base-parings in the ligand-free and ligand-bound states of the riboswitch. b) Activation of gene expression through transcription termination mechanism. In the absence of metabolite, the riboswitch forms transcription terminator. In the presence of metabolite, a fragment participating in the formation of the terminator forms alternative pairing within P1 that locks the metabolite-bound aptamer. c) Repression of translation by metabolite binding. The metabolite-free state allows ribosome binding to the Shine–Dalgarno sequence (square with SD) and translation initiation. Upon ligand binding, the SD sequence is sequestered in the hairpin structure and translation cannot initiate. d) Activation of translation. Metabolite binding leads to a regulatory response opposite to one in panel (c).
The first two mechanisms count on the ability of the RNA polymerase to “look back” at the transcribed mRNA and terminate transcription after synthesizing an intrinsic terminator, a self-complementary RNA region, which folds into a hairpin followed by a stretch of uridines (Fig. 2, a and b). Such intrinsic transcription terminators can form in the presence or absence of a metabolite, leading to the transcription repression or activation, respectively. Premature transcription termination prevents production of mRNA for translation and therefore saves nucleotides and other resources required for making mRNA. Not surprisingly, transcription termination is the prevailing mechanism of riboswitches and is often used for the negative feedback control. Interestingly, the riboswitch-mediated transcriptional mechanism does not involve true “switching” of the riboswitch and could be described better as a “molecular fuse” [27]. Indeed, as soon as the RNA commits to a regulatory pathway, the process cannot be reverted. The RNA polymerase cannot extend the terminated RNA and cannot stop transcription after passing through the terminator sequence, if the terminator hairpin is not formed.
The other two classical riboswitch mechanisms are based on the fact that the 30S ribosomal subunit requires base-pairing between 16S rRNA and a ribosome-binding site (Shine–Dalgarno or SD sequence) and unpaired initiation codon, for efficient translation initiation (Fig. 2, c and d). Engaging the SD sequence and the translation initiation codon in base-pairing, for example, within a hairpin, would prevent ribosome loading and translation of the mRNA. Releasing the sequestered SD sequence would allow the ribosome to bind and translate mRNA. Regulation on the level of translation requires transcription of at least the 5′ part of mRNA and appears less economical than transcriptional control. However, translational regulation has its own advantages. First, it could allow controlling an individual gene in the polycistronic mRNA, if the gene is translated from its own SD sequence. Second, the response time for the translational control could be very fast since the mRNA is already synthesized. In the case of activation, translation can begin instantly after releasing the SD sequence. Lastly, translational riboswitches could be true switches [28] binding and releasing the metabolite and activating or repressing translation of the same mRNA molecule depending on fluctuations of the metabolite concentration. However, riboswitches appear not to use this option routinely because most riboswitches are kinetically and not thermodynamically driven; in other words, they require the metabolite concentration higher than the KD for activation [27]. Kinetically driven translational riboswitches can choose the regulatory path as soon as the 5′ portion of the mRNA has been synthesized. On the contrary, reaching thermodynamic equilibrium could be a long process, approaching or exceeding the lifetime of mRNA and making the response obsolete. Since transcription and translation are coupled in bacteria, translation is initiated soon after transcription initiation, prior to completion of transcription.
TWEAKS IN CANONICAL RIBOSWITCH MECHANISMS
Many riboswitches that employ canonical regulatory mechanisms contain a separate aptamer domain and an expression platform, each capable of folding alternatively dependent on the availability of the riboswitch ligand. Such double domain arrangement is not absolutely required for efficient regulation as demonstrated by some riboswitches that combine both elements while keeping the “two state” regulatory response intact.
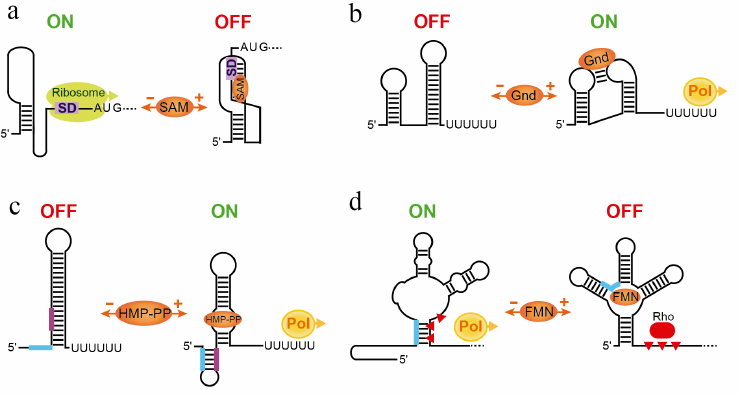
Because translation initiation does not require a structure present in the mRNA, many translational repressors are streamlined riboswitches, which lack a dedicated hairpin sequestering or exposing the SD sequence. In these riboswitches, the expression platform does not exist and the SD sequence could be considered an integral part of the ligand-bound aptamer domain. For example, in the SAM-III riboswitch [29], the ligand binding engages the SD sequence in base-pairing in the junctional core [30]. The SD sequence becomes inaccessible for ribosome loading and translation cannot initiate.
Involvement of the SD sequence in the folding of the ligand-bound riboswitch is especially important for aptamers adopting the so-called pseudoknot structure. In several riboswitches, such as SAM-II, biochemical and structural studies confirmed the bioinformatically predicted long-range base-pairing between the SD sequence and an internal loop in the middle of the aptamer [31, 32]. This tertiary pairing result in the formation of the pseudoknot structure that comprises two coaxially stacked helices connected by two loops (Fig. 3a). Other translational repressors predicted to form a similar long-range pairing likely adopt pseudoknot structures upon ligand binding as well [33].
Fig. 3. Streamlined and non-canonical mechanisms of riboswitches. a) Regulation of translation initiation by the SAM-II riboswitch [31, 101]. SAM binding stabilizes pseudoknot-based fold of the aptamer. The SD sequence participates in the formation of the pseudoknot and cannot be accessed by the ribosome. In the absence of SAM, the pseudoknot is not formed and the ribosome can bind to the SD sequence and initiate translation of the mRNA. b) A putative mechanism of transcription activation of the guanidine-IV riboswitch involving tertiary interactions between aptamer domain and expression platform [34]. Upon ligand binding, a transcription terminator structure is disrupted by long-range base-paring essential for the formation of the “aptamer” structure. Loops contain conserved nucleotides involved in tertiary interactions. The transcription terminator normally folds in the absence of guanidine. c) Transcriptional activation by the HMP-PP riboswitch [35]. In the absence of the ligand, the riboswitch folds into canonical transcription terminator. Ligand binding promotes formation of the alternative conformation that disrupts the terminator. Thick lines indicate key fragments involved in the formation of alternative structures. d) Putative mechanism of the Rho-dependent transcription repression of the E. coli FMN riboswitch [36]. In the FMN-bound conformation, factor Rho facilitates termination of transcription in the C-rich region downstream of the aptamer. In the absence of the ligand, a fragment in the 5′ half of the aptamer pairs with the C-rich region thereby precluding transcription termination. Red triangles indicate a region recognized by Rho.
In contrast to translational riboswitches, examples of streamlined transcriptional riboswitches are limited [34, 35]. The guanidine-IV riboswitch exploits the loop of an intrinsic terminator hairpin to form an aptamer structure by the long-range base-pairing with the internal loop in the body of the aptamer (Fig. 3b) [34]. Therefore, ligand binding disrupts the terminator and switches transcription on. In the absence of guanidine, formation of the transcription terminator is not impeded and transcription is aborted prematurely.
