REVIEW: Natural Activators of Autophagy
Julia A. Pavlova1,2,3,#a*, Ekaterina A. Guseva1,2,3#, Olga A. Dontsova1,2,3,4, and Petr V. Sergiev1,2,3,5,b*
1Center of Life Sciences, Skolkovo Institute of Science and Technology, 143025 Skolkovo, Russia2Belozersky Institute of Physico-Chemical Biology, Lomonosov Moscow State University, 119991 Moscow, Russia
3Department of Chemistry, Lomonosov Moscow State University, 119991 Moscow, Russia
4Shemyakin–Ovchinnikov Institute of Bioorganic Chemistry, 117997 Moscow, Russia
5Institute of Functional Genomics, Lomonosov Moscow State University, 119991 Moscow, Russia
* To whom correspondence should be addressed.
# These authors contributed equally to the study.
Received November 1, 2023; Revised November 25, 2023; Accepted November 29, 2023
Autophagy is the process by which cell contents, such as aggregated proteins, dysfunctional organelles, and cell structures are sequestered by autophagosome and delivered to lysosomes for degradation. As a process that allows the cell to get rid of non-functional components that tend to accumulate with age, autophagy has been associated with many human diseases. In this regard, the search for autophagy activators and the study of their mechanism of action is an important task for treatment of many diseases, as well as for increasing healthy life expectancy. Plants are rich sources of autophagy activators, containing large amounts of polyphenolic compounds in their composition, which can be autophagy activators in their original form, or can be metabolized by the intestinal microbiota to active compounds. This review is devoted to the plant-based autophagy activators with emphasis on the sources of their production, mechanism of action, and application in various diseases. The review also describes companies commercializing natural autophagy activators.
KEY WORDS: autophagy activators, mitophagy, lipophagy, mTOR, urolithin A, resveratrol, spermidine, curcuminDOI: 10.1134/S0006297924010012
Abbreviations: Akt, RAC-α serine/threonine protein kinase; AMPK, AMP-activated protein kinase; BECN1, beclin-1; LC3, microtubule-associated protein light chain 3; LKB1, liver kinase B1; mTOR, mammalian target of rapamycin; mTORC1, mammalian target of rapamycin complex 1; p62/SQSTM1, sequestosome 1; p70S6K, ribosomal protein S6 kinase; PI3K, phosphatidylinositol-3-kinase; PINK1, PTEN-induced protein kinase 1; ROS, reactive oxygen species; SIRT1, sirtuin 1; TFE3, transcription factor E3; TFEB, transcription factor EB; UA, urolithin A; Ub, ubiquitin; ULK1, unc-51-like autophagy-activating kinase 1.
INTRODUCTION
Autophagy is one of the fundamental molecular processes facilitating removal of damaged or non-operating properly cell components. Three different types of autophagy are recognized: microautophagy, macroautophagy, and autophagy associated with chaperons.
Microautophagy involves non-selective capture of macromolecules and parts of membranes followed by their digestion. Hence, the cell could receive additional energy and material for biosynthetic activity, which is essential, for example, under starvation conditions. However, it is important to note that similar processes could occur under normal conditions [1]. It should be mentioned that in this review microautophagy will not be discussed in detail, because it represents non-specific type of autophagy. More emphasis will be on macroautophagy that plays an important physiological role in human health, and dysfunction of which is a cause of numerous diseases.
In autophagy associated with chaperone activity, denatured proteins are transported to lysosomes, where they are digested. This process requires participation of cytoplasmic chaperons from the Hsp70–Hsp90 family, as well as membrane receptors recognizing complexes of proteins with chaperons. This type of autophagy has been observed only in mammalian cells and only under stress condition [2].
The last and selective type of autophagy, macroautophagy, seems the most interesting. In this process the organelles carrying the autophagosome label are recognized by the membrane receptors (LC3/GABARAP). Ubiquitin (Ub) is the most often found label in this case, but polysaccharide-based signals, lipids, Ub-like proteins could also be used [3]. Recognition of the signalling molecule results in separation of the part of cytoplasm from the rest by two membranes leading to formation of autophagosome. Next, autophagosome is fused with lysosome forming autophagolysosome, where digestion of the content occurs [4].
One of the features of macroautophagy is its fast and dynamic regulation. Under normal conditions the level of autophagy is rather low in the cell, but under starvation and under effect of specific signaling compounds it increases several folds. Continuous or periodic decrease in caloric intake without malnutrition is a promising strategy increasing life expectancy of organisms [5]. However, strict adherence to such diets is complicated, which facilitated development of the calorie restriction mimetics (CRM). CRMs are compounds that artificially induce pathways of calorie restriction thus facilitating autophagy. Potential of the use of calorie restriction mimetics in treatment of age-related diseases has been considered previously in the review devoted to the mechanisms of their action and possible therapeutic effects [6]. Some of the compounds described in this publication and recognized as calorie restriction mimetics (resveratrol, spermidine, quercetin, curcumin, and others) will be analyzed in the review predominantly from the point of view of the mechanism of their action.
Macroautophagy plays an important role in such physiological processes as adaptation to starvation, quality control of cellular organelles, elimination of intracellular parasites, and regulation of innate and acquired immunity [7].
Among the diverse macroautophagy processes autophagy of mitochondria (mitophagy) should be highlighted, because in the course of this process redundant and damaged mitochondria, which potentially are highly cytotoxic, are eliminated [8].
Balanced operation of the mitophagy systems is extremely important, and its disruption results in the development of cancer, cardiovascular, and neurodegenerative diseases [9]. It was demonstrated in the murine models that activation of mitophagy and autophagy processes with the help of pharmaceutical preparations results in improvement in the state of model animals with myocardium infarction, various types of cardiomyopathy, and atherosclerosis [8]. This fact is especially significant, because up to now cardiovascular diseases are the leading cause of death not only in Russia, but also worldwide [10].
It was shown with different animal models that the compounds initiating activation of mitophagy promote increase in life expectancy by preventing accumulation of damaged mitochondria. Moreover, consumption of such compounds prolongs normal activity of the organism by preventing age-related decrease of muscle strength and decrease of the speed of walking [11]. This could be explained by the fact that the level of mitophagy in the skeletal muscle cells decreases with age, which, in turn, results in the decrease of energy supply in the muscles [12].
Another interesting mechanism of targeted autophagy is lipophagy – a pathway of degradation of lipid droplets (LD), which participate in various biological processes in the cells. Excessive accumulation of LD is closely associated with various diseases including metabolic diseases. Targeted degradation of lipid droplets is a potential strategy for treatment obesity [13], diabetes [14], non-alcoholic steatohepatitis [15, 16], and other diseases associated with excessive accumulation of lipids.
