REVIEW: Encapsulins: Structure, Properties, and Biotechnological Applications
Nelly S. Chmelyuk1,2, Vera V. Oda1, Anna N. Gabashvili1, and Maxim A. Abakumov1,2,a*
1National University of Science and Technology “MISIS”, 119049 Moscow, Russia2Pirogov Russian National Research Medical University, Ministry of Health of the Russian Federation, 117977 Moscow, Russia
* To whom correspondence should be addressed.
Received September 16, 2022; Revised November 4, 2022; Accepted November 7, 2022
In 1994 a new class of prokaryotic compartments was discovered, collectively called “encapsulins” or “nanocompartments”. Encapsulin shell protomer proteins self-assemble to form icosahedral structures of various diameters (24-42 nm). Inside of nanocompartments shells, one or several cargo proteins, diverse in their functions, can be encapsulated. In addition, non-native cargo proteins can be loaded into nanocompartments, and shell surfaces can be modified via various compounds, which makes it possible to create targeted drug delivery systems, labels for optical and MRI imaging, and to use encapsulins as bioreactors. This review describes a number of strategies of encapsulins application in various fields of science, including biomedicine and nanobiotechnologies.
KEY WORDS: encapsulins, self-organization, nanocontainer systems, targeted delivery, genetic tags, cell trackingDOI: 10.1134/S0006297923010042
Abbreviations: CLP, cargo-loading peptide; DyP, dye de-coloring peroxidase; eMIONs, encapsulin-produced magnetic iron oxide nanoparticles; FLP, ferritin-like protein; miniSOG, mini singlet oxygen generator; OVA, ovalbumin; T, triangulation number.
INTRODUCTION
It is known that homeostasis in mammals is maintained via different mechanisms: for example, iron transport in cells is performed by transferrins, and its accumulation including defense against oxidative stress occurs with the help of ferritins; Na/K-ATPase controls amount of sodium and potassium ions in the cell. Practically all reactions in live organisms occur with participation of various enzymes, which could have metal ions in their structure such as Fe, Zn, Cu, and others, however, content of these metals in organism is rather low, and their large excess or deficiency under changing external factors could lead to serious, often irreversible consequences. The main depot for the excess of salts are blood and liver. Prokaryotic cells usually lack membrane compartments typical for eukaryotes, but instead contain numerous protein compartments that are capable of accumulating large number of molecules. For the first time protein nanocompartments, later named encapsulins, were found in 1994 in the supernatant fluid of the Brevibacterium linens culture, which exhibited bacteriostatic activity against different strains of Arthrobacter, Bacillus, Brevibacterium, Corynebacterium, and Listeria [1]. Later such protein complexes were detected in supernatants of Mycobacterium tuberculosis (Myc. tuberculosis) [2] and Thermotoga maritima [3] cultures; moreover, it was found that these structures contain proteolytic enzymes. In a number of studies encapsulins were found in bacteria Mycobacterium leprae, Streptomyces, and later – in Quasibacillus thermotolerans [4-9]. However, proteolytic activity was not detected in the later studies, and, at present, it is generally recognized that encapsulins play mainly structural function [10]. It was established in mid-2000s that the observed structures with high molecular mass comprise capsid-like protein complexes [10-12].
Over time, studies of nanocompartments have been continued. Part of the studies were devoted to the search of new protein nanocompartments and their properties, as well as to elucidation of physiological role of encapsulins and their protein cargo in the natural bacterial context. Another group of the studies were devoted to application of such structures as bioreactors, drug-delivery systems, as well as endogenous labels [13-20]. Bioinformatic analysis of the sequenced genomes allowed identifying thousands of nanocompartment structures both in bacteria and in archaea, which are loaded with a large variety of cargo proteins [19, 21-24]. Later, based on the conducted studies the database of bacterial nanocompartments was constructed that included also description and various characteristics of encapsulins [25].
STRUCTURE AND PROPERTIES OF THE NANOCOMPARTMENT SHELLS
Encapsulin shells comprise icosahedral complexes (12 vertices, 20 faces, 30 edges) formed as a result of self-assembly of protomers, which are proteins structurally homologous to gp5 – major capsid protein of the HK97 bacteriophage [26]. Similar to the case of viral capsids, proteins of the encapsulin shells assemble into icosahedra of different size. At present, three different types of encapsulins are known, which are characterized with the help of triangulation number (T) defined as a quotient obtained as a result of division of the number of protomers in the encapsulin shell by 60 [27]. For example, encapsulins with T = 1 are T. maritima [10], Myc. tuberculosis [22], and Rhodococcus jostii [21] encapsulins (shell diameter is 20-24 nm, it contains 60 identical protomer subunits). Nanocompartments from the bacteria Pyrococcus furiosus [12], Kuenenia stuttgartiensis [28, 29], and Myxococcus xanthus (Myx. xanthus) [23] are examples of encapsulins with T = 3 (shell diameter is 30-32 nm containing 180 protomers). And finally, the largest encapsulins found so far are encapsulins from bacteria Q. thermotolerans with T = 4 (shell diameter is 42 nm containing 240 protomers) [30].
There are three conserved domains in the structure of encapsulin shell protomer, similar to the capsomere gp5 of the HK97 bacteriophage. The first one is a peripheral domain (P) containing several α-helices and β-pleated sheets, the second one – an axial domain (A), which also contains several α-helices and β-pleated sheets and forms a 5-fold symmetry axis, and, finally, the third one – an elongated loop (E), which is important for encapsulin assembly and defines symmetry of the entire complex [4, 10, 13, 27, 30]. Structural homology between the capsomere of HK97 bacteriophage (Fig. 1a) and protomer of the T. maritima encapsulin shell (Fig. 1b) can be clearly seen in Fig. 1.
Fig. 1. a) Structure of the major capsid protein of HK97 bacteriophage. Capsid has diameter of 65 nm and consists of 420 capsomer proteins (PDB-ID 2FT1). Below the structure of a single capsomere is presented (gp5). b) Structure of the encapsulin shell form the T. maritima bacteria. T. maritima encapsulin is formed by 60 identical protomer proteins, diameter of the encapsulin shell is 24 nm (PDB-ID 3DKT). Below the structure of protomer protein of the T. maritima encapsulin is presented. Three conserved domains are present both in the structure of the encapsulin protomer protein and in the structure of gp5 capsomer: peripheral domain (P), axial domain (A), and elongated loop (E).
Despite high structural homology between the encapsulin protomers and capsomers of the HK97 bacteriophage, amino acid homology between these proteins is low. Hence, according to classification of the Pfam protein families database, encapsulins and HK97 are the members of the same clan (CL0373), but belong to different individual families (encapsulins belong to the PF04454 family, and bacteriophage HK97 – to the PF05065 family) [31].
Multiple pores with diameter 3-7 Å exist at the junctions of the shell protomers [10, 12]. These pores are likely serve as a permeability barrier for larger molecules allowing transport of only small molecules and ions. For example, low molecular weight substrates of encapsulated enzymes, such as hydrogen peroxide and two-valent iron, are capable of penetration through the shell, while proteins or other large molecules – cannot [16, 17, 21, 24]. Cargo proteins with catalytic activity are present in encapsulins, which could indicate selectivity of the substrate transport through the shell [10], moreover, the transport selectivity would depend on the type of encapsulated enzyme [32]. It is assumed that the most probable transport channels are located along the pores formed at the sites where five protomers are connected (5-fold pores), because these pores are the largest [10, 30].
It was shown recently that the pores could differ in electrostatic properties. For example, the 5-fold pores in the T. maritima encapsulins contain five histidine residues and carry positive charge, and the pores at the sites of connection of three protomers contain three phenylalanine residues and do not carry any charge [33]. And all pores in the Q. thermotolerans encapsulins carry negative charge, which allows transport of the positively charged substrates [30]. Moreover, it is known that there are many negatively charged amino acid residues on the inner surface of encapsulin shells, which indicates existence of a certain pathway of iron ions to the ferritin-like protein (FLP) after they enter encapsulin [34].
Pores of the Synechococcus elongatus (Syn. elongatus) encapsulin differ from the pores in the T. maritima encapsulin, they have positively charged amino acid residues outside [32]. This, likely, represents certain adaptation allowing transport of the negatively charged deprotonated substrate (L-cysteine). Pores of the Mycobacterium smegmatis encapsulins also have positively charged histidines, which facilitates selective transport of the negatively charged substrates of the cargo protein – peroxidase enzyme [34].
Pores in the encapsulin shells could have not only different charges, but also different conformations. It was shown in an interesting study that the pores in the Haliangium ochraceum encapsulin could exist in two different conformations: open and closed [35]. This allows suggesting that pores could respond to external stimuli.