A riboswitch responding to the thiamin pyrophosphate (TPP) precursor HMP-PP exhibits even more unusual architecture wherein a very small ligand-sensing aptamer is almost entirely embedded within an otherwise classic intrinsic transcription terminator stem (Fig. 3c) [35]. The riboswitch uses the same RNA sequence to fold into two mutually exclusive structures, the ligand-free terminator configuration, aborting transcription prematurely, and the ligand-bound aptamer configuration, preventing early transcription termination.
Some riboswitches identified in Gram-negative bacteria, unlike their counterparts in Bacillus subtilis, appear to lack intrinsic terminators. Such riboswitches were shown to control gene expression on the transcriptional level but with the help of the termination factor Rho [36]. Rho is a general termination factor that travels with RNA polymerase and is responsible for termination of ~20% of transcriptional units in Escherichia coli [37]. Rho interrupts transcription of unprotected RNAs at sites that are generally rich in C residues and depleted in G residues. In the E. coli flavin mononucleotide (FMN) riboswitch, stabilization of the FMN-bound aptamer induces Rho-dependent transcription termination downstream of the aptamer and upstream of the protein-coding region (Fig. 3d) [36]. Disruption of the aptamer in the absence of FMN prompts an alternative conformation of the untranslated region that eliminates Rho-dependent termination and allows transcription to proceed to the end of the gene.
MULTI-LIGAND SENSING BY RIBOSWITCHES
Sensing more than one molecule of the ligand by a single sensor. Many observations that a sensing domain of the riboswitch binds a single metabolite molecule have never excluded the possibility that the riboswitch can sense two or more molecules of the ligand.
One of the first evidences emerged after biochemical and structural characterization of the M-box (Mg-I) riboswitch responding to Mg2+ cations [38, 39]. This large RNA binds several Mg2+ cations for compaction and genetic response but other cations can replace Mg2+ cations to stimulate RNA folding. A more selective NiCo riboswitch responds to Ni2+ or Co2+ cations [40] and possibly to Fe2+ and Mn2+ [41]. The structure and biochemical data show that the riboswitch cooperatively binds four Ni2+ or Co2+ cations although it is unclear whether all of these sites are relevant at physiological metal concentrations [40]. A distinct metal sensor, a Mn2+ riboswitch, contains two adjacent metal binding sites, one for Mn2+ and another for Mg2+ [42]. Although Mn2+ cation at millimolar concentrations can replace Mg2+ in its binding site in vitro, this situation is not likely biologically relevant since E. coli maintains intracellular concentration of Mn2+ at the low micromolar levels to avoid toxicity.
Multi-ligand binding is not restricted to metal cations, which, because of small size, may have difficulty in directing RNA folding if bound as a single molecule. The THF-I riboswitch binds two tetrahydrofolate (THF) molecules [43]. The riboswitch strategically places one THF molecule in the hinged region to ensure parallel alignment of the main stem with a branched off stem-loop structure while the second THF molecule binds at the tip of the stem-loop and mediates long-range contacts with the main stem. These tertiary interactions stabilize the P1 helix for the regulatory response.
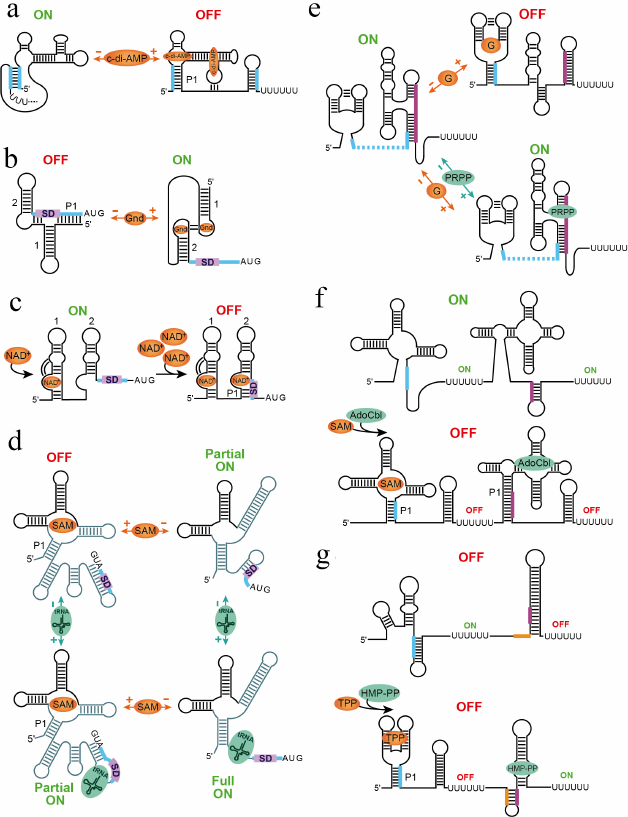
The cyclic di-adenosine monophosphate (c-di-AMP) riboswitch interacts with two molecules of c-di-AMP in two binding sites positioned across a pseudosymmetric square-shaped aptamer [44, 45] (Fig. 4a). Despite similarity, the sites are not equal for ligand binding and while disruption of one site eliminates binding to both sites, mutation of another site only decreases binding to the first site [44]. Depending on the expression platform, the riboswitch represses transcription or translation.
Fig. 4. Mechanisms of multi-ligand sensing and regulation. a) Putative mechanism of the transcriptional control by c-di-AMP riboswitch [44, 102]. The riboswitch requires binding of two c-di-AMP molecules for adopting a ligand-bound aptamer structure and terminating transcription [44, 45]. b) Mechanism of translation activation of guanidine-II riboswitch [60-62]. Binding of two ligands to conserved apical loops promotes formation of the head-to-head dimer that releases SD sequence for ribosome binding. c) Putative step-wise mechanism of translational repression by the NAD+-I riboswitch [66]. Since NAD+ is always present in cells, one NAD+ molecule can bind to a nascent transcript as soon as aptamer 1 is transcribed. High NAD+ concentrations would prompt binding of NAD+ to aptamer 2, which has lower affinity for the ligand, and sequestration of SD sequence within the aptamer structure. d) Hypothetical mechanism of the X. campestris SAM-I–tRNA riboswitch [67]. Upon sensing various concentrations of SAM and tRNAfMet, the riboswitch could adopt four different states corresponding to four regulatory responses. The RNA regions involved in most extensive conformational rearrangements are shown in lighter color. e) Regulatory responses for the tandem guanine-PRPP riboswitch [68]. In the ligands-free form, the riboswitch activates transcription. Binding of guanine in the absence of PRPP disrupts the PRPP aptamer and causes premature transcription termination. PRPP binding alone or in the presence of guanine stabilizes the PRPP aptamer and prevents guanine binding, leading to uninterrupted transcription. f) Regulatory responses of the SAM-I and AdoCbl riboswitch system [73]. The system turns on transcription when both ligands are absent. Binding of either ligand or both ligands simultaneously aborts transcription and turns gene expression off. g) Regulatory responses of the TPP and HMP-PP riboswitch system [35]. The TPP riboswitch represses transcription upon ligand binding while the HMP-PP riboswitch activates transcription in the ligand-bound form. Therefore, the gene is expressed only in the absence of TPP and presence of HMP-PP. The ligand-free, TPP-bound and TPP-bound/HMP-PP-bound states turn gene expression off.
A number of ligands need assistance from other small molecules, typically metal cations, for efficient binding to riboswitches. Practically all metabolites containing phosphates neutralize these moieties by Mg2+ cations, which remain bound to a ligand and mediate its interactions with the RNA [46-48]. Three Mg2+ cations encapsulate a fluoride anion and help this small ligand direct folding of the fluoride riboswitch [49]. K+ and Mg2+ cations facilitate binding of the carboxylate moiety of lysine and glycine, respectively, to their cognate riboswitches [50, 51].