Hence, investigation of the mechanisms of autophagy is a promising research area aiming at extending life expectancy, and, which is no less important, improving quality of life for the elderly. Moreover, activators of autophagy have been considered more often as potential therapeutics for treatment of such diseases as diabetes [17], infection diseases [18], oncological [19, 20], autoimmune [21], cardiovascular [22], neurodegenerative diseases [23], and others. Preparations approved by the US Food and Drug Administration (FDA), for which the ability to activate autophagy was demonstrated, have been described in detail in the review devoted to the synthetic activators of autophagy [24].
This review is devoted to the natural activators of autophagy and mitophagy with emphasis on the mechanism of their action, origins, as well as potential of their use for treatment of different diseases.
MECHANISMS OF ACTIVATION OF AUTOPHAGY
A significant progress was achieved in investigation of the process of autophagy in the recent decade. The evolutionary conserved genes regulating autophagy have been identified, which facilitated investigation of their functions associated with maintenance of homeostasis and total physiological state. Moreover, a vast amount of data has been accumulated indicating association between the disruption of macroautophagy and various diseases. Considering that it is macroautophagy that represents a selective type of autophagy and plays the most important physiological role in the development of different diseases, in this review we will describe mechanism of this particular process and omit discussion of the non-selective type of autophagy, microautophagy.
Macroautophagy. Numerous proteins participate in autophagy that are required for fusion of autophagosomes and lysosomes, acidification of lysosomes, and lysosomal digestion, as well as for transmission of the regulatory signals that link state of the environment and autophagy system. General scheme of autophagy regulation is presented in Fig. 1.
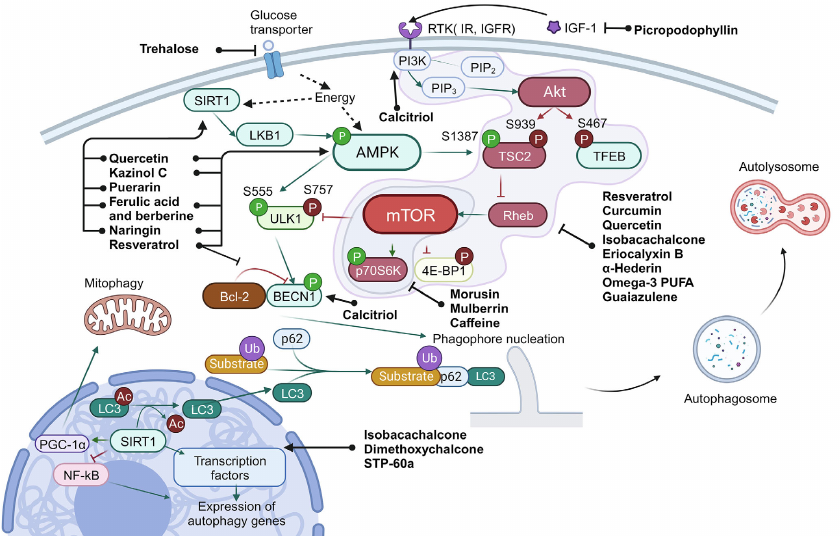
Fig. 1. Scheme of autophagy regulation. Ub, ubiquitination; P, phosphorylation (inhibitory phosphorylation is shown in red, activating – in green). Green arrows show interactions that activate target, and red arrows – interactions that inhibit target. Black arrows are used to show sequence of the stages in the process of autophagy, as well as to show targets of the compounds. Black dashed lines show effect of intracellular conditions/factors on the target.
The main signals of autophagy initiation are energy and amino acids starvation in the cell, as well as hypoxia and oxidative stress. The main sensor of energy starvation is the AMP-activated protein kinase (AMPK). Activity of this enzyme is also regulated by sirtuin 1 (SIRT1), NAD+-dependent deacetylase, and ADP-ribosyl transferase. Accumulation of NAD+ in the cell serves as a signal for SIRT1 activation, which, in turn, indicates exhaustion of energy sources and decrease of the mitochondrial membrane potential. In the activated state SIRT1 exhibits deacetylase activity, which, in turn, results in activation of the liver kinase B1 (LKB1) and AMPK, as well as in deacetylation of LC3 (microtubule-associated protein light chain 3), triggering its translocation from the nucleus to cytoplasm. SIRT1 also can control autophagy through the transcription factors FOXO1 and NF-κB (nuclear factor κB). Moreover, by maintaining deacetylated state of the peroxisome proliferator-activated receptor gamma co-activator 1-alpha (PGC-1α), SIRT1 participates in regulation of the mitochondrial function and oxidative stress [25].
The main signals for cell survival and growth are PI3K (phosphatidylinositol-3-kinase) and Akt (RAC-α serine/threonine protein kinase). Activation of this signaling pathway results in suppression of autophagy via inhibition of the tuberous sclerosis protein 2 (TSC2), which stops switching off the RheB regulator due to GTP hydrolysis. In addition, Akt also phosphorylate TFEB – main transcription factor regulating biogenesis of mitochondria. The phosphorylated state of TFEB is translocated from the nucleus to cytoplasm, which results in the decrease of autophagy level in the cells [26].
Serine/threonine protein kinase mTOR (mammalian target of rapamycin) is the central regulator involved in the vast majority of the pathways associated with both biogenesis and autophagy. mTOR controls cell growth and survival by recognizing intercellular signals and information on metabolism such as, for example, availability of amino acids. Activity of mTOR is also regulated via the AMPK- and Akt-dependent pathways.
In cell mTOR is in composition of two kinase complexes (mTORC1 and mTORC2), which have different functions. The role of mTORC2 involves regulation of growth factor signaling pathways by phosphorylation of the Akt and SGK kinases, which affect cell survival and suppress apoptosis [27]. At the same time, mTORC1 in active state inhibits autophagy by suppressing activity of the Unc-51-like kinase 1 (ULK1) and initiates protein biosynthesis via phosphorylation of the main mRNA translation regulators, such as proteins of the 4E-BP group inhibiting protein synthesis initiation factor eIF4E [27]. In order to be activated mTORC1 should be translocated to the lysosome surface, where it can be activated by amino acids [7]. In the case of mTOR inhibition by starvation or by the autophagy inducers, ULK1 is autophosphorylated and undergoes conformational changes, thus initiating assembly of the platform for further formation of autophagosome [28]. Beclin 1 (BECN1) is the main effector of this platform, which in composition of different complexes activates assembly of autophagophore, the two-membrane vesicle containing the eliminated organelles and part of cytoplasm.
The process of autophagophore assembly includes two main stages: nucleation and elongation of the isolating membrane. Complex of class III PI3K is required at the stage of nucleation of the phagophore membrane, which also includes BECN1. Regulation of activity of this complex is mediated by BECN1 with the help of ULK complex and also Bcl-2, anti-apoptotic protein, which inhibits autophagy by binding BECN1 under condition of excess of nutrients in the cell. Dissociation of BECN1 and Bcl-2 is required for autophagy induction [7].