It is also known that the pore size could be increased artificially. In particular, the probability of increasing pore size in the shell of the T. maritima encapsulin from 3 to 11 Å, i.e., 3.7-fold, was reported in one of the studies [36]. The authors of the study investigated effects of amino acid substitutions and deletions in the pore-forming loop at the site of connection of five protomer proteins on the structural integrity of the encapsulin shell. This loop in the T. maritima encapsulin consists of 13 aa. The authors constructed several mutant strains of T. maritima with different substitutions/deletions of amino acids in this loop. According to the obtained data the mutant with deletion of 7 aa in the loop was the optimal one. Pore diameter in the shell of this encapsulin increased to 11 Å, while integrity of the shell was not affected.
CARGO PROTEINS IN ENCAPSULINS
For the first time the mechanism of encapsulation of a cargo protein was suggested during investigation of the crystal structure of T. maritima encapsulin. It was established using X-ray diffraction method that there is a small additional electron density in the hydrophobic pocket on the luminal surface of the encapsulin shell, which corresponds to the short (around 10 aa) C-terminal sequence of FLP, found close to the gene of the encapsulin gene in the T. maritima genome [10]. It was shown using bioinformatics approaches that this C-terminal sequence is conserved in different species of bacteria, in which the genes of cargo proteins and the genes of encapsulin shell are located close together in a particular operon. Examples of such “predicted” cargo proteins include: FLP, dye-decolorizing peroxidase (DyP), hemerythrin, and rubrerythrin [10, 19].
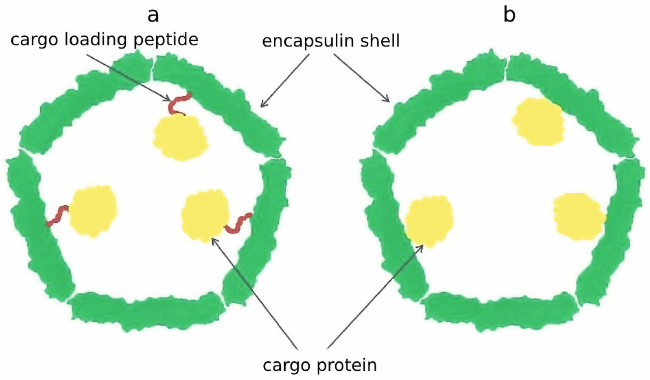
It was demonstrated that this C-terminal sequence, later termed cargo loading peptide (CLP), is sufficient for encapsulation of the cargo protein into the shell of a nanocompartment (Fig. 2a). The sequence encoding CLP could be located both at the 3′-end and at the 5′-end of the gene encoding cargo protein. Deletion of the CLP sequence disrupt encapsulation of the cargo protein, while fusion of CLP with the C-end of heterologous proteins such as green fluorescent protein or luciferase results in their loading [10, 14, 16, 17, 19, 22]. However, there are alternative models describing interaction between the cargo proteins and encapsulin shell proteins. In some cases, as in the case of P. furiosus encapsulin, CLP is absent, but the gene encoding shell protein is fused with the gene encoding cargo protein resulting in production of one polypeptide chain (Fig. 2b) [12]. Encapsulins found in Firmicute bacteria exhibit one particular feature: loading of cargo proteins can occur either through C-terminus or N-terminus of CLP. Such nanocompartment contains two cargo proteins: C-terminus of CLP is linked to the iron-mineralizing encapsulin-associated Firmicute (IMEF) protein participating in iron mineralization, and at the N-terminus of CLP there is an iron-sequestering ferredoxin [19].
Fig. 2. Cargo protein can be encapsulated into a nanocompartment shell via a cargo-loading peptide (a). In some cases, the gene encoding cargo protein and the gene encoding encapsulin shell are fused, which nullifies the need for loading peptide (b).
It is known that several cargo proteins could be encapsulated into one shell of a nanocompartment. For example, it was shown that the Myx. xanthus encapsulins contain three different cargo proteins: EncB, EncC, and EncD [23], and the Myc. tuberculosis encapsulins contain Mt-DyP, Mt-BfrB (bacterioferritin), and Mt-FolB (enzyme of folic acid synthesis) [22].
Another important issue associated with loading is the volume occupied by the cargo protein in the nanocompartment. Each protomer protein of the shell in encapsulin has the CLP binding site, but obviously, the cargo amount is limited by the shell volume. It is important to take into account that stoichiometry depends not only of the size of the encapsulin shell, but also on the state of oligomerization of the cargo protein(s). The modeling studies demonstrated that it is impossible to reach the ratio of cargo protein to protomer protein higher than 1 : 1 due to the steric hindrance [10]. The B. linens nanocompartment could be presented as such example that contains the previously mentioned DyP enzyme as a cargo, which is assembled into a hexamer (trimer of dimers) with diameter 89 Å. It was predicted that steric hindrance should limit load to one hexamer per nanocompartment [10], and measurements with the help of native mass spectrometry confirmed the presence of 6 DyP in the nanocompartment with T = 1, which corresponds to the ratio of cargo protein to protomer protein 1 : 10 [37]. The data on the load size of various cargo proteins in nanocompartments are presented in the table.
Load of native cargo proteins into different encapsulin systems

Notes. Fd, ferredoxin; CyD, cysteine desulfurase, EM, electron
microscopy, MS, mass spectrometry.
* Load determined as a ratio between the number of monomeric units of
the cargo protein and number of protomers in the encapsulin shell.
** Cargo load determined by disassembly and assembly of encapsulin shell
under acidic conditions.
PHYSIOLOGICAL FUNCTIONS OF NANOCOMPARTMENTS
Role of encapsulins in bacteria and archaea metabolism is poorly understood. At present the most informative results were obtained in investigation of encapsulins containing FLP. The available data allow suggesting that nanocompartments could accumulate iron, thus decreasing oxidative stress, which was demonstrated using Q. thermotolerans bacteria that do not have ferritins in their genome [27]. It was found in another study investigating Myx. xanthus cells under conditions of amino acid starvation that the expression of genes of the encapsulin shell protomers (EncA) and its three cargo FLP proteins (EncB, EncC, and EncD) was significantly upregulated [23]. The authors hypothesized that encapsulin could play a role of the secondary ferritin-like system with large capacity that is capable of accumulating iron under stress initiated by starvation or sequestering iron during oxidative stress. When bivalent iron (Fe2+) is subjected to the action of reactive oxygen species, Fenton reaction occurs and Fe2+ is oxidized to Fe3+ with formation of the side product – hydroxyl radical [43, 44]; ferritins protect cells against the toxic effects of this product. For example, it was demonstrated in the study by McHugh et al. [23] that the mutant strain of Myx. xanthus with deletion in the gene sequence encoding encapsulin shells was more sensitive to addition of hydrogen peroxide than the wild type Myx. xanthus. Viability of the mutant strain of Myx. xanthus under conditions of oxidative stress caused by incubation of bacteria with 0.5 mM solution of hydrogen peroxide was significantly lower than viability of the wild type Myx. xanthus (25% and 75% of viable bacteria, respectively). The same feature was observed in Myc. tuberculosis [22]: each of the three cargo proteins in Myc. tuberculosis encapsulins mentioned above (BfrB, FolB, and DyP) exhibit antioxidant activity [45-48].
In addition to their potential role in decreasing oxidative stress, DyP-containing encapsulins also participate in catabolism [21]. For example, the mutant strain of R. jostii RHA1 bacteria with deletion of the gene encoding DyP, is not capable of lignin degradation [49], while the wild type bacteria catabolize lignin very actively. It was also shown in the studies that activity of the encapsulin–DypB complex in degradation of nitrated lignin is 8-fold higher in comparison with the non-encapsulated DypB enzyme [21]. Increase of enzymatic activity upon encapsulation allows suggesting that nanocompartment could produce this effect either via stabilization of the cargo protein, or via increase of the local concentration of the enzyme substrate, which results in the increase of enzymatic activity [50].
For example, it has been assumed that encapsulation of such cargo protein as DyP is not required for performing its functions [10]. However, observations support the hypothesis that encapsulation allows enhancing stability and/or lifetime of cargo proteins through, for example, increasing their resistance to proteases. It was shown, for example, that T. maritima encapsulins are extremely resistant to the effects of high temperature and denaturing agents [10, 17], and B. linens encapsulins are stable in a wide range of pH [38]. Similar to the capsids of virus particles, nanocompartment shells demonstrate minimal degradation following treatment with non-specific proteases [16, 17]. Resistance to proteases also applies to cargo proteins, for example, firefly luciferase packed into Rhodococcus erythropolis N771 encapsulins was not degraded after exposure to trypsin, while the non-encapsulated luciferase was completely degraded [16].
Encapsulins are often found in supernatants of bacterial cultures [1, 2, 21], which led to suggestion that nanocompartments are products of bacterial secretion. This was partially confirmed while observing localization of encapsulins on the cell membranes [2], but such localization has not been observed in all prokaryotes, in particular, Streptomyces griseus nanocompartments are located in cytoplasm [9]. Moreover, no mechanism is known for secretion of intact 24-42-nm protein complex [51]. Considering extremely high chemical stability and resistance of encapsulins to proteases, it seems more logical to suggest that encapsulins are accumulated in supernatants of bacterial cultures after the cell lysis [52].