While examples listed above are consistent with a single-input signaling model, there are riboswitches whose control depends or involves a dual input. A recent study [52] showed that responsiveness of the B. subtilis lysC lysine riboswitch to lysine heavily depends on subtle changes in intracellular levels of Mg2+ cations. At the low end of the physiological Mg2+ concentration range (<1 mM), the aptamer remain unstructured and requires higher lysine concentration for response. 1 mM increase in the Mg2+ concentration pre-organizes the structure for lysine binding so that the ligand can interact with the riboswitch at two orders of magnitude lower concentration. Thus, changes in the levels of Mg2+ under, for example, normal growth conditions and in stressed environments can determine activity of the riboswitch.
Riboswitch specificity often means that a riboswitch binds to a cognate metabolite with the affinity higher than to similar molecules. In cells, similar non-cognate ligands are thought to be rejected by the riboswitch and the regulatory response is ascribed to the best riboswitch binder. Nevertheless, at high concentrations, non-cognate ligands can compete with a cognate ligand for riboswitch binding. When bound, a non-cognate ligand may not advance riboswitch folding to the ligand-bound state and exert the corresponding regulatory response. An in vivo study showed that activity of the B. subtilis glmS riboswitch-ribozyme is modulated by both the cognate ligand glucosamine-6-phosphate (GlcN6P), activating the response, and its several precursors, inhibiting the response [53]. Thus, in cells, even single-input riboswitches may integrate information from an array of chemical signals and respond based on the concentrations of several compounds.
Sensing same ligands by multiple sensors. While most riboswitches utilize a single aptamer to bind a ligand, a few riboswitch classes regulate gene expression by using multiple adjacent aptamers, each with a ligand-binding site, followed by a single expression platform. Multiple aptamer arrangements can achieve sharper, more digital responses to changing ligand concentrations. The first example of such riboswitches dates back to the early 2000s [54]. This study identified glycine-specific riboswitches composed of two similar sensing domains in tandem arrangement and suggested cooperative binding of two glycine molecules, one molecule per sensor. Structural and biochemical studies identified intermolecular interactions between two modules [51, 55]. Although binding cooperativity was challenged [56], a recent study confirmed existence of cooperativity that could be masked by the difference in binding affinity of two sensors [57].
Several more riboswitches use multiple aptamers followed by a single expression platform for regulation. Over half of the glnA glutamine riboswitches (glutamine-I) are arranged in tandem orientations, where two or three aptamers are separated by small linker regions [58]. Many glutamine-I riboswitches have similar linker sequences, which are predicted to base-pair with the sequence downstream of the 3′ domain. The X-ray crystal structure showed ligand binding in the junction connecting riboswitch stems and their large rearrangement upon ligand binding [59]. Biochemical experiments did not find evidence of cooperativity [58] and, while structural studies predict similar ligand binding to all aptamers of the tandem riboswitches, benefits of having more than one aptamer are not clear.
The guanidine-II riboswitch is composed of two hairpins containing similar and highly conserved apical loops, which bind free guanidine in cooperative manner [60] (Fig. 4b). Structural studies of individual aptamers revealed that each loop houses a guanidine binding site and that guanidine binding facilitates homo-dimerization of two aptamer domains via tertiary base pairing between the loops [61, 62]. Formation of this ligand-dependent head-to-head dimer may explain the cooperative binding of guanidine seen in biochemical experiments with a full-length riboswitch [60] and suggests the regulatory mechanism based on dimerization of two aptamers within the same riboswitch [62, 63]. Guanidine-II appears to act as a translational ON riboswitch. In the absence of guanidine, the SD sequence is sequestered by base-pairing with a linker region and nucleotides at the 5′-end of the first aptamer thereby preventing translation initiation. Guanidine binding promotes head-to-head dimerization of two aptamers and enforces the spatial separation of the SD sequence from the complementary anti-SD sequence. The unpaired SD sequence can bind the ribosome and initiate translation.
As the guanidine-II riboswitch, the nicotinamide adenine dinucleotide (NAD+) class I riboswitch is formed by two hairpins; however, similar conserved regions are located in the bulged loops (Fig. 4c) [64]. Structural studies revealed that NAD+ indeed binds the RNA in this bulge [65, 66]. Bioinformatics revealed small differences between two aptamers in the central regions and the original study did not report NAD+ binding to the second aptamer [64]. In the later work, binding to the second aptamer was detected but with much lower affinity than to the first aptamer [66]. The structures revealed that ligand binding stabilizes a tertiary contact between the ligand-binding site and the middle irregular region of the RNA. This contact is missing in the second aptamer. Interestingly, both aptamers recognize only the adenosine moiety of the ligand, not the nicotinamide moiety, suggesting that the structures of the individual domains may not have captured all interactions with the ligand. Perhaps the nicotinamide moiety of the ligand bound to aptamer 1 could be recognized by aptamer 2. Since NAD+ is always present in the cells, there is an intriguing possibility that the riboswitch acts as a two-concentration sensor [66]. At low concentrations, NAD+ bind to the first aptamer, leaving aptamer 2 unbound and unstructured and the SD sequence exposed for translation. At higher concentrations, NAD+ binds to aptamer 2 as well, promoting base-pairing of the SD sequence and triggering translational repression.
Sensing different ligands by multiple sensors. Among the most exciting findings in the riboswitch field are discoveries of the two-input riboswitches, which possess separate binding sites for different ligands but work in concert through a single expression platform. These findings are especially interesting from the evolutionary perspective. Although gene duplication can position similar aptamers next to each other, it is more difficult to imagine how sensors with different specificities have evolved to join and control the same expression unit.
An interesting dual-input mechanism was recently proposed for a SAM-I riboswitch that regulates methionine synthesis in Xanthomonas campestris (Fig. 4d) [67]. This riboswitch primarily responds to cellular S-adenosylmethionine (SAM) at the translational level; however, the expression platform also contains a binding site for uncharged initiator Met tRNA. Binding of tRNA and SAM appears to promote several alternative base pairings and produce a number of regulatory responses. While SAM binding sequesters the SD sequence and abolishes translation, simultaneous tRNA binding sequesters the anti-SD sequence and partially releases the SD sequence for ribosome binding. Partial gene expression can also be observed in the absence of both ligands while full derepression apparently requires binding of tRNA in the absence of SAM.
A complex regulatory outcome was also observed in the riboswitch comprising a guanine sensor followed by a sensor for phosphoribosyl pyrophosphate (PRPP) [68] (Fig. 4e). The intervening sequence between these aptamers is short and the 5′ region of the PRPP sensor overlaps with the 3′ region of the preceding guanine sensor. Therefore, guanine binding precludes formation of the PRPP sensor and facilitates formation of the transcription terminator, thereby shutting down transcription. On the other hand, PRPP aptamer is “preformed” in the absence of both guanine and PRPP [69, 70]. This conformation of the aptamer precludes formation of the terminator and allows transcription to proceed to the end. In the presence of both ligands, PRPP dominates binding to the riboswitch and prevents guanine binding, ensuring uninterrupted transcription of the gene [68].
A special mechanism was demonstrated for a cyclic di-GMP riboswitch, which is located upstream of group I self-splicing intron [71]. This RNA regulator combines self-splicing and translation activation to regulate downstream genes. C-di-GMP binding to its aptamer potentiates folding of the intron core, binding of GTP to the intron’s 5′ splice site, and self-excising of the intron. Removal of the intron brings together two distantly positioned parts of the SD sequence to form a complete SD sequence for translation initiation [71, 72]. In the absence of c-di-GMP, GTP binds in the alternative site and a new cleavage results in a truncated SD sequence, which does not efficiently bind the ribosome and initiate translation.