PI3K complex also initiates assembly of the platform required for attracting, modification, and anchoring of LC3 in the phagophore membrane. Initially LC3 is present in the cell in the form of precursor – proLC3. In the first step of modification a small fragment is cleaved from the C-end. A soluble form of LC3, LC3-I, is produced as a result of this reaction [29]. In LC3-I the C-terminal glycine is exposed on the surface. In the next stage Atg7 (E1) and Atg3 (E2) carry reaction similar to ubiquitination [29]. As a result, a bond is formed between the C-terminal glycine and phosphatidylethanolamine leading to production of the protein isoform associated with the membrane – LC3-II. In the process LC3-II binds both to the outer and inner membranes of the autophagosome.
Special adapter systems exist in the cells that connect LC3 with the autophagy targets. The most thoroughly investigated among them is p62, also known as SQSTM1/sequestosome 1. p62 is a selective receptor of autophagy. Five domains are in composition of this protein: N-terminal domains Phox and Bem1p (PB1), zinc finger domain of ZZ-type, PEST site (containing presumed phosphorylation sites), and C-terminal domain binding polyubiquitin (UBA). The N-terminal domain PB1 is used for p62 polymerization and binding to other proteins containing PB1 domains [30]. It has been suggested that p62 facilitates binding of polyubiquitinated protein aggregates with LC3, which results in their incorporation into autophagosome and degradation. Hence, the cell destroys the incorrectly folded proteins together with p62 [30].
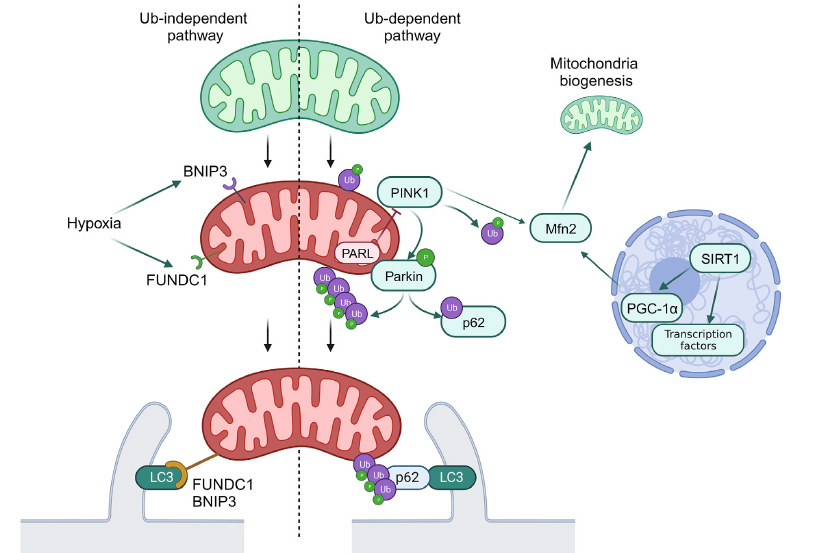
Mitophagy. Mitophagy is a selective form of macroautophagy, which is required for elimination of damaged or redundant mitochondria and, hence, is one of the most important processes maintaining cellular homeostasis. Disruption of mitophagy could result in the development of a number of diseases including cancer, cardiovascular and neurodegenerative diseases, as well as liver diseases [8, 31-35].
Mechanisms of mitophagy can be classified as Ub-dependent and Ub-independent pathways. The Ub-dependent mitophagy pathway is based on ubiquitination of the surface proteins in mitochondria to be recognized as damaged organelles. The most investigated pathway of mitophagy activations is the PINK1/Parkin system (see Fig. 2).
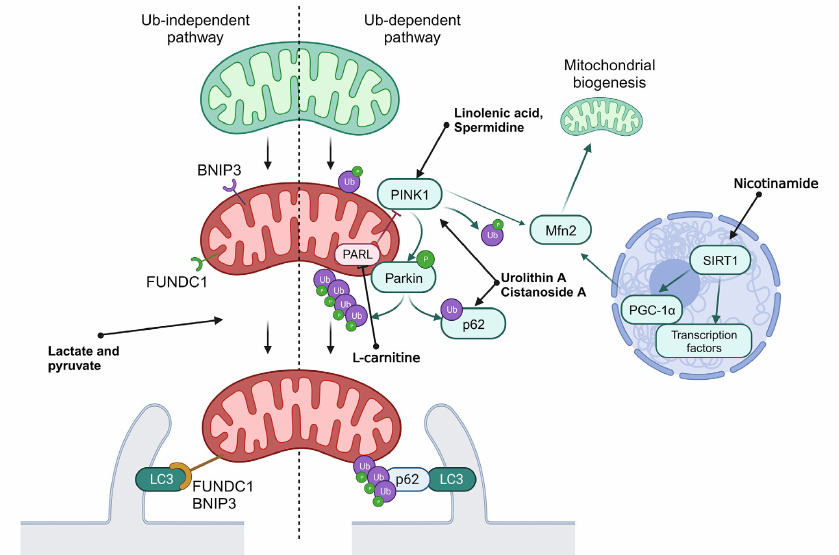
Fig. 2. General scheme of mitophagy. Normal mitochondria are shown in green, and mitochondria with disrupted functioning – in red. Ub, ubiquitination; P, activating phosphorylation. Green arrows show interactions activating the target. Red arrows show interactions inhibiting the target. Black arrows used to show sequence of stages in mitophagy.
PINK1 is a PTEN-inducible Ser/Thr kinase 1, which is accumulated at the outer mitochondrial membrane in response to disruption of membrane potential. PINK1 phosphorylates ubiquitin ligase Parkin and its substrate, ubiquitin, which results in the labeling of damaged mitochondria with ubiquitin labels. Ubiquitin ligases SMURF1 and Gp78B also participate in this process. Ubiquitin labels on mitochondria are recognized by the autophagy receptors OPTN and NDP52, which initiates formation of autophagosomes close to mitochondria, and also play a role of adapters in binding of mitochondria with LC3-II [9]. It was also discovered that p62 participated in mitophagy via the PINK1/Parkin-mediated pathway. The phosphorylated Parkin ubiquitinates p62 and facilitates its binding with LC3-II, which, in turn, facilitates transport of mitochondria to autophagosome [36].
Regulation of deacetylation of transcription factors mediated by SIRT1 could result in activation of mitophagy via the SIRT1/PGC1α/Mfn2/Parkin pathway [37]. The PINK1/Parkin system catalyzes phosphoubiquitination of Mfn2, thus initiating the p97-dependent dissociation of the Mfn2 complexes from the outer mitochondrial membrane. This results in separation of mitochondria from endoplasmic reticulum and their fragmentation, which facilitates mitophagy [38].