APPLICATIONS OF NANOCOMPARTMENTS IN BIOTECHNOLOGY
Summarizing all the above, it can be stated that encapsulins are special protein particles with two major advantageous features. Firstly, similar to all nanoparticles, encapsulins have very developed surface area (Ssp >> Vsp) and, most importantly, have two surfaces of their shell – inner and outer. Secondly, encapsulins are encoded in genome (i.e., they are produced via biosynthesis) and, hence, can be produced in biological systems with 100% repeatability, which is not achievable with any physical or chemical methods of synthesis of nanoparticles of any composition. Contrary to the chemical of physical synthesis, biosynthesis is more economical and, which no less important, is safe for environment as no toxic side products are formed during biosynthesis, which must be utilized with special procedures.
Encapsulins as a platform for drug delivery. It is known that during last decades numerous nanosize systems have been developed for targeted drug delivery based on micelles [53, 54], liposomes [55], inorganic [56] and polymeric [57] nanoparticles, as well as protein compartments [58, 59]. All these particles loaded with medicinal preparations are used to enhance efficiency of delivery, increase level of accumulation of preparations in comparison with non-encapsulated therapeutics, increase drug circulation time in blood, and decrease the number of side effects [60]. In addition, targeted delivery preparations could play a role in disease diagnostics through interaction with specific molecular targets expressed in one or another pathology. As has been mentioned above, encapsulins are very stable and sturdy structures, which facilitates their use as a platform for solving various tasks of biomedicine.
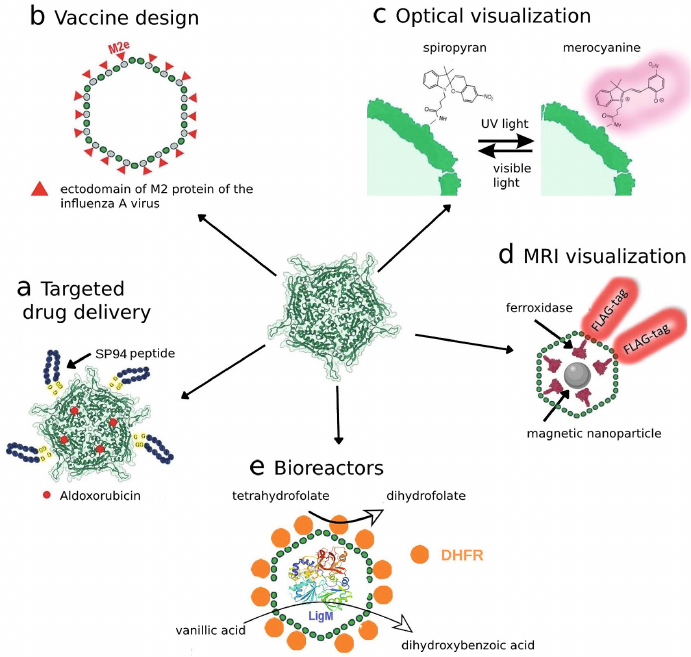
In particular, in one of the studies [13] the T. maritima encsapsulins were used for targeted delivery of fluorophore labels and therapeutics. The shells of nanocompartments were modified by addition of the SP94-peptide to it, which specifically binds to the 78-kDa glucose-regulated protein (GRP78) overexpressed in different tumor cells including the cells of HepG2 cell line [61, 62]. Shell protein of the T. maritima encapsulin (Encap) contains two cysteine residues (C123 and C197) with C123 located at the outer surface of Encap, which allows conjugation of the SP94 peptide and fluorescent probe (fluorescein) on the surface of the encapsulin shell. Hence, the produced “nanodevice” was able to specifically bind to HepG2 cells facilitating their visualization due to the presence of fluorophore (Fig. 3a). It was shown later that aldoxorubicin (6-maleimidocaproyl), which is pro-drug releasing doxorubicin at acidic pH inside tumor cells, can be loaded into this construct. Dose-dependent cytotoxic effect of this preparation on HepG2 cells was demonstrated.
Fig. 3. Use of encapsulins for different biotechnological tasks: a) nanocontainer systems for targeted drug delivery; b) new types of vaccines; c) photoswitchable labels for optical visualization; d) genetically encoded labels for MRI visualization; e) biosynthesis with the help of encapsulated enzymes.
It was shown in the study by Putri et al. [38] that B. linens encapsulins loaded with TFP (teal fluorescent protein,) were successfully captured by the mouse macrophages J774 in in vitro culture, which resulted in the cell fluorescence. In the process, encapsulins remained in the macrophage cytoplasm and were not found in the nucleus. The authors of the study note that the selected model cargo protein with fluorescent phenotype can be replaced with a therapeutic agent and encapsulin-based technology could be used for its delivery.
In one study [63] a biological photosensitizer miniSOG (mini singlet oxygen generator) was loaded into the T. maritima encapsulins by fusing CLP with the C-terminus of the miniSOG protein. The produced photosensitive nanoreactor was called Enc-mSOG. The authors tested formation of reactive oxygen species in the lung cancer cells A549 after illumination with blue light. Prior to that the cells were incubated with free miniSOG or Enc-mSOG for 7 h. Cells without added photosensitizer were used as a control. The study showed that the highest level of reactive oxygen species, release of which was induced by illumination with blue light, was observed in the cells incubated with Enc-mSOG [63]. The similar systems were produced for the targeted therapy of HER2-positive breast carcinoma [64]. In this study the authors also used the genetically encoded T. maritima encapsulin shell as a platform that contained inside it a miniSOG photosensitizer produced by fusion of the C-terminus of the protein with CLP. The encapsulin surface was saturated with the DARPin9.29 (Designed Ankyrin repeat protein) capable of selective binding to the receptor of epidermal growth factor overexpressed on the surface of breast carcinoma cells. DARPin9.29 was cloned into the reading frame of the T. maritima encapsulin gene in order to produce the fusion protein TmEnc-DARPin-STII. The obtained system demonstrated high activity towards the selected cancer model and low selectivity towards the control cells not expressing HER2n protein on the surface. The authors of this study showed that the modified encapsulins are capable of selective binding to the cells of SK-BR-3 cell line (human breast adenocarcinoma), be internalized and, hence, deliver the encapsulated form of miniSOG into the cells. Illumination of the SK-BR-3 with blue light resulted in the significant decrease of the cell viability (up to 48%) due to the action of miniSOG. Interestingly enough, the human mesenchymal stem cells used as a control in this study were not affected and retain their viability. The main advantage of this system is the possibility to construct effective encapsulin-based preparation using only one plasmid encoding all three components: carrier (encapsulin shell), delivery vector (DARPin9.29), and active ingredient (miniSOG).
The encapsulin-based delivery systems also have found application in the immunotherapy of cancer. For example, the T. maritima encapsulins were used in one of studies [18] as nanocontainers for the delivery of antigen (OT-1-peptide, epitope of ovalbumin protein, OVA) to the dendrite cells. Presentation of the OT-1-peptide by dendrite cells results in activation of the T-cell-mediated immune response leading to generation OT-1-specific cytotoxic CD8 T-lymphocytes. It was demonstrated using the in vivo mouse model of subcutaneous tumors produced by transplantation of transgenic mouse melanoma cells B16 expressing OT-1 (B16-OVA) that vaccination of mice with encapsulins containing OT-1 significantly hampered tumor development. Moreover, it was shown that the tumors were infiltrated significantly with the CD8 T-cells.
It was also demonstrated that the T. maritima encapsulins could be genetically modified in such a way to be able to express the IgG Fc domain-binding peptide (FcBP) [65] that exhibits high affinity to the Fc-fragment of rabbit Immunoglobulin G [66]. Using real-time surface plasmon resonance technique and piezoelectric microweighing it was shown that FcBP was indeed present on the outer surface of encapsulin and was accessible for binding to the rabbit IgG Fc fragment. The constructed vector was able to selectively bind to the cells of SCC-7 cell line (mouse squamous cells carcinoma). Moon et al. [65] suggested that the obtained construct could be used as a nanoplatform for the development of multifunctional system for theranostic of squamous cells carcinoma.
It was also shown that the surface of encapsulin from the R. erythropolis N771 bacteria, mentioned above, could be successfully coated with polyethylene glycol (PEG). It is well-known that PEG is a biocompatible modifier of drug carriers. Its presence hinders recognition of the carriers by the cells of monocyte-macrophage system, as well as decreases aggregation. Moreover, the authors of this study demonstrated that PEGylation did not prevent self-assembly of the nanocompartment [20].