COOPERATION OF INDEPENDENT RIBOSWITCHES
Although the majority of riboswitches function as a single regulatory unit, which comprises one or more aptamers and a single expression platform or an equivalent mRNA region, some riboswitches contain two independent regulatory units in tandem arrangement [35, 73-75]. Most often, these riboswitches contain sensors responding to the same metabolite, for example, TPP, followed by the same type of the expression platform, such as transcription terminator [73]. Composite switches reminiscent of the double TPP riboswitch likely enable more digital response or greater responsiveness to changes in metabolite concentration compared with lone riboswitches. In some cases, sensors respond to different ligands.
In the tandem SAM-I and AdoCbl riboswitch system, binding of SAM or AdoCbl to their respective sensors causes transcription termination in the individual riboswitch that abolishes transcription of the gene. Respectively, the gene is turned on only when both ligands are present at the under-threshold concentrations in cells (Fig. 4f).
The tandem TPP and HMP-PP riboswitch system contains a repressing TPP riboswitch followed by an activating HMP-PP riboswitch. This system controls expression of the gene, which codes for an enzyme needed to synthesize another precursor on the pathway to make TPP [35]. Bacteria do not need to make TPP when it is abundant; therefore, binding of TPP to its aptamer shuts down expression of the downstream gene regardless of the concentration of HMP-PP (Fig. 4g). When TPP is absent, the response could follow two scenarios. If HMP-PP is also absent in cells, the gene is not expressed. High concentrations of HMP-PP activate expression of the gene.
Existence of riboswitch systems that join individual regulatory units with different specificities into tandem arrangements offers the possibility of integrating various signals from the environment and intracellular milieu. Each riboswitch unit senses one signal; however, the regulatory response is combinatorial. Such complex systems could allow accurate regulation that depends on the presence of the end-products as well as metabolic state of the cell or the presence of precursor molecules.
RNA DEGRADATION AS AN ADDITIONAL MECHANISM OF RIBOSWITCHES
Undoubtedly, transcriptional and translational control play very important roles in gene regulation. In addition, changes in the lifetime of mRNA can greatly affect production of proteins. In bacteria, mRNA stability is governed by several ribonucleases and associated factors, which vary across species [76]. In E. coli and other Gram-negative bacteria, RNA degradation typically begins with internal cleavage by RNase E, followed by 3′-end-dependent exonuclease activity. In B. subtilis and other Firmicutes, mRNA degradation involves a different endonuclease RNase Y and the 5′ exonuclease RNase J. Contribution of ribonucleases to riboswitch mechanisms was noted long time ago but only recent studies revealed that RNA degradation could be a major mechanism of riboswitch action [77, 78] and not only the means for RNA removal after completion of the regulatory response [79].
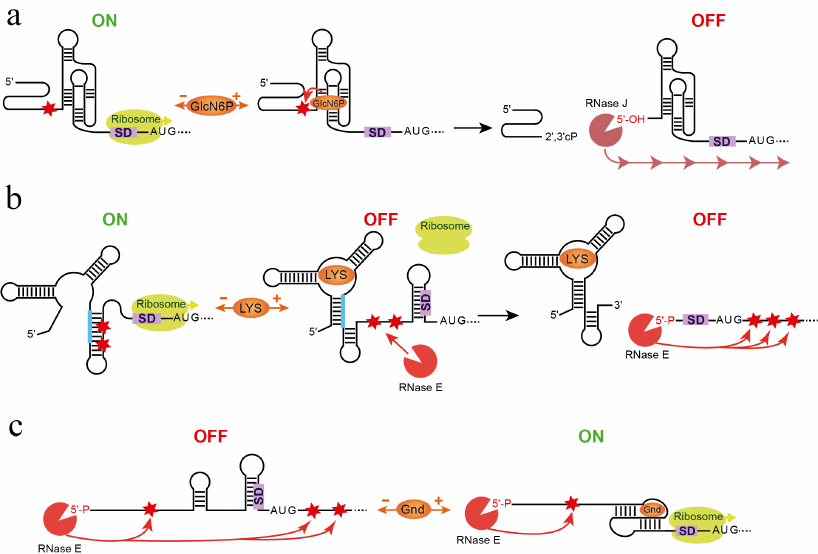
The first association between a riboswitch and RNA degradation machinery was established after the discovery of the glmS riboswitch-ribozyme (Fig. 5a) [80, 81]. In B. subtilis, glmS mRNA encodes a GlcN6P synthase. At high concentrations, the end-product of the reaction, GlcN6P, initiates negative feedback regulation [80]. Unlike in other riboswitches, GlcN6P binds to the preformed aptamer [82, 83] and, instead of allosterically modulating its structure, catalyzes mRNA cleavage reaction upstream of the aptamer [80, 84]. The resulting RNA product is no longer protected by the triphosphate on the 5′-end and undergoes rapid degradation by exonuclease RNase J [81].
Fig. 5. Involvement of RNA degradation in riboswitch response. a) Mechanism of the glmS riboswitch-ribozyme [80, 81]. In the absence of GlcN6P, the gene is transcribed and translated. GlcN6P binding induces cleavage (arrow) in the site (star) upstream of the aptamer. Exonuclease RNase J binds to the dephosphorylated 5′ end and digests the mRNA (line with arrows), turning gene expression off. b) Dual mechanism of the E. coli lysC riboswitch controlling translation initiation and mRNA decay [85]. In the absence of lysine, the riboswitch folds into the ON state that exposes the SD sequence for translation initiation and sequesters the RNase E cleavage sites. Upon ligand binding, the aptamer domain adopts the OFF state that sequesters the SD sequence and exposes the RNase E sites for cleavage. Consecutively, the RNase E is recruited to the 5′-monophosphorylated end of the RNA and makes further endonucleolytic cleavages (long arrows), followed by the 3′-end-dependent exonucleolytic digestion (not shown). c) Involvement of RNA degradation in the mechanism of the Legionella pneumophila sugE guanidine-III riboswitch. In the absence of the ligand, the 5′-end-bound RNase E can make endonucleolytic cleavages both upstream and downstream of the aptamer because the ligand-free riboswitch does not stop the 5′-to-3′ search of the cleavage sites. The mRNA degrades and the protein is not produced. Formation of the ligand-bound pseudoknot in the aptamer domain releases the SD sequence for translation and impedes scanning by RNase E, preventing cleavages downstream of the aptamer domain. Therefore, the ligand-bound aptamer protects translating ribosome from degradation, ensuring the ON status of the gene expression.
Another example illustrates a different involvement of an RNase to the riboswitch mechanism [85]. Upon lysine binding to the sensing domain of the E. coli lysC riboswitch, the expression platform folds into the SD sequestering hairpin that turns off translation and exposes two RNase E cleavage sites located in the expression platform (Fig. 5b). These cleavages produce RNAs with 5′-monophosphorylated ends that appear to trigger accelerated degradation of the lysC coding region by RNase E. In the absence of lysine, the riboswitch forms a SD anti-sequestering hairpin that also sequesters RNase E cleavage sites, allowing translation initiation to proceed without mRNA degradation. Thus, the lysine riboswitch is a dual action regulator: two different mechanisms, translational suppression and RNA degradation, inhibit expression of lysC gene. A similar involvement of RNase might contribute to genetic responses of other riboswitches [86].
In another system, metabolite binding causes an opposite effect on RNA stability [87, 88]. Cyclic di-GMP-dependent (c-di-GMP) Vc2 riboswitch appears to downregulate Vibrio cholerae tfoY gene expression in response to ligand binding through sequestering the SD sequence and inhibiting translation [88-90]. c-di-GMP binding to Vc2 also leads to the accumulation of the upstream untranslated RNAs, probably functioning as sRNAs in the control of cell motility [87]. Although the molecular basis for stability of putative sRNAs is unclear, stabilization of the aptamer by the bound c-di-GMP likely engages the 3′-end in base-pairing and prevents access of the 3′-end-dependent exonucleases.