In addition to the described signaling system, mitophagy receptors BNIP3, NIX/BNIP3L10, and FUNDC1B are localized on the outer mitochondrial membrane, which interact directly with LC3 through typical or atypical LIR motifs (LC3-interacting region) independent on ubiquitination. Initially NIX was identified as a key mediator of mitochondria degradation during erythrocyte maturation, but it has been shown recently that it is also activated in response to disruption of mitochondrial membrane potential. It was also demonstrated that expression of NIX and BNIP3 is under partial control of HIF-1 (hypoxia inducible factor 1), and their activation during hypoxia facilitates elimination of the damaged mitochondria. Hence, BNIP3 together with FUNDC1 are considered as the main mediators of mitophagy induced by hypoxia. Unlike in the case of NIX and BNIP3, expression of FUNDC1 does not depend significantly on hypoxia or mitochondrial membrane potential, and its activity in controlled via posttranslational modifications. Under normal conditions interaction between FUNDC1 and LC3 is suppressed by phosphorylation of the LIR domain. However, in hypoxia or in the case of disruption of mitochondrial membrane potential Ser/Thr phosphatase PGAM5 dephosphorylates FUNDC1, which increases its affinity for LC3 and promotes attachment of mitochondria to the membrane of the forming autophagosome [9].
NATURAL ACTIVATORS OF AUTOPHAGY AND MITOPHAGY
Complex system of regulation of autophagy in general and mitophagy in particular explains the fact that a rather diverse set of natural and synthetic compounds could affect this process. Considering that activation or inhibition of autophagy are important for maintenance of the balance between the monomeric blocks such as amino acids from the one side and macromolecules and organelles from the other side, search for and investigation of the compounds regulating autophagy is an important task of the basic and applied science.
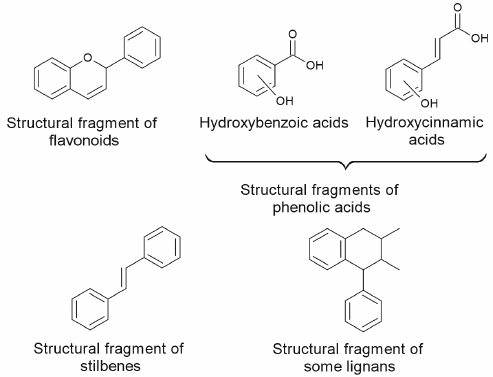
Sources of natural activators of autophagy and mitophagy. Secondary metabolites of plants. Plants contain large amounts of polyphenolic compounds with main structures presented in Fig. 3. These compounds affect human health exhibiting antioxidant, anti-inflammatory, antitumor, anti-adaptogenic, and neuroprotective properties [39-42]. Role of polyphenolic compounds in the processes of autophagy and apoptosis, as well as their possible use in chemotherapy of oncological diseases have been described in detail in numerous reviews [43-50].
Fig. 3. Main structural fragments in the composition of polyphenolic compounds.
Among the phytocompounds with anticarcinogenic properties and phytocompounds affecting processes of autophagy and apoptosis representatives of all classes of polyphenolic compounds such as flavonoids (flavonols, flavones, flavanones, isoflavones, flavan-3-ols, anthocyanins) and non-flavonoids (coumarins, curcuminoids, phenolic acids, lignans, stilbenes, xanthones) have been described [43, 48]. Majority of flavonoids are found in plants in the form bound to sugars, such as O- or C-glycosides, which interferes with their absorption in small intestine. The reactions of sugar hydrolysis leading to formation flavonoid aglycones increases their bioavailability for the organism.
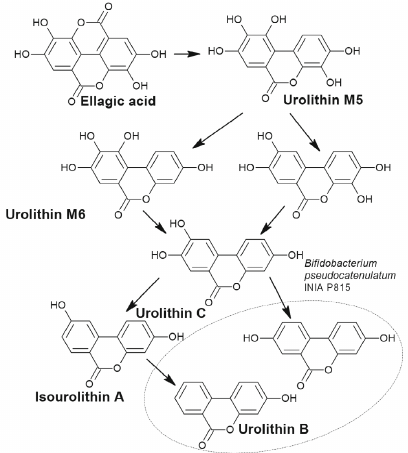
Below we present examples of some polyphenolic phytocompounds; metabolism of these compounds by microorganisms results in formation of products activating autophagy. Ellagitannins found in many plant species belong to such compounds. Due to their molecular weight these compounds have very low bioavailability and are not absorbed in the intestine until they are hydrolyzed by the gut microflora to urolithins [51, 52]. Bacteria transformation of ellagitannins involves reduction of one of the two lactone groups followed by decarboxylation and sequential reduction to tetrahydroxy (urolithin D), trihydroxy (urolithin C), dihydroxy (urolithin A and isourolithin A), and monohydroxy (urolithin B) dibenzopyranones.
The ability to transform ellagitannins to urolithins was found in Gordonibacter urolithinfaciens and Gordonibacter pamelaeae (Coriobacteriaceae family), as well as in Ellagibacter isourolithinifaciens (Eggerthellaceae family), and in the strain Bifidobacterium pseudocatenulatum INIA P815 (Fig. 4) [53, 54].
Fig. 4. Simplified scheme of formation of urolithins from ellagic acid by the bacterium Bifidobacterium pseudocatenulatum INIA P815.
Three metabolome phenotype groups (A, B, and 0) were observed in humans depending on the type of formed of urolithins [55]. In the group A (25-80%) only urolithin A is formed; in the group B (10-50%) – isourolithin A and/or B and urolithin A are detected. In the group 0 (5-25%) urolithins are not formed.
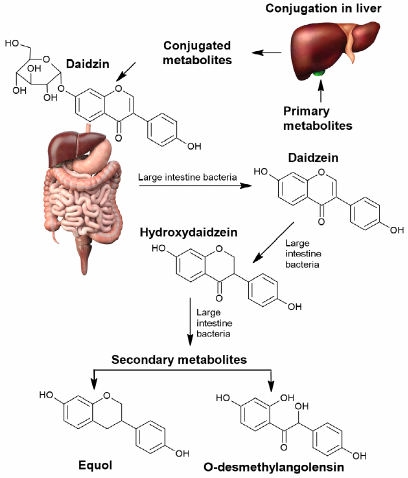
Daidzein is an example of another phytocompound transformation of which by the gut microbiota results in formation of more active and bioavailable metabolites than the initial isoflavonoid. Transformation of daidzein by the gut microbiota could result in formation of (S)-equol (isoflavandiol estrogen) or O-desmethylangolensin (O-DMA) depending on the microbiota composition (Fig. 5).