Encapsulins as a platform for development of vaccines. It is not a secret that the development of novel safe vaccines is one of the most important tasks of humankind. Encapsulins could be used as a platform for vaccine development due to possibilities of simultaneous synthesis of the encapsulin protomers and other proteins. For example, one interesting study has been devoted to the use of T. maritima encapsulins as carriers of the ectodomain of the M2 protein of the influenza A virus (M2e-epitope) [67] (Fig. 3b). M2 protein forms ion channels on the virion surface required for transporting viral ribonucleoproteins into cytoplasm of the host cell. Its amino acid sequence is highly conserved, and immunogenicity in the process of natural infection is rather low [68]. These features make M2e good candidate for the development of vaccine with broad activity spectrum. In this study, presence of antibodies specific against the M2e-epitope was demonstrated in the serum of mice immunized using produced constructs.
Encapsulation of fluorophores. Photoswitchable fluorescence labels such as spiropyrans represent an important tool used in super-resolution microscopy. The process of probe switching itself is necessary for mapping of fluorescent points, which is achieved through stochastic activation of a certain number of fluorophores at the certain time point, while all other fluorophores are “switched off”. Next the super-resolution images can be reconstructed from the multiple fluorescent images produced. In addition, the possibility of switching fluorescence on and off could be used to avoid overlapping of the fluorescent signals in the presence of different labeled structures. Encapsulins can be modified with the help of photoswitchable fluorophores [69], in particular, the B. linens encapsulin was modified using carbodiimide technique with the spiropyran-based fluorophores – organic compounds with photochromic properties. When illuminated with visible and ultraviolet light spiropyrans are able to switch reversibly between the fluorescent photoisomer and non-radiative isomer, which allows switching fluorescence on and off [70] (Fig. 3c). It is important to mention that encapsulins preserved their structural integrity for at least five cycles of photoswitching.
Accumulation of inorganic nanoparticles in encapsulins. The possibility of controlled disassembly of encapsulins into protein subunits by treatment with acid and addition of the targeted cargo protein with affinity to some shell protein sequences followed by reconstruction of the nanocompartment structure on restoring pH to neutral levels must mentioned. Using this approach molecules/particles with size larger than the pore size could be loaded into the encapsulin shell. The study by Künzle et al. [71] is worth mentioning, in which authors used gold nanoparticles with 13 ± 1-nm diameter coated with (11-mercaptoundecyl)-N,N,N-trimethylammonium bromide as a cargo. These particles were loaded into the shell of T. maritima encapsulins via partial replacement of the stabilizing ligand of the nanoparticle with the mentioned above CLP. This approach could be potentially used for photothermal cancer therapy that requires introduction of stable and biocompatible gold nanoparticles to the specified area followed by irradiation with predetermined wavelength causing death of tumor cells, which are more sensitive to higher temperature than the healthy ones.
Encapsulin systems capable of biomineralization of iron could be used in the therapy based on magnetic hyperthermia (MH). In the study by Zhang et al. [72] encapsulins from the Myx. xanthus bacteria were used for synthesis of magnetic nanoparticles that were called encapsulin-produced magnetic iron oxide nanoparticles (eMIONs). It was shown that the particles are monodisperse, resistant to extreme pH levels and proteolytic cleavage. When exposed to alternating magnetic field eMIONs effectively absorbed magnetic energy, which resulted in the significant increase of temperature in vitro and in vivo. Moreover, eMIONs are capable of decomposition of H2O2 with formation of O2, causing specific death of tumor cells due to increased level of H2O2 in them. Considering high permeability of magnetic fields, eMIONs seems very promising therapeutic agents for treatment of solid tumors.
In addition to iron oxide nanoparticles, nanocompartments are capable of synthesizing silver nanoparticles. For example, the T. maritima encapsulins were used as a platform for production of monodisperse silver nanoparticles with average diameter 13.5 nm [19].
It is worth mentioning that the ability of encapsulins of sequestering metal ions in them could be used for water purification. It is known that, for example, in many cases industrial waste contains significant amounts of Zn2+, which could be harmful to the environment. Wang et al. [73] suggested a system consisting of modified Escherichia coli bacteria carrying a gene of encapsulin cEnc from Candidatus Brocadia fulgida, which could sequester and reduce extracellular concentration of zinc ions. The obtained results provide effective strategy for the increase of bacteria tolerance to toxic metals and reduction of Zn2+ in the medium, which could be potentially used for remediation of the environment.
Encapsulins as genetically coded labels. In addition to the studies devoted to accumulation of inorganic nanoparticles for the purpose of reducing concentration of metals outside of encapsulin shells, the studies devoted to cell monitoring with the help of magnetic resonance imaging (MRI) due to accumulation of iron-containing encapsulins in eukaryotic cells should be mentioned. The first such approach involves direct labeling in which exogenous labels are added to the growth medium of the cultured cells. In this case magnetic nanoparticles, radioisotopes, as well as low-molecular-weight fluorophores could be used as labels [74, 75]. The second approach is based on introduction of genetic sequences into the cell genome that encode endogenous labels such as luciferase and fluorescent proteins [76]. For optical monitoring in the first approach the most popular labels are quantum dots and fluorophores [77, 78], for monitoring based on single-photon emission computed tomography (SPECT) and positron emission tomography (PET) radionuclides are used as labels, and superparamagnetic iron oxide nanoparticles (SPIONPs) are used as labels in MRI-based monitoring [79-84]. The important drawback of these approaches is that accumulation of the probe in the cell decreases with cell division resulting in the decrease of the signal intensity, that is why labeling with exogenous agents is not appropriate for long-term monitoring of the cells.
Indirect specific labelling technique using reporter genes is commonly used in the studies requiring long-term monitoring of the cells. Most often the genes encoding fluorescent proteins [85-93] or firefly luciferase [94] are used. There are also specific genetic labels used in such methods as SPECT and PET; these include magnetic resonance imaging reporter genes encoding intracellularly produced metalloproteins such as transferrin, ferritin, tyrosinase [95]. As has been mentioned above, some encapsulin systems are capable of sequestering iron. This property allows using nanocompartments as genetically encoded labels for MRI-based visualization of cells. Several studies have been published on heterologous expression of the genes encoding encapsulin systems in mammalian cells. In particular, expression of the Q. thermotolerans and Myx. xanthus encapsulin genes in the HEK293T cells was demonstrated in the study by Sigmund et al. [42] using the method of transient cotransfection. Cells were transfected with a plasmid DNA encoding proteins of encapsulin shells, its cargo protein (enzyme ferroxidase), as well as iron transporter required for more effective iron transport into the cell. Iron ammonium sulfate was added to the cells 24 h after transfection, which served as a source of two-valent iron that was oxidized under the action of ferroxidase with formation of magnetic nanoparticles inside encapsulins (Fig. 3d). It was shown using cryogenic electron microscopy that in the case of Q. hermotolerans encapsulins the shells are self-assembled into nanocompartments with icosahedral symmetry with T = 4 diameter ~42 nm. Furthermore, it was established that the native cargo protein preserved its ferroxidase activity, which facilitated efficient iron biomineralization. In particular, it was shown that the ferritin-like protein of Myx. xanthus was able to mineralize up to 30,000 iron atoms per nanocompartment, which was one order of magnitude higher than the amount of iron that could be sequestered by ferritins.
In another study [96] it was demonstrated that two cargo proteins could be simultaneously loaded into the Myx. xanthus encapsulins: fluorescent protein PAmCherry and ferritin-like protein B. The authors also showed that the HEK293T expressing encapsulins accumulate iron so efficiently that could be sorted with the help of commercial magnetic cell separators (around 5% of the initial cell population) and even could be detected in vivo with MRI upon administration into the rat brain. It should be mentioned that in contrast to exogenous labels based on iron oxide nanoparticles, the nanoparticles formed in encapsulins are not released by the cells and, hence, are not phagocytized by the neighboring cells such as microglia cells. This represents a significant benefit allowing to avoid MRI signal distortion with time, as well as the errors in the result interpretation. Heterologous transient expression of encapsulin genes could be realized not only in the HEK293T cells, but also in other cell lines, such as, for example, cells of hepatocellular carcinoma HepG2 [97].
Moreover, there are studies demonstrating the possibility of constructing stable cells lines containing encapsulin systems sequestering iron. It was shown, for example, that the human mesenchymal stem cells are capable of stable expression of encapsulin genes of Myx. xanthus [98], and the cells of mouse breast carcinoma 4T1 can express the Q. thermotolerans genes [99]. It was shown both in the first and the second case that the presence of such genetic probe did not affect viability of the cells and their ability for proliferation, and T2 relaxation time for the genetically modified cells containing encapsulin was lower than for the control cells, which allowed detection of cells with MRI. This represents a very important advantage of encapsulins in comparison with other labels, which are often toxic and, hence, could affect cell proliferation. In addition, it was shown in the mentioned publications that formation of nanoparticles in encapsulins proceeded very fast and it took around 24 h in contrast to production of nanoparticles using chemical synthesis that could take several days considering isolation, purification, and surface functionalization, after which the particles should be characterized, and their toxicity tested using a cell line in order to ensure safety of their application [100].