A most recent study elegantly showed an entirely different mechanism employed by a riboswitch to regulate gene expression [78] (Fig. 5c). This mechanism is based on several findings related to the activity of RNase E. This endoribonuclease cuts RNA at specific locations in single-stranded regions [91] and its activity is greatly accelerated after recruitment to the 5′-monophosphorylated RNA substrates via a pocket specific for 5′-monophosphorylated RNA [92, 93]. Once recruited, RNase E searches for cleavage sites by scanning the mRNA from 5′- to 3′-ends. The enzyme by-passes small obstacles such as orthogonally base-paired stem-loops but stops at larger impediments such as bound proteins or structures containing base-paired elements coaxial with the path of scanning [78, 94]. Translation of the Legionella pneumophila sugE transcript is controlled by the guanidine-III riboswitch. Upon ligand binding, the aptamer domain adopts a compact pseudoknot structure while the SD-sequestering hairpin in the expression platform unfolds and frees the SD sequence for ribosome binding [78]. In addition, the ligand-bound pseudoknot in the aptamer presents a significant impediment to scanning by RNase E and protects downstream cleavage sites from the nuclease access. As a result, stability of the sugE mRNA increases. Since RNase E activity does not dependent on the sequence context, this mechanism could be common to many systems employing riboswitches, whose ligand-bound aptamers interfere with scanning.
PROSPECTIVES
Relatively simple riboswitch mechanisms discussed in this review do not provide a full picture of the multitude of mechanisms used by riboswitches. Riboswitch-mediated control can involve extra layers of sensing [95] or represent more sophisticated multi-component regulatory responses involving non-coding RNA and proteins [96, 97]. Given that bioinformatics searches have not identified expression platforms for many putative riboswitches, we can expect more riboswitch mechanisms to be discovered in future. Could these new mechanisms involve Rho-dependent transcription termination, RNA degradation, or other means? New methodologies, such as RNA-Seq, have already identified multiple small RNAs potentially associated with riboswitches [98-100] and one can expect a flow of studies deciphering sRNA-related riboswitch mechanisms. Identification of cognate ligands for many “orphan” riboswitches and discovery of new riboswitch classes will also enrich the mechanistic repertoire of riboswitches. These studies will further help us understand regulatory responses and major players involved in metabolic pathways and will provide the molecular basis for interfering with the most critical cellular processes in pathogenic bacteria.
Acknowledgments. This work was supported by the Ramon Areces Foundation grant (A. A.-M.) and the National Institute of Health grant 2R01GM112940 (A. S.).
Ethic declarations. The authors declare no conflict of interest in financial or any other sphere. This article does not contain any studies with human participants or animals performed by any of the authors.
REFERENCES
1.McNerney, M. P., and Styczynski, M. P. (2018) Small
molecule signaling, regulation, and potential applications in cellular
therapeutics, Wiley Interdiscip. Rev. Syst. Biol. Med.,
10, doi: 10.1002/wsbm.1405.
2.Hauryliuk, V., Atkinson, G. C., Murakami, K. S.,
Tenson, T., and Gerdes, K. (2015) Recent functional insights into the
role of (p)ppGpp in bacterial physiology, Nat. Rev. Microbiol.,
13, 298-309, doi: 10.1038/nrmicro3448.
3.Zimmermann, G. R., Jenison, R. D., Wick, C. L.,
Simorre, J. P., and Pardi, A. (1997) Interlocking structural motifs
mediate molecular discrimination by a theophylline-binding RNA, Nat.
Struct. Biol., 4, 644-649, doi: 10.1038/nsb0897-644.
4.Nahvi, A., Sudarsan, N., Ebert, M. S., Zou, X.,
Brown, K. L., and Breaker, R. R. (2002) Genetic control by a metabolite
binding mRNA, Chem. Biol., 9, 1043, doi:
10.1016/s1074-5521(02)00224-7.
5.Winkler, W., Nahvi, A., and Breaker, R. R. (2002)
Thiamine derivatives bind messenger RNAs directly to regulate bacterial
gene expression, Nature, 419, 952-956, doi:
10.1038/nature01145.
6.Mironov, A. S., Gusarov, I., Rafikov, R., Lopez, L.
E., Shatalin, K., et al. (2002) Sensing small molecules by nascent RNA:
a mechanism to control transcription in bacteria, Cell,
111, 747-756, doi: 10.1016/s0092-8674(02)01134-0.
7.Nudler, E., and Mironov, A. S. (2004) The
riboswitch control of bacterial metabolism, Trends Biochem.
Sci., 29, 11-17, doi: 10.1016/j.tibs.2003.11.004.
8.Pavlova, N., Kaloudas, D., and Penchovsky, R.
(2019) Riboswitch distribution, structure, and function in bacteria,
Gene, 708, 38-48, doi: 10.1016/j.gene.2019.05.036.
9.Serganov, A., and Patel, D. J. (2012) Metabolite
recognition principles and molecular mechanisms underlying riboswitch
function, Annu. Rev. Biophys., 41, 343-370, doi:
10.1146/annurev-biophys-101211-113224.
10.Nomura, M., Gourse, R., and Baughman, G. (1984)
Regulation of the synthesis of ribosomes and ribosomal components,
Annu. Rev. Biochem., 53, 75-117, doi:
10.1146/annurev.bi.53.070184.000451.
11.Serganov, A., Ennifar, E., Portier, C.,
Ehresmann, B., and Ehresmann, C. (2002) Do mRNA and rRNA binding sites
of E. coli ribosomal protein S15 share common structural
determinants? J. Mol. Biol., 320, 963-978, doi:
10.1016/s0022-2836(02)00553-3.
12.Cech, T. R., Zaug, A. J., and Grabowski, P. J.
(1981) In vitro splicing of the ribosomal RNA precursor of
Tetrahymena: involvement of a guanosine nucleotide in the
excision of the intervening sequence, Cell, 27, 487-496,
doi: 10.1016/0092-8674(81)90390-1.
13.Ellington, A. D., and Szostak, J. W. (1990) In
vitro selection of RNA molecules that bind specific ligands,
Nature, 346, 818-822, doi: 10.1038/346818a0.
14.Robertson, D. L., and Joyce, G. F. (1990)
Selection in vitro of an RNA enzyme that specifically cleaves
single-stranded DNA, Nature, 344, 467-468, doi:
10.1038/344467a0.
15.Tuerk, C., and Gold, L. (1990) Systematic
evolution of ligands by exponential enrichment: RNA ligands to
bacteriophage T4 DNA polymerase, Science, 249, 505-510,
doi: 10.1126/science.2200121.
16.Yanofsky, C. (1981) Attenuation in the control of
expression of bacterial operons, Nature, 289, 751-758,
doi: 10.1038/289751a0.
17.Miranda-Rios, J., Navarro, M., and Soberon, M.
(2001) A conserved RNA structure (thi box) is involved in
regulation of thiamin biosynthetic gene expression in bacteria,
Proc. Natl. Acad. Sci. USA, 98, 9736-9741, doi:
10.1073/pnas.161168098.
18.Nou, X., and Kadner, R. J. (1998) Coupled changes
in translation and transcription during cobalamin-dependent regulation
of btuB expression in Escherichia coli, J.
Bacteriol., 180, 6719-6728, doi:
10.1128/JB.180.24.6719-6728.1998.