Fig. 5. Transformation of daidzein by the human gut microbiota into bioavailable derivatives.
In the majority of individuals formation of O-desmethylangolensin occurs with participation of Clostridium species. In 30% of the tested individuals daidzein is transformed into S-equol, which exhibits stronger estrogenic effect, antioxidant activity, and bioavailability than daidzein. S-equol is formed by a wide spectrum of bacteria: Streptococcus intermedius, Bacteroides ovatus, Blautia producta, Eggerthella sp. Julong 732, Adlercreutzia equolifaciens, Slakia isoflavoniconvertens, Slakia equolifaciens, Asaccharobacter celatus, Enterorhabdus mucosicola (Actinobacteria), and Lactococcus garvieae (Firmicutes) [56, 57].
Secondary bacterial metabolites. Microbiome and products of its metabolism could potentially affect activity of autophagy, as has been shown for formation of urolithin. In general, it can be presumed that microbiome formed under conditions of starvation could potentially activate autophagy. Based on suggestion on the benefits of the process of autophagy under conditions of starvation both for the host organism and for its microbiota, in this review mostly metabolites are considered that are formed by commensal gut bacteria. It has been shown in the studies that gnotobiotic (germ-free without microbiota) mice, rats, and birds have higher mortality under nutrient deficiency than the normal animals, despite the fact that the weight loss rate is the same in both cases. This is explained by the ability of gut microbiota to use alternative energy sources (fatty acids, ketone bodies, host proteins), to stimulate immunity and other protective mechanisms in the cells [58]. Under conditions of hibernation and starvation, high molecular weight glycoproteins of the intestinal mucus, atrophic epithelial cells, and dead bacterial cells are the main sources of energy and carbon. This results in the changes of gut microbiota composition; fraction of the Bacteroidetes and Akkermansia bacteria that use endogenous glycosylated protein increases, and fraction of the bacteria of Firmicutes phylum that use plant polysaccharides for growth decreases [58]. Under conditions of complete starvation, bacteria that are capable of degradation of the host proteins receive competitive advantage [59, 60]. Metagenomic studies of the cecum contents in live hibernating or starving organisms demonstrated prevalence of Bacteroides (B. thetaiotaomicron, B. fragilis), certain Firmicutes (Ruminococcus gnavus), and Verrucomicrobiales (Akkermansia muciniphila). The bacteria A. muciniphila use exclusively mucins unlike the Bacteroides capable of consuming a wide spectrum of plant polysaccharides. The main metabolites of gut microflora in animals are short-chain (volatile) fatty acids formed as a result of fermentation of plant polysaccharides (cellulose, hemicellulose, resistant starch, pectins, dextrans, and oligosaccharides) indigestible by animal enzymes. The main products of fermentation (acetate, propionate, and butyrate) are formed, respectively, at a ratio from 75/15/10 to 40/40/20 (on average – 60/20/18) [61].
It was shown in the study by Iannucci et al. [62] that propionate and butyrate induced the UCP2/AMPK-dependent autophagy in the mouse liver cells via activation of PPARg (peroxisome proliferator-activated receptor gamma coactivator). Administration of antibiotics significantly decreased basal level of autophagy in the mouse liver cells.
Hence, under starvation conditions gut microbiota both in animals and in humans predominantly contained bacteria capable of using mucins and other endogenous host substrates. It is likely that starvation is a factor for selection of bacterial species that are capable of surviving during prolonged absence of nutrients by switching metabolism to the use of available endogenous mucins and/or recyclization of dead bacterial or epithelial cells.
Natural compounds inducing autophagy. Among the activators of autophagy there are quite a few natural compounds, which often could act on several targets simultaneously. This complicates significantly elucidation of the exact mechanism of their action and could explain the fact that the data on these compounds mostly involve results of the experiments with cell cultures [63].
Let us begin the review of natural activators of autophagy considering compounds with the mechanism of action associated with the mTOR-dependent signaling pathway, inhibition of which positively regulates autophagy. There are many examples of compounds with this mechanism of action.
The natural polyphenol resveratrol, derivative of trans-stilbene, exerts a wide range of effects including antiproliferative activity via stimulation of both the BECN1-independent autophagy and apoptosis of cancer cells [64]. It was also shown that resveratrol inhibits the PI3K/Akt/mTOR pathway and activates AMPK, which are essential events in the suppression of adenoma growth in the mouse model of colorectal cancer [65]. However, according to another study, resveratrol induces autophagy in the ovarian cancer cells via alternative mechanism – by disruption of the BECN1–Bcl-2 complex [66]. It has been also reported that the resveratrol-induced apoptosis leads to inhibition of autophagy in the endometrial cancer cells [67] and in the cells of esophageal squamous cell carcinoma [68]. In addition to that, it also enhances expression SIRT1, which could result in increase of autophagy [69]. It must be mentioned that the effects produced by resveratrol are multifaceted and significantly depend on the cell line and used concentrations.
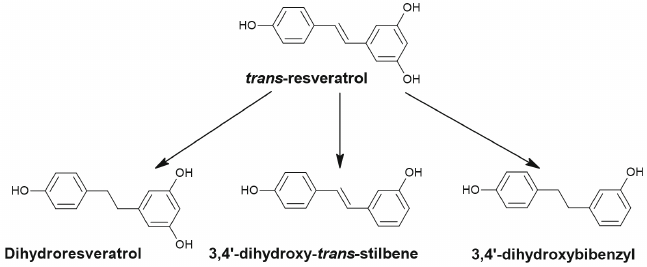
Resveratrol is quickly absorbed via passive diffusion in the intestine and next is metabolized through glucuronidation and sulfation in enterocytes and in liver or is reduced to dihydroresveratrol by the gut bacteria Sl. equolifaciens and Adl. equolifaciens [70, 71]. In addition to dihydroresveratrol (Fig. 6), 3,4′-dihydroxy-trans-stilbene and 3,4′-dihydroxybibenzyl (lunularin) are formed as a result of metabolism by the gut microbiota [70].
Fig. 6. Dehydrogenation of trans-resveratrol with formation of dihydroresveratrol, 3,4′-dihydroxy-trans-stilbene and 3,4′-dihydroxybibenzyl (lunularin) by the gut microbiota.
There are no studies devoted to antitumor effects and effects on autophagy of the microbial metabolites of resveratrol despite the large number of such studies investigating resveratrol itself and its metabolites produced in liver and enterocytes [72].