Encapsulins as bioreactors. Another method of application of encapsulins involves their use as bioreactors. It is known than fine chemical synthesis could be performed with the help of various enzymes, which allows significant simplification of the reaction conditions. Large number of reactions in chemistry and chemical industry are conducted in the presence of catalysts, and catalysts for heterophase reactions themselves are composed of different fine particles or are applied to a porous support with developed surface to increase efficiency of the process. In one of the studies the T. maritima encapsulins were used as such support, and the required enzymes were attached to the surface (inner and outer) of the protein compartment. The T. maritima encapsulins were modified as follows: C-terminus of the protomer on the outer side was conjugated with the FbaB58 fibronectin with the help of the SpyCatcher/SpyTag technology and then sequentially was attached to the N-terminus of the E. coli dihydrofolate reductase (DHFR), and a tetrahydrofolate-dependent enzyme (demethylase LigM) was loaded into the encapsulin. It was possible to attach around 60 copies of DHFR to the encapsulin shell (Fig. 3e).
Tetrahydrofolate produced on the outer side of the shell with the help of DHFR was capable of mediating demethylation of the aryl substrate produced from lignin by the encapsulated demethylases. It was shown that the introduction of the deletion causing increase of the pore size in the encapsulin shell gene enhances metabolite exchange. Hence, such encapsulin-based construct catalyzes the reaction with the same rate as two enzymes freely dispersed in the solution [101].
Due their inner size, these protein compartments perfectly mimic closed intracellular environment and allow investigating enzyme kinetics under conditions closer to natural ones [102, 103]. It was shown that the shell of the R. erythropolis encapsulin could be loaded with such proteins as GFP and firefly luciferase (Luc), which are not native for this bacterial strain [16]. In the process, GFP retained its ability to fluoresce and luciferase exhibited enzymatic activity with its substrate luciferin. The similar study was conducted with the B. linens encapsulins. C-terminal sequence of the native cargo protein DyP was fused with the C-terminus of TFP [104]. Following confirmation of the structural integrity of the isolated nanocompartments, it was determined that on average, each encapsulin contains 12 TFP molecules.
CONCLUSION
Without doubts, encapsulins currently are relatively new research objects, not all their properties and their physiological functions in prokaryotes have been identified and investigated. However, practical applications of encapsulins in biotechnology seems very promising. Such properties of nanocompartments as small size and possibility of functionalization of the surface for selective interaction with specific proteins allows creating various systems of targeted delivery of drugs or labels. Encapsulins could serve as nanoreactors, which facilitate production of very uniform nanoparticles or to use this effect for removing metal ions from the environment. Also, encapsulins could serve as a platform for conjugation with various enzymes, which allows investigating their kinetics under natural conditions. Encapsulin-based systems capable of iron biomineralization with formation of magnetic nanoparticles could be used as contrast agents in MRI for monitoring spread of the cells in an organism.
In comparison with other nanoparticles used in biotechnology, encapsulins have, in our opinion, an important advantage – high repeatability of encapsulin biosynthesis in cells. Both in prokaryotic and eukaryotic cells synthesis of encapsulins has genetic nature, while chemical synthesis is a multifactorial process, success of which depends on purity and quality of reagents, selected procedures, etc. Considering magnetic nanoparticles used as contrast agent in MRI in more detail, it should be noted that the nanoparticles formed in encapsulins are outcompeted by the exogenous nanoparticles in the intensity of generated MR signal. However, the exogenous magnetic nanoparticles have a significant drawback. The matter is that the iron nanoparticles captured by the cells during cultivation could be released from the cells later on and be next captured by other cells, such as macrophages, which could result in the distortion of the MR signal and hinder localization of introduced cells. Another important drawback of nanoparticles produced by chemical synthesis is their toxicity towards different cell cultures, especially primary stem cell cultures. In addition to magnetic nanoparticles, there are also genetically encoded labels for MRI such as, for example, ferritin-based labels. But these labels are less efficient than the iron-sequestering encapsulins in their capacity (ferritin is capable of sequestering around 3000 iron atoms, while the Q. thermotolerans encapsulin – by one order of magnitude more). Another issue related to application of different encapsulin-based nanocontainer systems is associated with their potential in vivo immunogenicity. It has been suggested that this problem could be alleviated by coating encapsulins with PEG. At present no evaluation of in vivo immunogenicity of encapsulins isolated from bacteria or eukaryotic cells has been conducted. However, it has been shown that the abovementioned malignant mouse breast carcinoma cells 4T1 stably expressing the genes of Q. thermotolerans encapsulin system successfully formed subcutaneous tumors following implantation into immunocompetent mice, and tumor growth dynamics was the same as growth dynamics of the tumors produced from 4T1 cells not containing any transgenes.
Despite the limitations described above, encapsulins already demonstrated high potential as an alternative to conventional nanoplatforms. Each year new types of encapsulins are being discovered in different strains of bacteria and archaea. Further investigation of these structures and their properties seems very promising for a wide range of biotechnological applications.
Contributions. N. S. Chmelyuk, V. V. Oda – writing the paper, preparation of figures; A. N. Gabashvili, M. A. Abakumov – editing of the text.
Funding. This work was financially supported by the Russian Science Foundation (grant no. 21-75-00096).
Ethics declarations. The authors declare no conflict of interest in financial or any other sphere. This article does not contain any studies with human participants or animals performed by any of the authors.
REFERENCES
1.Valdes-Stauber, N., and Scherer, S. (1994)
Isolation and characterization of linocin M18, a bacteriocin produced
by Brevibacterium linens, Appl. Environ.
Microbiol., 60, 3809-3814, doi:
10.1128/aem.60.10.3809-3814.1994.
2.Rosenkrands, I., Rasmussen, P. B., Carnio, M.,
Jacobsen, S., Theisen, M., et al. (1998) Identification and
characterization of a 29-kilodalton protein from Mycobacterium
tuberculosis culture filtrate recognized by mouse memory
effector cells, Infect. Immun., 66, 2728-2735, doi:
10.1128/iai.66.6.2728-2735.1998.
3.Hicks, P. M., Rinker, K. D., Baker, J. R., and
Kelly, R. M. (1998) Homomultimeric protease in the hyperthermophilic
bacterium Thermotoga maritima has structural and amino
acid sequence homology to bacteriocins in mesophilic bacteria, FEBS
Lett., 440, 393-398, doi: 10.1016/S0014-5793(98)01451-3.
4.Giessen, T. W., and Silver, P. A. (2017) Widespread
distribution of encapsulin nanocompartments reveals functional
diversity, Nat. Microbiol., 2, 17029, doi:
10.1038/nmicrobiol.2017.29.
5.Winter, N., Triccas, J. A., Rivoire, B., Pessolani,
M. C. V., Eiglmeier, K., et al. (1995) Characterization of the gene
encoding the immunodominant 35 kDa protein of Mycobacterium
leprae, Mol. Microbiol., 16, 865-876, doi:
10.1111/j.1365-2958.1995.tb02314.x.
6.Triccas, J. A., Roche, P. W., Winter, N., Feng, C.
G., Ruth Butlin, C., et al. (1996) A 35-kilodalton protein is a major
target of the human immune response to Mycobacterium
leprae, Infect. Immun., 64, 5171-5177, doi:
10.1128/iai.64.12.5171-5177.1996.
7.Kawamoto, S., Watanabe, M., Saito, N., Hesketh, A.,
Vachalova, K., et al. (2001) Molecular and functional analyses of the
gene (eshA) encoding the 52-kilodalton protein of
Streptomyces coelicolor A3(2) required for antibiotic
production, J. Bacteriol., 183, 6009-6016, doi:
10.1128/JB.183.20.6009-6016.2001.
8.Kwak, J., McCue, L. A., Trczianka, K., and
Kendrick, K. E. (2001) Identification and characterization of a
developmentally regulated protein, EshA, required for sporogenic hyphal
branches in Streptomyces griseus, J. Bacteriol.,
183, 3004-3015, doi: 10.1128/JB.183.10.3004-3015.2001.
9.Saito, N., Matsubara, K., Watanabe, M., Kato, F.,
and Ochi, K. (2003) Genetic and biochemical characterization of EshA, a
protein that forms large multimers and affects developmental processes
in Streptomyces griseus, J. Biol. Chem., 278,
5902-5911, doi: 10.1074/jbc.M208564200.
10.Sutter, M., Boehringer, D., Gutmann, S.,
Günther, S., Prangishvili, D., et al. (2008) Structural basis of
enzyme encapsulation into a bacterial nanocompartment, Nat. Struct.
Mol. Biol., 15, 939-947, doi: 10.1038/nsmb.1473.
11.Namba, K., Hagiwara, K., Tanaka, H., Nakaishi,
Y., Chong, K. T., et al. (2005) Expression and molecular
characterization of spherical particles derived from the genome of the
hyperthermophilic euryarchaeote Pyrococcus furiosus, J.
Biochem., 138, 193-199, doi: 10.1093/jb/mvi111.