19.Gelfand, M. S., Mironov, A. A., Jomantas, J.,
Kozlov, Y. I., and Perumov, D. A. (1999) A conserved RNA structure
element involved in the regulation of bacterial riboflavin synthesis
genes, Trends Genet., 15, 439-442, doi:
10.1016/s0168-9525(99)01856-9.
20.Perkins, J. B., and Pero, J. (2002) Biosynthesis
of riboflavin, biotin, folic acid, and cobalamin, in Bacillus
subtillis and Its Closest Relatives: from Genes to Cells
(Sonenshein, A. L., Hoch, J. A., and Losick, R., eds.) ASM Press,
Washington, pp. 271-286.
21.Stormo, G. D., and Ji, Y. (2001) Do mRNAs act as
direct sensors of small molecules to control their expression? Proc.
Natl. Acad. Sci. USA, 98, 9465-9467, doi:
10.1073/pnas.181334498.
22.Ravnum, S., and Andersson, D. I. (2001) An
adenosyl-cobalamin (coenzyme-B12)-repressed translational enhancer in
the cob mRNA of Salmonella typhimurium, Mol.
Microbiol., 39, 1585-1594, doi:
10.1046/j.1365-2958.2001.02346.x.
23.Nou, X., and Kadner, R. J. (2000)
Adenosylcobalamin inhibits ribosome binding to btuB RNA, Proc. Natl.
Acad. Sci. USA, 97, 7190-7195, doi:
10.1073/pnas.130013897.
24.Sudarsan, N., Barrick, J. E., and Breaker, R. R.
(2003) Metabolite-binding RNA domains are present in the genes of
eukaryotes, RNA, 9, 644-647, doi:
10.1261/rna.5090103.
25.Cheah, M. T., Wachter, A., Sudarsan, N., and
Breaker, R. R. (2007) Control of alternative RNA splicing and gene
expression by eukaryotic riboswitches, Nature, 447,
497-500, doi: 10.1038/nature05769.
26.Bocobza, S. E., and Aharoni, A. (2008) Switching
the light on plant riboswitches, Trends Plant Sci., 13,
526-533, doi: 10.1016/j.tplants.2008.07.004.
27.Wickiser, J. K., Winkler, W. C., Breaker, R. R.,
and Crothers, D. M. (2005) The speed of RNA transcription and
metabolite binding kinetics operate an FMN riboswitch, Mol.
Cell., 18, 49-60, doi: 10.1016/j.molcel.2005.02.032.
28.Smith, A. M., Fuchs, R. T., Grundy, F. J., and
Henkin, T. M. (2010) The SAM-responsive SMK box is a
reversible riboswitch, Mol. Microbiol., 78, 1393-1402,
doi: 10.1111/j.1365-2958.2010.07410.x.
29.Fuchs, R. T., Grundy, F. J., and Henkin, T. M.
(2006) The S(MK) box is a new SAM-binding RNA for translational
regulation of SAM synthetase, Nat. Struct. Mol. Biol.,
13, 226-233, doi: 10.1038/nsmb1059.
30.Lu, C., Smith, A. M., Fuchs, R. T., Ding, F.,
Rajashankar, K., et al. (2008) Crystal structures of the
SAM-III/SMK riboswitch reveal the SAM-dependent translation
inhibition mechanism, Nat. Struct. Mol. Biol., 15,
1076-1083, doi: 10.1038/nsmb.1494.
31.Gilbert, S. D., Rambo, R. P., Van Tyne, D., and
Batey, R. T. (2008) Structure of the SAM-II riboswitch bound to
S-adenosylmethionine, Nat. Struct. Mol. Biol., 15,
177-182, doi: 10.1038/nsmb.1371.
32.Corbino, K. A., Barrick, J. E., Lim, J., Welz,
R., Tucker, B. J., et al. (2005) Evidence for a second class of
S-adenosylmethionine riboswitches and other regulatory RNA motifs in
alpha-proteobacteria, Genome Biol., 6, R70, doi:
10.1186/gb-2005-6-8-r70.
33.Panchapakesan, S. S. S., Corey, L., Malkowski, S.
N., Higgs, G., and Breaker, R. R. (2021) A second riboswitch class for
the enzyme cofactor NAD+, RNA, 27, 99-105,
doi: 10.1261/rna.077891.120.
34.Salvail, H., Balaji, A., Yu, D., Roth, A., and
Breaker, R. R. (2020) Biochemical validation of a fourth guanidine
riboswitch class in Bacteria, Biochemistry, 59,
4654-4662, doi: 10.1021/acs.biochem.0c00793.
35.Atilho, R. M., Mirihana Arachchilage, G.,
Greenlee, E. B., Knecht, K. M., and Breaker, R. R. (2019) A bacterial
riboswitch class for the thiamin precursor HMP-PP employs a
terminator-embedded aptamer, Elife, 8, doi:
10.7554/eLife.45210.
36.Hollands, K., Proshkin, S., Sklyarova, S.,
Epshtein, V., Mironov, A., et al. (2012) Riboswitch control of
Rho-dependent transcription termination, Proc. Natl. Acad. Sci.
USA, 109, 5376-5381, doi: 10.1073/pnas.1112211109.
37.Peters, J. M., Mooney, R. A., Kuan, P. F.,
Rowland, J. L., Keles, S., and Landick, R. (2009) Rho directs
widespread termination of intragenic and stable RNA transcription,
Proc. Natl. Acad. Sci. USA, 106, 15406-15411, doi:
10.1073/pnas.0903846106.
38.Dann, C. E., 3rd, Wakeman, C. A., Sieling, C. L.,
Baker, S. C., Irnov, I., and Winkler, W. C. (2007) Structure and
mechanism of a metal-sensing regulatory RNA, Cell, 130,
878-892, doi: 10.1016/j.cell.2007.06.051.
39.Wakeman, C. A., Ramesh, A., and Winkler, W. C.
(2009) Multiple metal-binding cores are required for metalloregulation
by M-box riboswitch RNAs, J. Mol. Biol., 392, 723-735,
doi: 10.1016/j.jmb.2009.07.033.
40.Furukawa, K., Ramesh, A., Zhou, Z., Weinberg, Z.,
Vallery, T., et al. (2015) Bacterial riboswitches cooperatively bind
Ni2+ or Co2+ ions and control expression of heavy
metal transporters, Mol. Cell, 57, 1088-1098, doi:
10.1016/j.molcel.2015.02.009.
41.Xu, J., and Cotruvo, J. A., Jr. (2020) The czcD
(NiCo) riboswitch responds to Iron(II), Biochemistry, 59,
1508-1516, doi: 10.1021/acs.biochem.0c00074.
42.Price, I. R., Gaballa, A., Ding, F., Helmann, J.
D., and Ke, A. (2015) Mn2+-sensing mechanisms of
yybP-ykoY orphan riboswitches, Mol. Cell,
57, 1110-1123, doi: 10.1016/j.molcel.2015.02.016.
43.Trausch, J. J., Ceres, P., Reyes, F. E., and
Batey, R. T. (2011) The structure of a tetrahydrofolate-sensing
riboswitch reveals two ligand binding sites in a single aptamer,
Structure, 19, 1413-1423, doi:
10.1016/j.str.2011.06.019.
44.Gao, A., and Serganov, A. (2014) Structural
insights into recognition of c-di-AMP by the ydaO riboswitch,
Nat. Chem. Biol., 10, 787-792, doi:
10.1038/nchembio.1607.
45.Ren, A., and Patel, D. J. (2014) c-di-AMP binds
the ydaO riboswitch in two pseudo-symmetry-related pockets,
Nat. Chem. Biol., 10, 780-786, doi:
10.1038/nchembio.1606.