Curcumin, the most studied biologically active compound isolated from the Curcuma plant species, has the same mechanism of action. Based on the results of preclinical and clinical studies this compound is recognized as an agent sensitizing cancer cells to chemo- and radiation therapy [73]. In the stomach cancer cell lines SGC-7901 and BGC-823 this compound suppresses the PI3K/Akt/mTOR pathway, which causes increase of expression of BECN1, Atg3, Atg5, p53, and p21, increases the LC3-II/LC3-I ratio, and potentiates autophagy [74]. In the cells of castration-resistant prostate cancer curcumin induces protective type of autophagy [75]. Inhibitory action of curcumin on the processes of transcription of ribosomal genes and activation of autophagy in the mouse embryonic fibroblasts (MAFs) [76], in the glioma cells [77], and in the lung carcinoma [78] has been also established. It has been mentioned in many clinical studies that curcumin exhibits very low bioavailability due to low solubility and fast metabolism [79]. It has been suggested that the mechanism of positive effects of curcumin with its low bioavailability could be associated with its effect on the gut microbiota [80-83]. Transformation of curcumin by the gut microbiota could result in formation of compounds with high biological activity and improved bioavailability. One of the main metabolites of curcumin, tetrahydroxycurcumin, induced autophagy in the human leukemia cells HL-60 and lung cancer cells A549 NSCLC [84, 85].
Quercetin, natural flavonoid contained in many herbs, has the similar mechanism of action [86, 87]. Numerous preclinical studies have been devoted to investigation of its antiproliferative effects. Quercetin, similarly to curcumin, suppresses proliferation of the cells of stomach cancer [87], glioma [78], and lung cancer [88]. In the stomach cancer cells quercetin induces protective autophagy via the Akt/mTOR pathway and signal transduction mediated by hypoxia-inducible factor 1α (HIF-1α) [87]. In the study investigating human lung cancer cells A549 and H1299 quercetin increased the levels of SIRT1 protein and accumulation of the phosphorylated form of AMPK in dose-dependent manner [89].
There are other activators of autophagy among the flavonoids with mechanism of action associated with inhibition of mTOR. In particular, isobacachalcone from Ashitaba and Psoralea corylifolia inhibits activity of Akt and mTORC1, activates pro-autophagy transcription factors TFEB and TFE3 in vitro, as well as in vivo in the mouse model of fibrosarcoma [90]. It is important to clarify that in addition to activation of autophagy isobacachalcone also triggers endoplasmic reticulum stress manifested as endoplasmic reticulum PKR-like kinase (PERK)-dependent phosphorylation of eIF2α.
Representatives of the group of prenylated flavones, such as morusin and mulberrin, could slow down aging in the models of budding yeasts, worms, and human cells affecting the mTOR-Sch9/p70S6K system inducing autophagy. In the study with the HeLa human cells morusin and mulberrin inhibited phosphorylation of the β1 kinase of the ribosomal protein S6 (p70S6K), stimulated autophagy, and slowed down cell aging [91]. Moreover, the worms treated with morusin and mulberrin demonstrated additional benefits such as increase of reproduction without affecting the worms’ health.
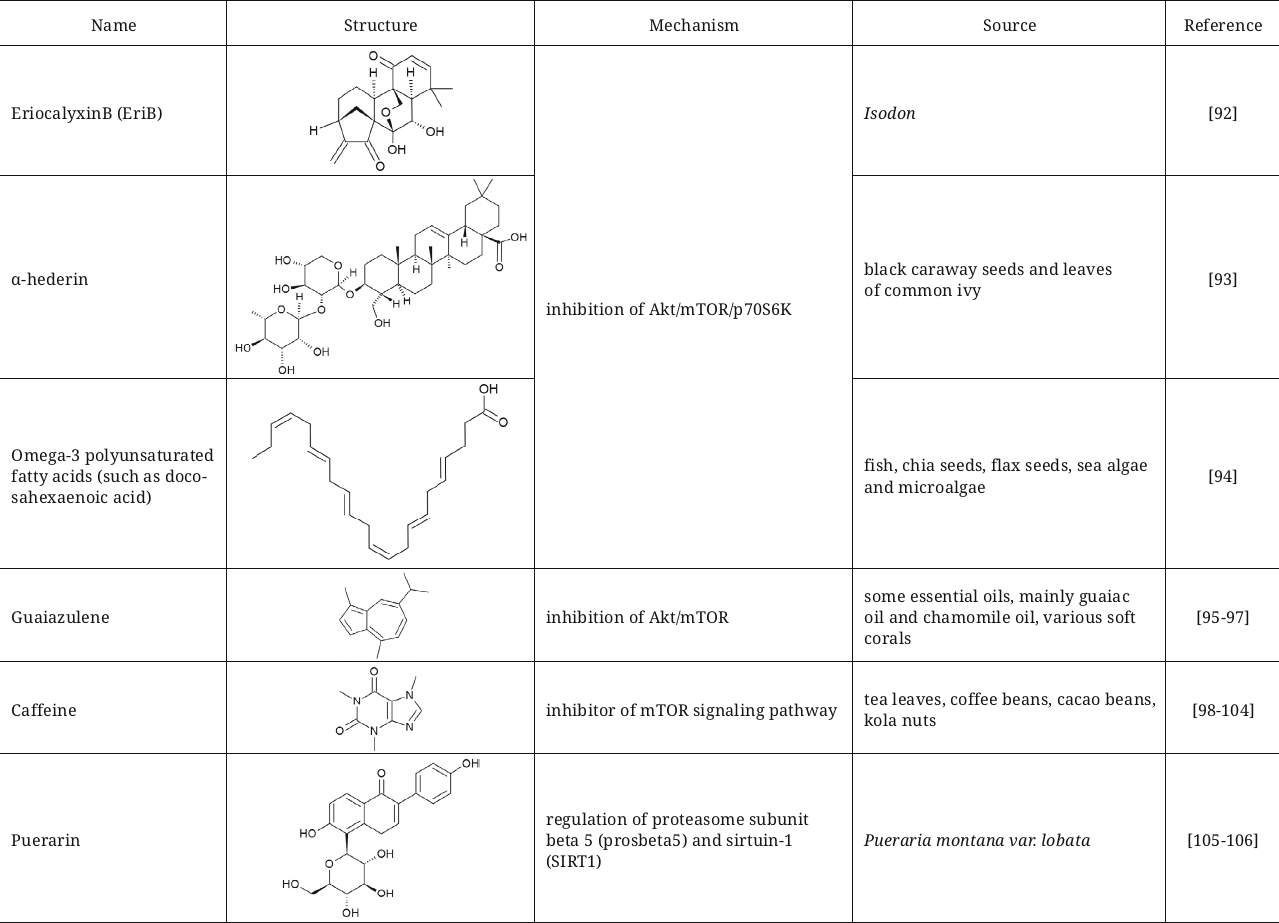
Another three compounds – eriocalyxin B (EriB), diterpenoid from isodon, α-hederin, triterpenoid saponin, and omega-3 polyunsaturated fatty acids – display similar mechanism of autophagy induction by inhibiting Akt/mTOR/p70S6K signaling and, simultaneously, initiating generation of reactive oxygen species (ROS) in the breast cancer [92], colorectal cancer [93], and prostate cancer [94] cells.