12.Akita, F., Chong, K. T., Tanaka, H., Yamashita,
E., Miyazaki, N., et al. (2007) The crystal structure of a virus-like
particle from the hyperthermophilic archaeon Pyrococcus furiosus
provides insight into the evolution of viruses, J. Mol. Biol.,
368, 1469-1483, doi: 10.1016/j.jmb.2007.02.075.
13.Moon, H., Lee, J., Min, J., and Kang, S. (2014)
Developing genetically engineered encapsulin protein cage nanoparticles
as a targeted delivery nanoplatform, Biomacromolecules,
15, 3794-3801, doi: 10.1021/bm501066m.
14.Rurup, W. F., Snijder, J., Koay, M. S. T., Heck,
A. J. R., and Cornelissen, J. J. L. M. (2014) Self-sorting of foreign
proteins in a bacterial nanocompartment, J. Am. Chem. Soc.,
136, 3828-3832, doi: 10.1021/ja410891c.
15.Snijder, J., Van De Waterbeemd, M., Damoc, E.,
Denisov, E., Grinfeld, D., et al. (2014) Defining the stoichiometry and
cargo load of viral and bacterial nanoparticles by orbitrap mass
spectrometry, J. Am. Chem. Soc., 136, 7295-7299, doi:
10.1021/ja502616y.
16.Tamura, A., Fukutani, Y., Takami, T., Fujii, M.,
Nakaguchi, Y., et al. (2015) Packaging guest proteins into the
encapsulin nanocompartment from Rhodococcus erythropolis N771,
Biotechnol. Bioeng., 112, 13-20, doi:
10.1002/bit.25322.
17.Cassidy-Amstutz, C., Oltrogge, L., Going, C. C.,
Lee, A., Teng, P., et al. (2016) Identification of a minimal peptide
tag for in vivo and in vitro loading of encapsulin,
Biochemistry, 55, 3461-3468, doi:
10.1021/acs.biochem.6b00294.
18.Choi, B., Moon, H., Hong, S. J., Shin, C., Do,
Y., et al. (2016) Effective delivery of antigen-encapsulin nanoparticle
fusions to dendritic cells leads to antigen-specific cytotoxic T cell
activation and tumor rejection, ACS Nano, 10, 7339-7350,
doi: 10.1021/acsnano.5b08084.
19.Giessen, T. W., and Silver, P. A. (2016)
Converting a natural protein compartment into a nanofactory for the
size-constrained synthesis of antimicrobial silver nanoparticles,
ACS Synth. Biol., 5, 1497-1504, doi:
10.1021/acssynbio.6b00117.
20.Sonotaki, S., Takami, T., Noguchi, K., Odaka, M.,
Yohda, M., et al. (2017) Successful PEGylation of hollow encapsulin
nanoparticles from: Rhodococcus erythropolis N771 without
affecting their disassembly and reassembly properties, Biomater.
Sci., 5, 1082-1089, doi: 10.1039/c7bm00207f.
21.Rahmanpour, R., and Bugg, T. D. H. (2013)
Assembly in vitro of Rhodococcus jostii RHA1
encapsulin and peroxidase DypB to form a nanocompartment, FEBS
J., 280, 2097-2104, doi: 10.1111/febs.12234.
22.Contreras, H., Joens, M. S., McMath, L. M., Le,
V. P., Tullius, M. V., et al. (2014) Characterization of a
Mycobacterium tuberculosis nanocompartment and its
potential cargo proteins, J. Biol. Chem., 289,
18279-18289, doi: 10.1074/jbc.M114.570119.
23.McHugh, C. A., Fontana, J., Nemecek, D., Cheng,
N., Aksyuk, A. A., et al. (2014) A virus capsid-like nanocompartment
that stores iron and protects bacteria from oxidative stress, EMBO
J., 33, 1896-1911, doi: 10.15252/embj.201488566.
24.He, D., Hughes, S., Vanden-Hehir, S., Georgiev,
A., Altenbach, K., et al. (2016) Structural characterization of
encapsulated ferritin provides insight into iron storage in bacterial
nanocompartments, Elife, 5, e18972, doi:
10.7554/eLife.18972.
25.Ochoa, J. M., Bair, K., Holton, T., Bobik, T. A.,
and Yeates, T. O. (2021) MCPdb: The bacterial microcompartment
database, PLoS One, 16, e0248269, doi:
10.1371/journal.pone.0248269.
26.Wikoff, W. R., Liljas, L., Duda, R. L., Tsuruta,
H., Hendrix, R. W., et al. (2000) Topologically linked protein rings in
the bacteriophage HK97 capsid, Science, 289, 2129-2133,
doi: 10.1126/science.289.5487.2129.
27.Caspar, D. L., and Klug, A. (1962) Physical
principles in the construction of regular viruses, Cold Spring Harb.
Symp. Quant. Biol., 27, 1-24, doi:
10.1101/SQB.1962.027.001.005.
28.Almeida, A. V., Carvalho, A. J., and Pereira, A.
S. (2021) Encapsulin nanocages: protein encapsulation and iron
sequestration, Coord. Chem. Rev., 448, 214188, doi:
10.1016/j.ccr.2021.214188.
29.Tracey, J. C., Coronado, M., Giessen, T. W., Lau,
M. C. Y., Silver, P. A., et al. (2019) The discovery of twenty-eight
new encapsulin sequences, including three in anammox bacteria, Sci.
Rep., 9, 20122, doi: 10.1038/s41598-019-56533-5.
30.Giessen, T. W., Orlando, B. J., Verdegaal, A. A.,
Chambers, M. G., Gardener, J., et al. (2019) Large protein organelles
form a new iron sequestration system with high storage capacity,
Elife, 8, e46070, doi: 10.7554/eLife.46070.
31.El-Gebali, S., Mistry, J., Bateman, A., Eddy, S.
R., Luciani, A., et al. (2019) The Pfam protein families database in
2019, Nucleic Acids Res., 47, D427-D432, doi:
10.1093/nar/gky995.
32.Nichols, R. J., LaFrance, B., Phillips, N. R.,
Radford, D. R., Oltrogge, L. M., et al. (2021) Discovery and
characterization of a novel family of prokaryotic nanocompartments
involved in sulfur metabolism, Elife, 10, e59288, doi:
10.7554/eLife.59288.
33.Wiryaman, T., and Toor, N. (2021) Cryo-EM
structure of a thermostable bacterial nanocompartment, IUCrJ,
8, 342-350, doi: 10.1107/S2052252521001949.
34.Wiryaman, T., and Toor, N. (2022) Recent advances
in the structural biology of encapsulin bacterial nanocompartments,
J. Struct. Biol. X, 6, 100062, doi:
10.1016/j.yjsbx.2022.100062.
35.Ross, J., McIver, Z., Lambert, T., Piergentili,
C., Bird, J. E., et al. (2022) Pore dynamics and asymmetric cargo
loading in an encapsulin nanocompartment, Sci. Adv., 8,
eabj4461, doi: 10.1126/sciadv.abj4461.
36.Williams, E. M., Jung, S. M., Coffman, J. L., and
Lutz, S. (2018) Pore engineering for enhanced mass transport in
encapsulin nanocompartments, ACS Synth. Biol., 7,
2514-2517, doi: 10.1021/acssynbio.8b00295.
37.Snijder, J., Kononova, O., Barbu, I. M.,
Uetrecht, C., Rurup, W. F., et al. (2016) Assembly and mechanical
properties of the cargo-free and cargo-loaded bacterial nanocompartment
encapsulin, Biomacromolecules, 17, 2522-2529, doi:
10.1021/acs.biomac.6b00469.
38.Putri, R. M., Allende-Ballestero, C., Luque, D.,
Klem, R., Rousou, K. A., et al. (2017) Structural characterization of
native and modified encapsulins as nanoplatforms for in
vitro catalysis and cellular uptake, ACS Nano, 11,
12796-12804, doi: 10.1021/acsnano.7b07669.
39.Kirykowicz, A. M., and Woodward, J. D. (2020)
Shotgun EM of mycobacterial protein complexes during stationary phase
stress, Curr. Res. Struct. Biol., 2, 204-212, doi:
10.1016/j.crstbi.2020.09.002.
40.Tang, Y., Mu, A., Zhang, Y., Zhou, S., Wang, W.,
et al. (2021) Cryo-EM structure of Mycobacterium smegmatis
DyP-loaded encapsulin, Proc. Natl. Acad. Sci., 118,
e2025658118, doi: 10.1073/pnas.2025658118.
41.LaFrance, B. J., Cassidy-Amstutz, C., Nichols, R.
J., Oltrogge, L. M., Nogales, E., et al. (2021) The encapsulin from
Thermotoga maritima is a flavoprotein with a symmetry
matched ferritin-like cargo protein, Sci. Rep., 11,
22810, doi: 10.1038/s41598-021-01932-w.