46.Serganov, A., Polonskaia, A., Phan, A. T.,
Breaker, R. R., and Patel, D. J. (2006) Structural basis for gene
regulation by a thiamine pyrophosphate-sensing riboswitch,
Nature, 441, 1167-1171, doi: 10.1038/nature04740.
47.Serganov, A., Huang, L., and Patel, D. J. (2009)
Coenzyme recognition and gene regulation by a flavin mononucleotide
riboswitch, Nature, 458, 233-237, doi:
10.1038/nature07642.
48.Edwards, T. E., and Ferre-D’Amare, A. R.
(2006) Crystal structures of the thi-box riboswitch bound to thiamine
pyrophosphate analogs reveal adaptive RNA-small molecule recognition,
Structure, 14, 1459-1468, doi:
10.1016/j.str.2006.07.008.
49.Ren, A., Rajashankar, K. R., and Patel, D. J.
(2012) Fluoride ion encapsulation by Mg2+ ions and
phosphates in a fluoride riboswitch, Nature, 486, 85-89,
doi: 10.1038/nature11152.
50.Serganov, A., Huang, L., and Patel, D. J. (2008)
Structural insights into amino acid binding and gene control by a
lysine riboswitch, Nature, 455, 1263-1267, doi:
10.1038/nature07326.
51.Huang, L., Serganov, A., and Patel, D. J. (2010)
Structural insights into ligand recognition by a sensing domain of the
cooperative glycine riboswitch, Mol. Cell, 40, 774-786,
doi: 10.1016/j.molcel.2010.11.026.
52.McCluskey, K., Boudreault, J., St-Pierre, P.,
Perez-Gonzalez, C., Chauvier, A., et al. (2019) Unprecedented
tunability of riboswitch structure and regulatory function by
sub-millimolar variations in physiological Mg2+, Nucleic
Acids Res., 47, 6478-6487, doi: 10.1093/nar/gkz316.
53.Watson, P. Y., and Fedor, M. J. (2011) The
glmS riboswitch integrates signals from activating and
inhibitory metabolites in vivo, Nat. Struct. Mol. Biol.,
18, 359-363, doi: 10.1038/nsmb.1989.
54.Mandal, M., Lee, M., Barrick, J. E., Weinberg,
Z., Emilsson, G. M., et al. (2004) A glycine-dependent riboswitch that
uses cooperative binding to control gene expression, Science,
306, 275-279, doi: 10.1126/science.1100829.
55.Butler, E. B., Xiong, Y., Wang, J., and Strobel,
S. A. (2011) Structural basis of cooperative ligand binding by the
glycine riboswitch, Chem. Biol., 18, 293-298, doi:
10.1016/j.chembiol.2011.01.013.
56.Sherman, E. M., Esquiaqui, J., Elsayed, G., and
Ye, J. D. (2012) An energetically beneficial leader-linker interaction
abolishes ligand-binding cooperativity in glycine riboswitches,
RNA, 18, 496-507, doi: 10.1261/rna.031286.111.
57.Torgerson, C. D., Hiller, D. A., and Strobel, S.
A. (2020) The asymmetry and cooperativity of tandem glycine riboswitch
aptamers, RNA, 26, 564-580, doi:
10.1261/rna.073577.119.
58.Ames, T. D., and Breaker, R. R. (2011) Bacterial
aptamers that selectively bind glutamine, RNA Biol., 8,
82-89, doi: 10.4161/rna.8.1.13864.
59.Ren, A., Xue, Y., Peselis, A., Serganov, A.,
Al-Hashimi, H. M., and Patel, D. J. (2015) Structural and dynamic basis
for low-affinity, high-selectivity binding of L-glutamine by the
glutamine riboswitch, Cell Rep., 13, 1800-1813, doi:
10.1016/j.celrep.2015.10.062.
60.Sherlock, M. E., Malkowski, S. N., and Breaker,
R. R. (2017) Biochemical validation of a second guanidine riboswitch
class in Bacteria, Biochemistry, 56, 352-358, doi:
10.1021/acs.biochem.6b01270.
61.Huang, L., Wang, J., and Lilley, D. M. J. (2017)
The structure of the guanidine-II riboswitch, Cell. Chem. Biol.,
24, 695-702 e692, doi: 10.1016/j.chembiol.2017.05.014.
62.Reiss, C. W., and Strobel, S. A. (2017)
Structural basis for ligand binding to the guanidine-II riboswitch,
RNA, 23, 1338-1343, doi: 10.1261/rna.061804.117.
63.Wuebben, C., Vicino, M. F., Mueller, M., and
Schiemann, O. (2020) Do the P1 and P2 hairpins of the Guanidine-II
riboswitch interact? Nucleic Acids Res., 48, 10518-10526,
doi: 10.1093/nar/gkaa703.
64.Malkowski, S. N., Spencer, T. C. J., and Breaker,
R. R. (2019) Evidence that the nadA motif is a bacterial
riboswitch for the ubiquitous enzyme cofactor NAD+,
RNA, 25, 1616-1627, doi: 10.1261/rna.072538.119.
65.Huang, L., Wang, J., and Lilley, D. M. J. (2020)
Structure and ligand binding of the ADP-binding domain of the
NAD+ riboswitch, RNA, 26, 878-887, doi:
10.1261/rna.074898.120.
66.Chen, H., Egger, M., Xu, X., Flemmich, L.,
Krasheninina, O., et al. (2020) Structural distinctions between
NAD+ riboswitch domains 1 and 2 determine differential
folding and ligand binding, Nucleic Acids Res., 48,
12394-12406, doi: 10.1093/nar/gkaa1029.
67.Tang, D. J., Du, X., Shi, Q., Zhang, J. L., He,
Y. P., et al. (2020) A SAM-I riboswitch with the ability to sense and
respond to uncharged initiator tRNA, Nat. Commun., 11,
2794, doi: 10.1038/s41467-020-16417-z.
68.Sherlock, M. E., Sudarsan, N., Stav, S., and
Breaker, R. R. (2018) Tandem riboswitches form a natural Boolean logic
gate to control purine metabolism in bacteria, Elife, 7,
doi: 10.7554/eLife.33908.
69.Peselis, A., and Serganov, A. (2018) ykkC
riboswitches employ an add-on helix to adjust specificity for
polyanionic ligands, Nat. Chem. Biol., 14, 887-894, doi:
10.1038/s41589-018-0114-4.
70.Knappenberger, A. J., Reiss, C. W., and Strobel,
S. A. (2018) Structures of two aptamers with differing ligand
specificity reveal ruggedness in the functional landscape of RNA,
Elife, 7, doi: 10.7554/eLife.36381.
71.Lee, E. R., Baker, J. L., Weinberg, Z., Sudarsan,
N., and Breaker, R. R. (2010) An allosteric self-splicing ribozyme
triggered by a bacterial second messenger, Science, 329,
845-848, doi: 10.1126/science.1190713.
72.Chen, A. G., Sudarsan, N., and Breaker, R. R.
(2011) Mechanism for gene control by a natural allosteric group I
ribozyme, RNA, 17, 1967-1972, doi:
10.1261/rna.2757311.
73.Sudarsan, N., Hammond, M. C., Block, K. F., Welz,
R., Barrick, J. E., et al. (2006) Tandem riboswitch architectures
exhibit complex gene control functions, Science, 314,
300-304, doi: 10.1126/science.1130716.
74.Stoddard, C. D., and Batey, R. T. (2006)
Mix-and-match riboswitches, ACS Chem. Biol., 1, 751-754,
doi: 10.1021/cb600458w.
75.Stav, S., Atilho, R. M., Mirihana Arachchilage,
G., Nguyen, G., Higgs, G., and Breaker, R. R. (2019) Genome-wide
discovery of structured noncoding RNAs in bacteria, BMC
Microbiol., 19, 66, doi: 10.1186/s12866-019-1433-7.