The FDA approved natural compound guaiazulene (1,4-dimethyl-7-isoprpylazulene) also could be assigned to the group of Akt/mTOR inhibitors [95]; previously it was used in ophthalmic preparations and later became popular as an ingredient of cosmetic products, toothpastes, and pharmaceutical preparations. This compound exhibits anticancer properties, and also can be used for pain treatment during inflammation in combination with diclofenac [96]. Guaiazulene suppresses non-small cell lung cancer (NCSLC) by initiating apoptosis induced by ROS. In the process, induction of autophagy was demonstrated in the NCSLC cells mediated by inhibition of the Akt/mTOR signaling pathway [97].
Caffeine is another interesting example of the inhibitor of mTOR signalling pathway, which initiates autophagy both in vitro and in vivo [98-100]. Caffeine decreases liver hepatosis in mice with non-alcoholic fatty liver disease [101] and protects against the prion-mediated neurotoxicity [102]. It is also worth mentioning that it was demonstrated in the studies that the coffee beans containing in addition to caffeine large amounts of polyphenols cause enhancement of autophagy, inhibition of mTORC1, decrease of the level of p70S6K phosphorylation, and translocation of LC3B into autophagosomes [103]. It could be suggested that the caffeine-induced autophagy is the cause underlying the fact that coffee decreases mortality among the middle-age individuals [104].
Autophagy induction through inhibition of the PI3K/Akt/mTOR signaling pathway could be used as an approach for obesity treatment. This has been corroborated in the study demonstrating that the flavonoids from Pueraria montana improved the state of obese mice with non-alcoholic fatty acid disease due to this mechanism [105]. It was also shown that puerarin, one of the isoflavones from P. montana, could significantly extend lifespan of Drosophila melanogaster via regulation of the proteasome subunit beta 5 (prosbeta5) and SIRT1 [106].
Another mechanism of autophagy activation is inhibition of the receptor of insulin-like growth factor-1 (IGF1), which regulates the PI3K/AKT/mTOR pathway. This mechanism has been observed for picropodophyllin (PPP), cyclolignan alkaloid isolated from the mayapple plant. PPP has been described as a powerful inducer of autophagy realizing its effect by inhibition of the insulin-like growth factor-1 receptor (IGF1R). Administration of PPP to mice with tumors increased efficiency of immunogenic chemotherapy, which was based on the induction of autophagy in the malignant cells [107].
Despite the fact that mTOR plays an important role in the cell metabolism participating in sensing of nutrients, there is a wide variety of mTOR-independent pathways, which are targets of other autophagy activators. Trehalose, natural disaccharide, stimulates autophagy and decrease protein aggregation by inhibiting glucose transporters [108, 109]. It was shown that trehalose increases conversion of LC3-I into LC3-II via the mTOR-independent pathway, similarly to sucrose [110]. Hence, trehalose could be used for inducing autophagy in the Parkinson’s [111] and Alzheimer’s [112] diseases, as well as in prion diseases [113]. Despite this, trehalose is not used in clinical practice, because it has low bioavailability due to fast metabolism in the organism. That is why more stable analogs of trehalose, lenztrehaloses A, B, and C, were synthesized, which were shown to be active activators of autophagy [114].
The hormonally-active form of vitamin D (calcitriol) inhibits replication of HIV-1 [115] and of mycobacterium tuberculosis in human macrophages [18] and kills breast cancer cells [116] due to induction of autophagy mediated by PI3K, ATG5, and BECN1. This explains the fact that the vitamin D deficit could be the cause of enhanced susceptibility to various cancer types and infectious diseases [117].
Among the natural autophagy activators, the AMPK kinase activators could be highlighted, which positively regulate autophagy by phosphorylating the ULK1 kinase at Ser-317, Ser-555, and Ser-777. Kazinol C from the Broussonetia kazinoki plant is one of such compounds, which induces apoptosis at high concentrations and autophagy at low concentration through activation of AMPK [118]. Hence, owing to these properties kazinol C could serve as an effective therapeutic agent for treatment of both cancer and metabolic diseases.
Regulation of AMPK could be realized through, for example, the SIRT1/LKB1 pathway [119]. SIRT1 activation results in deacetylation and increase of kinase activity of the LKB1 protein, which could activate AMPK by phosphorylation. Examples of double activators of SIRT1 and AMPK include combination of ferulic acid with berberine, which promotes autophagy, mitophagy, mitochondrial biogenesis, and DNA repair; this process facilitates cell survival due to suppression of apoptosis, aging, and inflammation caused by NF-κB [120].