42.Sigmund, F., Pettinger, S., Kube, M., Schneider,
F., Schifferer, M., et al. (2019) Iron-sequestering nanocompartments as
multiplexed electron microscopy gene reporters, ACS Nano,
13, 8114-8123, doi: 10.1021/acsnano.9b03140.
43.Imlay, J. A., Chin, S. M., and Linn, S. (1988)
Toxic DNA damage by hydrogen peroxide through the fenton reaction in
vivo and in vitro, Science, 240, 640-642, doi:
10.1126/science.2834821.
44.Andrews, S. C. (1998) Iron storage in bacteria,
Adv. Microb. Physiol., 40, 281-351, doi:
10.1038/279015a0.
45.Goulding, C. W., Apostol, M. I., Sawaya, M. R.,
Phillips, M., Parseghian, A., et al. (2005) Regulation by
oligomerization in a mycobacterial folate biosynthetic enzyme, J.
Mol. Biol., 349, 61-72, doi: 10.1016/j.jmb.2005.03.023.
46.Sugano, Y., Muramatsu, R., Ichiyanagi, A., Sato,
T., and Shoda, M. (2007) DyP, a unique dye-decolorizing peroxidase,
represents a novel heme peroxidase family: ASP171 replaces the distal
histidine of classical peroxidases, J. Biol. Chem., 282,
36652-36658, doi: 10.1074/jbc.M706996200.
47.Pandey, R., and Rodriguez, G. M. (2012) A
ferritin mutant of Mycobacterium tuberculosis is highly
susceptible to killing by antibiotics and is unable to establish a
chronic infection in mice, Infect. Immun., 80, 3650-3659,
doi: 10.1128/IAI.00229-12.
48.Reddy, P. V., Puri, R. V., Khera, A., and Tyagi,
A. K. (2012) Iron storage proteins are essential for the survival and
pathogenesis of Mycobacterium tuberculosis in THP-1
macrophages and the guinea pig model of infection, J.
Bacteriol., 194, 567-575, doi: 10.1128/JB.05553-11.
49.Ahmad, M., Roberts, J. N., Hardiman, E. M.,
Singh, R., Eltis, L. D., et al. (2011) Identification of DypB from
Rhodococcus jostii RHA1 as a lignin peroxidase,
Biochemistry, 50, 5096-5107, doi: 10.1021/bi101892z.
50.Bobik, T. A., Lehman, B. P., and Yeates, T. O.
(2015) Bacterial microcompartments: widespread prokaryotic organelles
for isolation and optimization of metabolic pathways, Mol.
Microbiol., 98, 193-207, doi: 10.1111/mmi.13117.
51.Green, E. R., and Mecsas, J. (2016) Bacterial
secretion systems: an overview, Microbiol. Spectr., 4,
doi: 10.1128/microbiolspec.vmbf-0012-2015.
52.Wang, P., Robert, L., Pelletier, J., Dang, W. L.,
Taddei, F., et al. (2010) Robust growth of Escherichia coli,
Curr. Biol., 20, 1099-1103, doi:
10.1016/j.cub.2010.04.045.
53.Gong, J., Chen, M., Zheng, Y., Wang, S., and
Wang, Y. (2012) Polymeric micelles drug delivery system in oncology,
J. Control. Release, 159, 312-323, doi:
10.1016/j.jconrel.2011.12.012.
54.Rösler, A., Vandermeulen, G. W. M., and
Klok, H. A. (2012) Advanced drug delivery devices via self-assembly of
amphiphilic block copolymers, Adv. Drug Deliv. Rev., 53,
95-108, doi: 10.1016/j.addr.2012.09.026.
55.Allen, T. M., and Cullis, P. R. (2013) Liposomal
drug delivery systems: from concept to clinical applications, Adv.
Drug Deliv. Rev., 65, 36-48, doi:
10.1016/j.addr.2012.09.037.
56.Wang, A. Z., Langer, R., and Farokhzad, O. C.
(2012) Nanoparticle delivery of cancer drugs, Annu. Rev. Med.,
63, 185-198, doi: 10.1146/annurev-med-040210-162544.
57.Haag, R., and Kratz, F. (2006) Polymer
therapeutics: Concepts and applications, Angew. Chemie Int. Ed.,
45, 1198-1215, doi: 10.1002/anie.200502113.
58.Ma, Y., Nolte, R. J. M., and Cornelissen, J. J.
L. M. (2012) Virus-based nanocarriers for drug delivery, Adv. Drug
Deliv. Rev., 64, 811-825, doi:
10.1016/j.addr.2012.01.005.
59.MaHam, A., Tang, Z., Wu, H., Wang, J., and Lin,
Y. (2009) Protein-based nanomedicine platforms for drug delivery,
Small, 5, 1706-1721, doi: 10.1002/smll.200801602.
60.Brigger, I., Dubernet, C., and Couvreur, P.
(2002) Nanoparticles in cancer therapy and diagnosis, Adv. Drug
Deliv. Rev., 54, 631-651, doi:
10.1016/S0169-409X(02)00044-3.
61.Toita, R., Murata, M., Tabata, S., Abe, K.,
Narahara, S., et al. (2012) Development of human hepatocellular
carcinoma cell-targeted protein cages, Bioconjug. Chem.,
23, 1494-1501, doi: 10.1021/bc300015f.
62.Toita, R., Murata, M., Abe, K., Narahara, S.,
Piao, J. S., et al. (2013) A nanocarrier based on a genetically
engineered protein cage to deliver doxorubicin to human hepatocellular
carcinoma cells, Chem. Commun., 49, 7442-7444, doi:
10.1039/c3cc44508a.
63.Diaz, D., Vidal, X., Sunna, A., and Care, A.
(2021) Bioengineering a light-responsive encapsulin nanoreactor: a
potential tool for in vitro photodynamic therapy, ACS Appl.
Mater. Interfaces, 13, 7977-7986, doi:
10.1021/acsami.0c21141.
64.Van de Steen, A., Khalife, R., Colant, N.,
Mustafa Khan, H., Deveikis, M., et al. (2021) Bioengineering bacterial
encapsulin nanocompartments as targeted drug delivery system, Synth.
Syst. Biotechnol., 6, 231-241, doi:
10.1016/j.synbio.2021.09.001.
65.Moon, H., Lee, J., Kim, H., Heo, S., Min, J., et
al. (2014) Genetically engineering encapsulin protein cage nanoparticle
as a SCC-7 cell targeting optical nanoprobe, Biomater. Res.,
18, 21, doi: 10.1186/2055-7124-18-21.
66.Jung, Y., Kang, H. J., Lee, J. M., Jung, S. O.,
Yun, W. S., et al. (2008) Controlled antibody immobilization onto
immunoanalytical platforms by synthetic peptide, Anal. Biochem.,
374, 99-105, doi: 10.1016/j.ab.2007.10.022.
67.Lagoutte, P., Mignon, C., Stadthagen, G.,
Potisopon, S., Donnat, S., et al. (2018) Simultaneous surface display
and cargo loading of encapsulin nanocompartments and their use for
rational vaccine design, Vaccine, 36, 3622-3628, doi:
10.1016/j.vaccine.2018.05.034.
68.Cho, K. J., Schepens, B., Seok, J. H., Kim, S.,
Roose, K., et al. (2015) Structure of the extracellular domain of
matrix protein 2 of influenza A virus in complex with a protective
monoclonal antibody, J. Virol., 89, 3700-3711, doi:
10.1128/jvi.02576-14.
69.Putri, R. M., Fredy, J. W., Cornelissen, J. J. L.
M., Koay, M. S. T., and Katsonis, N. (2016) Labelling bacterial
nanocages with photo-switchable fluorophores, ChemPhysChem,
17, 1815-1818, doi: 10.1002/cphc.201600013.
70.Klajn, R. (2014) Spiropyran-based dynamic
materials, Chem. Soc. Rev., 43, 148-184, doi:
10.1039/c3cs60181a.
71.Künzle, M., Mangler, J., Lach, M., and Beck,
T. (2018) Peptide-directed encapsulation of inorganic nanoparticles
into protein containers, Nanoscale, 10, 22917-22926, doi:
10.1039/c8nr06236f.
72.Zhang, Y., Wang, X., Chu, C., Zhou, Z., Chen, B.,
et al. (2020) Genetically engineered magnetic nanocages for cancer
magneto-catalytic theranostics, Nat. Commun., 11, 5421,
doi: 10.1038/s41467-020-19061-9.
73.Wang, Q., Zhou, Y.-M., Xing, C.-Y., Li, W.-C.,
Shen, Y., et al. (2022) Encapsulins from Ca. Brocadia
fulgida: An effective tool to enhance the tolerance of
engineered bacteria (pET-28a-cEnc) to Zn2+, J. Hazard.
Mater., 435, 128954, doi: 10.1016/j.jhazmat.2022.128954.
74.Kim, T., Momin, E., Choi, J., Yuan, K., Zaidi,
H., et al. (2011) Mesoporous silica-coated hollow manganese oxide
nanoparticles as positive T1 contrast agents for labeling and MRI
tracking of adipose-derived mesenchymal stem cells, J. Am. Chem.