76.Hui, M. P., Foley, P. L., and Belasco, J. G.
(2014) Messenger RNA degradation in bacterial cells, Annu. Rev.
Genet., 48, 537-559, doi:
10.1146/annurev-genet-120213-092340.
77.Richards, J., and Belasco, J. G. (2021)
Riboswitch control of bacterial RNA stability, Mol. Microbiol.,
doi: 10.1111/mmi.14723.
78.Richards, J., and Belasco, J. G. (2021)
Widespread protection of RNA cleavage sites by a riboswitch aptamer
that folds as a compact obstacle to scanning by RNase E, Mol.
Cell, 81, 127-138.e124, doi:
10.1016/j.molcel.2020.10.025.
79.Shahbabian, K., Jamalli, A., Zig, L., and Putzer,
H. (2009) RNase Y, a novel endoribonuclease, initiates riboswitch
turnover in Bacillus subtilis, EMBO J., 28,
3523-3533, doi: 10.1038/emboj.2009.283.
80.Winkler, W. C., Nahvi, A., Roth, A., Collins, J.
A., and Breaker, R. R. (2004) Control of gene expression by a natural
metabolite-responsive ribozyme, Nature, 428, 281-286,
doi: 10.1038/nature02362.
81.Collins, J. A., Irnov, I., Baker, S., and
Winkler, W. C. (2007) Mechanism of mRNA destabilization by the
glmS ribozyme, Genes Dev., 21, 3356-3368, doi:
10.1101/gad.1605307.
82.Klein, D. J., and Ferre-D’Amare, A. R.
(2006) Structural basis of glmS ribozyme activation by
glucosamine-6-phosphate, Science, 313, 1752-1756, doi:
10.1126/science.1129666.
83.Cochrane, J. C., Lipchock, S. V., and Strobel, S.
A. (2007) Structural investigation of the GlmS ribozyme bound to its
catalytic cofactor, Chem. Biol., 14, 97-105, doi:
10.1016/j.chembiol.2006.12.005.
84.Davis, J. H., Dunican, B. F., and Strobel, S. A.
(2011) glmS Riboswitch binding to the glucosamine-6-phosphate
alpha-anomer shifts the pKa toward neutrality,
Biochemistry, 50, 7236-7242, doi: 10.1021/bi200471c.
85.Caron, M. P., Bastet, L., Lussier, A.,
Simoneau-Roy, M., Masse, E., and Lafontaine, D. A. (2012) Dual-acting
riboswitch control of translation initiation and mRNA decay, Proc.
Natl. Acad. Sci. USA, 109, E3444-3453, doi:
10.1073/pnas.1214024109.
86.Takemoto, N., Tanaka, Y., and Inui, M. (2015) Rho
and RNase play a central role in FMN riboswitch regulation in
Corynebacterium glutamicum, Nucleic Acids Res.,
43, 520-529, doi: 10.1093/nar/gku1281.
87.Pursley, B. R., Fernandez, N. L., Severin, G. B.,
and Waters, C. M. (2019) The Vc2 cyclic di-GMP-dependent riboswitch of
Vibrio cholerae regulates expression of an upstream putative
small RNA by controlling RNA stability, J. Bacteriol.,
201, doi: 10.1128/JB.00293-19.
88.Pursley, B. R., Maiden, M. M., Hsieh, M. L.,
Fernandez, N. L., Severin, G. B., and Waters, C. M. (2018) Cyclic
di-GMP regulates TfoY in Vibrio cholerae to control motility by
both transcriptional and posttranscriptional mechanisms, J.
Bacteriol., 200, doi: 10.1128/JB.00578-17.
89.Inuzuka, S., Nishimura, K., Kakizawa, H., Fujita,
Y., Furuta, H., et al. (2016) Mutational analysis of structural
elements in a class-I cyclic di-GMP riboswitch to elucidate its
regulatory mechanism, J. Biochem., 160, 153-162, doi:
10.1093/jb/mvw026.
90.Inuzuka, S., Kakizawa, H., Nishimura, K. I.,
Naito, T., Miyazaki, K., et al. (2018) Recognition of cyclic-di-GMP by
a riboswitch conducts translational repression through masking the
ribosome-binding site distant from the aptamer domain, Genes
Cells, 23, 435-447, doi: 10.1111/gtc.12586.
91.Kaberdin, V. R. (2003) Probing the substrate
specificity of Escherichia coli RNase E using a novel
oligonucleotide-based assay, Nucleic Acids Res., 31,
4710-4716, doi: 10.1093/nar/gkg690.
92.Mackie, G. A. (1998) Ribonuclease E is a
5′-end-dependent endonuclease, Nature, 395,
720-723, doi: 10.1038/27246.
93.Callaghan, A. J., Marcaida, M. J., Stead, J. A.,
McDowall, K. J., Scott, W. G., and Luisi, B. F. (2005) Structure of
Escherichia coli RNase E catalytic domain and implications for
RNA turnover, Nature, 437, 1187-1191, doi:
10.1038/nature04084.
94.Richards, J., and Belasco, J. G. (2019) Obstacles
to scanning by RNase E govern bacterial mRNA lifetimes by hindering
access to distal cleavage sites, Mol. Cell, 74, 284-295
e285, doi: 10.1016/j.molcel.2019.01.044.
95.Reining, A., Nozinovic, S., Schlepckow, K., Buhr,
F., Furtig, B., and Schwalbe, H. (2013) Three-state mechanism couples
ligand and temperature sensing in riboswitches, Nature,
499, 355-359, doi: 10.1038/nature12378.
96.Mellin, J. R., Koutero, M., Dar, D., Nahori, M.
A., Sorek, R., and Cossart, P. (2014) Sequestration of a two-component
response regulator by a riboswitch-regulated noncoding RNA,
Science, 345, 940-943, doi: 10.1126/science.1255083.
97.DebRoy, S., Gebbie, M., Ramesh, A., Goodson, J.
R., Cruz, M. R., et al. (2014) A riboswitch-containing sRNA controls
gene expression by sequestration of a response regulator,
Science, 345, 937-940, doi: 10.1126/science.1255091.
98.Karunker, I., Rotem, O., Dori-Bachash, M.,
Jurkevitch, E., and Sorek, R. (2013) A global transcriptional switch
between the attack and growth forms of Bdellovibrio
bacteriovorus, PLoS One, 8, e61850, doi:
10.1371/journal.pone.0061850.
99.Weber, L., Thoelken, C., Volk, M., Remes, B.,
Lechner, M., and Klug, G. (2016) The conserved Dcw gene cluster of
R. sphaeroides is preceded by an uncommonly extended 5′
leader featuring the sRNA UpsM, PLoS One, 11, e0165694,
doi: 10.1371/journal.pone.0165694.
100.Soutourina, O. A., Monot, M., Boudry, P.,
Saujet, L., Pichon, C., et al. (2013) Genome-wide identification of
regulatory RNAs in the human pathogen Clostridium difficile,
PLoS Genet., 9, e1003493, doi: 10.1371/journal.pgen.
1003493.
101.Haller, A., Rieder, U., Aigner, M., Blanchard,
S. C., and Micura, R. (2011) Conformational capture of the SAM-II
riboswitch, Nat. Chem. Biol., 7, 393-400, doi:
10.1038/nchembio.562.
102.Nelson, J. W., Sudarsan, N., Furukawa, K.,
Weinberg, Z., Wang, J. X., and Breaker, R. R. (2013) Riboswitches in
eubacteria sense the second messenger c-di-AMP, Nat. Chem.
Biol., 9, 834-839, doi: 10.1038/nchembio.1363.