Soc., 133, 2955-2961, doi: 10.1021/ja1084095.
75.Di Corato, R., Gazeau, F., Le Visage, C., Fayol,
D., Levitz, P., et al. (2013) High-resolution cellular MRI: gadolinium
and iron oxide nanoparticles for in-depth dual-cell imaging of
engineered tissue constructs, ACS Nano, 7, 7500-7512,
doi: 10.1021/nn401095p.
76.Schrepfer, S., Deuse, T., Reichenspurner, H.,
Fischbein, M. P., Robbins, R. C., et al. (2007) Stem cell
transplantation: the lung barrier, Transplant. Proc., 39,
573-576, doi: 10.1016/j.transproceed.2006.12.019.
77.Michalet, X., Pinaud, F. F., Bentolila, L. A.,
Tsay, J. M., Doose, S., et al. (2005) Quantum dots for live cells,
in vivo imaging, and diagnostics, Science, 307,
538-544, doi: 10.1126/science.1104274.
78.Kalchenko, V., Shivtiel, S., Malina, V., Lapid,
K., Haramati, S., et al. (2006) Use of lipophilic near-infrared dye in
whole-body optical imaging of hematopoietic cell homing, J. Biomed.
Opt., 11, 050507, doi: 10.1117/1.2364903.
79.Himes, N., Min, J. Y., Lee, R., Brown, C., Shea,
J., et al. (2004) In vivo MRI of embryonic stem cells in a mouse
model of myocardial infarction, Magn. Reson. Med., 52,
1214-1219, doi: 10.1002/mrm.20220.
80.Arbab, A. S., Bashaw, L. A., Miller, B. R.,
Jordan, E. K., Bulte, J. W. M., et al. (2003) Intracytoplasmic tagging
of cells with ferumoxides and transfection agent for cellular magnetic
resonance imaging after cell transplantation: methods and techniques,
Transplantation, 76, 1123-1130, doi:
10.1097/01.TP.0000089237.39220.83.
81.Frank, J. A., Miller, B. R., Arbab, A. S.,
Zywicke, H. A., Jordan, E. K., et al. (2003) Clinically applicable
labeling of mammalian and stem cells by combining superparamagnetic
iron oxides and transfection agents, Radiology, 228,
480-487, doi: 10.1148/radiol.2281020638.
82.Bos, C., Delmas, Y., Desmoulière, A.,
Solanilla, A., Hauger, O., et al. (2004) In vivo MR imaging of
intravascularly injected magnetically labeled mesenchymal stem cells in
rat kidney and liver, Radiology, 233, 781-789, doi:
10.1148/radiol.2333031714.
83.Di Tucci, A. A., Matta, G., Deplano, S., Gabbas,
A., Depau, C., et al. (2008) Myocardial iron overload assessment by T2*
magnetic resonance imaging in adult transfusion dependent patients with
acquired anemias, Haematologica, 93, 1385-1388, doi:
10.3324/haematol.12759.
84.Ittrich, H., Lange, C., Tögel, F., Zander,
A. R., Dahnke, H., et al. (2007) In vivo magnetic resonance
imaging of iron oxide-labeled, arterially-injected mesenchymal stem
cells in kidneys of rats with acute ischemic kidney injury: detection
and monitoring at 3T, J. Magn. Reson. Imaging, 25,
1179-1191, doi: 10.1002/jmri.20925.
85.Massoud, T. F., and Gambhir, S. S. (2003)
Molecular imaging in living subjects: seeing fundamental biological
processes in a new light, Genes Dev., 17, 545-580, doi:
10.1101/gad.1047403.
86.Wu, J. C., Tseng, J. R., and Gambhir, S. S.
(2004) Molecular imaging of cardiovascular gene products, J. Nucl.
Cardiol., 11, 491-505, doi:
10.1016/j.nuclcard.2004.04.004.
87.Contag, C. H., Jenkins, D., Contag, P. R., and
Negrin, R. S. (2000) Use of reporter genes for optical measurements of
neoplastic disease in vivo, Neoplasia, 2, 41-52,
doi: 10.1038/sj.neo.7900079.
88.Herschman, H. R. (2004) Noninvasive imaging of
reporter gene expression in living subjects, Adv. Cancer Res.,
92, 29-80, doi: 10.1016/S0065-230X(04)92003-9.
89.Inubushi, M., and Tamaki, N. (2007) Radionuclide
reporter gene imaging for cardiac gene therapy, Eur. J. Nucl. Med.
Mol. Imaging, 34 Suppl 1, S27-S33, doi:
10.1007/s00259-007-0438-x.
90.Joo, H. K., and Chung, J. K. (2008)
Molecular-genetic imaging based on reporter gene expression, J.
Nucl. Med., 49 Suppl 2, 164S-179S, doi:
10.2967/jnumed.107.045955.
91.Gambhir, S. S., Barrio, J. R., Phelps, M. E.,
Iyer, M., Namavari, M., et al. (1999) Imaging adenoviral-directed
reporter gene expression in living animals with positron emission
tomography, Proc. Natl. Acad. Sci. USA, 96, 2333-2338,
doi: 10.1073/pnas.96.5.2333.
92.Phelps, M. E. (2000) Positron emission tomography
provides molecular imaging of biological processes, Proc. Natl.
Acad. Sci. USA, 97, 9226-9233, doi:
10.1073/pnas.97.16.9226.
93.Zhang, S. J., and Wu, J. C. (2007) Comparison of
imaging techniques for tracking cardiac stem cell therapy, J. Nucl.
Med., 48, 1916-1919, doi: 10.2967/jnumed.107.043299.
94.Okada, S., Ishii, K., Yamane, J., Iwanami, A.,
Ikegami, T., et al. (2005) In vivo imaging of engrafted neural
stem cells: its application in evaluating the optimal timing of
transplantation for spinal cord injury, FASEB J., 19,
1839-1841, doi: 10.1096/fj.05-4082fje.
95.Gilad, A. A., Winnard, P. T., van Zijl, P. C. M.,
and Bulte, J. W. M. (2007) Developing MR reporter genes: promises and
pitfalls, NMR Biomed., 20, 275-290, doi:
10.1002/nbm.1134.
96.Sigmund, F., Massner, C., Erdmann, P., Stelzl,
A., Rolbieski, H., et al. (2018) Bacterial encapsulins as orthogonal
compartments for mammalian cell engineering, Nat. Commun.,
9, 1990, doi: 10.1038/s41467-018-04227-3.
97.Efremova, M. V., Bodea, S.-V., Sigmund, F.,
Semkina, A., Westmeyer, G. G., et al. (2021) Genetically encoded
self-assembling iron oxide nanoparticles as a possible platform for
cancer-cell tracking, Pharmaceutics, 13, 397, doi:
10.3390/pharmaceutics13030397.
98.Gabashvili, A. N., Vodopyanov, S. S., Chmelyuk,
N. S., Sarkisova, V. A., Fedotov, K. A., et al. (2021) Encapsulin based
self-assembling iron-containing protein nanoparticles for stem cells
MRI visualization, Int. J. Mol. Sci., 22, 12275, doi:
10.3390/ijms222212275.
99.Gabashvili, A. N., Efremova, M. V., Vodopyanov,
S. S., Chmelyuk, N. S., Oda, V. V., et al. (2022) New approach to
non-invasive tumor model monitoring via self-assemble iron containing
protein nanocompartments, Nanomaterials, 12, 1657, doi:
10.3390/nano12101657.
100.Roy, N., Gaur, A., Jain, A., Bhattacharya, S.,
and Rani, V. (2013) Green synthesis of silver nanoparticles: an
approach to overcome toxicity, Environ. Toxicol. Pharmacol.,
36, 807-812, doi: 10.1016/j.etap.2013.07.005.
101.Jenkins, M. C., and Lutz, S. (2021) Encapsulin
nanocontainers as versatile scaffolds for the development of artificial
metabolons, ACS Synth. Biol., 10, 857-869, doi:
10.1021/acssynbio.0c00636.
102.Maity, B., Fujita, K., and Ueno, T. (2015) Use
of the confined spaces of apo-ferritin and virus capsids as
nanoreactors for catalytic reactions, Curr. Opin. Chem. Biol.,
25, 88-97, doi: 10.1016/j.cbpa.2014.12.026.
103.Safont-Sempere, M. M., Fernández, G.,
and Würthner, F. (2011) Self-sorting phenomena in complex
supramolecular systems, Chem. Rev., 111, 5784-5814, doi:
10.1021/cr100357h.
104.Ai, H. W., Henderson, J. N., Remington, S. J.,
and Campbell, R. E. (2006) Directed evolution of a monomeric, bright
and photostable version of Clavularia cyan fluorescent protein:
structural characterization and applications in fluorescence imaging,
Biochem. J., 400, 531-540, doi: 10.1042/BJ20060874.