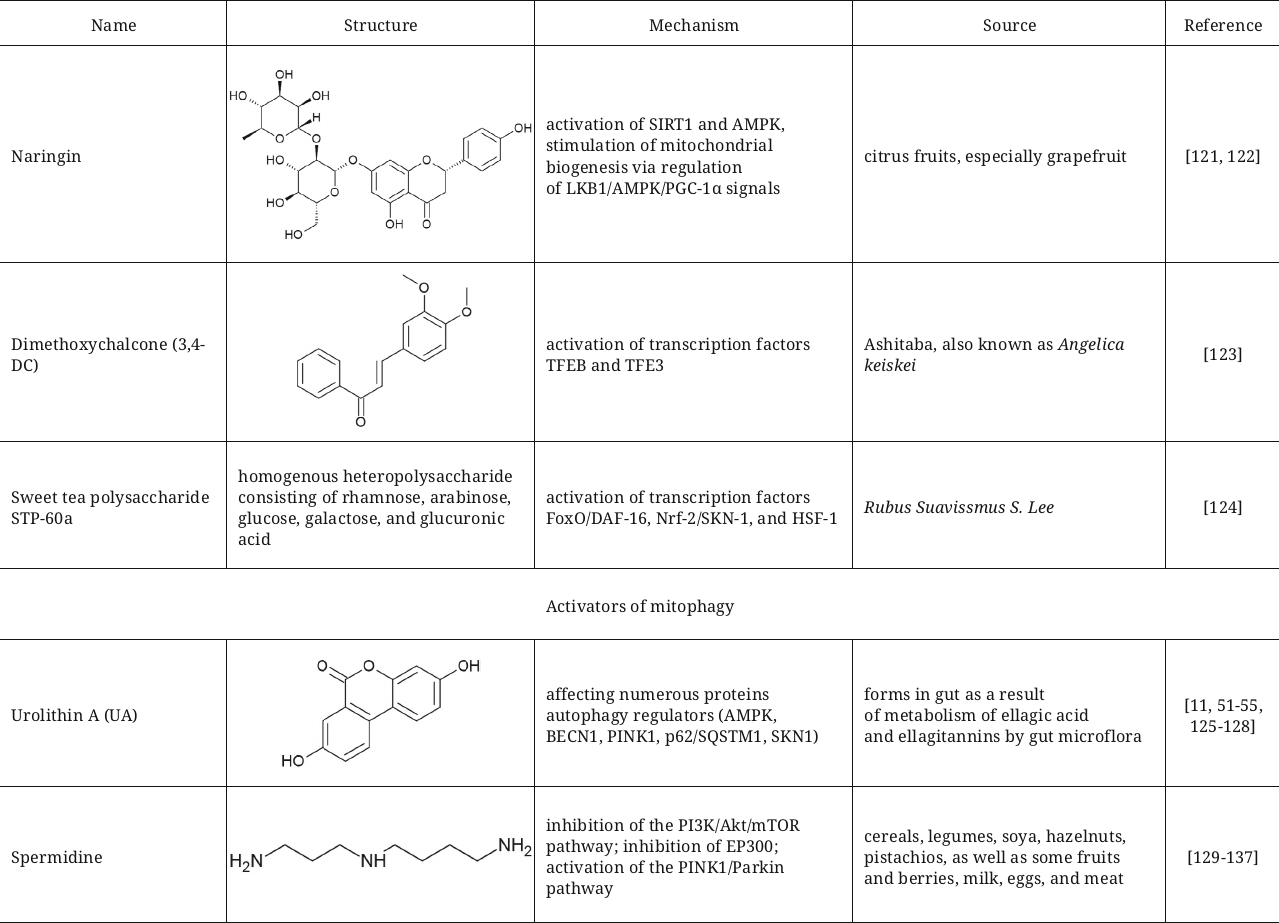
Dihydroflavonoid compound naringin, which is abundant in citrus fruits and used in traditional Chinese phytotherapy, slows down aging in the Caenorhabditis elegans model, decreases fat accumulation, and facilitates autophagy. Transcriptome analysis showed changes in the levels of transcription of the genes associated with regulation of biosynthesis and metabolism of lipids and autophagy (such as genes skn-1, hlh-30, lgg-1, unc-51, and pha-4) [121]. According to the results of the study, naringin significantly activates SIRT1 and AMPK and stimulates mitochondrial biogenesis pathway through regulation of the LKB1/AMPK/PGC-1α signals [122].
Regulation of autophagy at the level of transcription is another possible mechanism of action of some activators. In particular, 3,4-dimethoxychalcone (3,4-DC), which has been described as a calorie restriction mimetic, induces autophagy by activation transcription factors TFEB and TFE3 [123]. Sweet tea (Rubus suavissmus S. Lee) polysaccharide STP-60a in the study with C. elegans extended lifespan of the animals through the insulin-dependent and mitochondrial pathways that depend on autophagy. It activates three transcription factors (FoxO/DAF-16, Nrf-2/SKN-1, and HSF-1) that operate downstream of the insulin signaling pathway [124].
Hence, natural activators of autophagy are very diverse in their mode of action, which in the majority of cases is associated simultaneously with several targets. The scheme summarizing targets of the natural activators of autophagy is presented in Fig. 7.
Fig. 7. Mechanism of action of natural activators of autophagy. Ub, ubiquitination; P, phosphorylation (inhibitory phosphorylation shown in red, and activating – in green). Green arrows show interaction activating the target, and red – interactions inhibiting the target. Black arrows used for presenting sequence of the stages in the process of autophagy, as well as to show targets of the compounds. Black dashed lines reflect effects of intracellular conditions/factors on the target.
It must be emphasized that on the summarizing scheme only major targets are presented, which are affected by the considered compounds; however, for numerous compounds alternative mechanisms of action have been described in the literature, as noted in this review. Many molecules affect the process of autophagy by interacting with proteins (AMPK, ULK1, SIRT1, and others) and positively regulate this process. However, quite a few molecules activate autophagy by suppressing the AKT/mTOR pathway, which inhibits autophagy. Obviously, there are compounds affecting both of the indicated pathways, such as, for example, resveratrol and quercetin. However, it must be understood that the effects of many described molecules also include interactions with other targets mentioned above depending on the cell type and concentration. Hence, common feature of the majority of natural activators of autophagy is absence of unique mechanisms of action and, as a consequence, existence of a large number of side effects that limit their application.
Natural activators of mitophagy. Many natural compounds discussed in the previous section also can trigger mitophagy, such as spermidine and resveratrol. It seems logical to elaborate on the most known mitophagy activator – urolithin A (UA). Urolithin A belongs to the class of benzocoumarins, which is formed as a result of metabolism of ellagic acid and ellagitannins by the gut microflora [125]. No foods contain urolithin, and its bioavailability depends of the individual composition of microbiota. Mechanism of mitophagy induction with the help of UA is still poorly understood, but it is clear that it maintains mitochondrial functions with the help of several proteins regulators of mitophagy such as AMPK, BECN1, PINK1, p62/SQSTM1, and others. UA can activate SKN1 (homolog of NRF2 in C. elegans nematode), which extends lifespan and improves mitochondrial biogenesis thus preserving population and activity of mitochondria. It was shown in the experiments with nematodes that the long-term administration of UA results in activation of mitochondrial biogenesis, which, in turn, results in the improvement of mobility, activity of pharyngeal pumping and respiration processes; moreover, these effects were shown to be independent on age and diet [11]. It was also demonstrated in the mouse models that prolonged oral administration of urolithin A results in the increase of physical endurance in the young mice and both muscle strength and endurance increase in the old mice. These results highlight therapeutic potential of urolithin A in improvement of muscle system functioning and mobility [126]. The results of in vivo studies did not reveal any toxic or specific side effects [127], moreover, elderly tolerate well treatment with UA [128]. In 2018 FDA included urolithin A in the list of safe food supplements.
Another natural activator of autophagy used at present as a food supplement for maintenance of mitochondrial functions is spermidine, which is a natural endogenous polyamine synthesized from diamine putrescine. Cereals, legumins, soya, hazelnuts, pistachios, as well as some fruits and berries are natural sources of spermidine [129]. Milk, eggs, as well as meat contain spermidine in their composition. Spermidine is considered to exert such properties as slowing down aging, cardio protective effects [130], improvement of cognitive functions [131], protection against neurodegenerative disorders [132]; there are quite a few mechanisms suggested for possible effects of spermidine on autophagy. It was demonstrated in one study that spermidine, similarly to curcumin and quercetin, exerts properties of epigenetic regulator and induces autophagy in vitro by inhibiting histone acetyltransferase p300 (EP300), which suppresses function of several autophagy proteins [133]. In the female germline stem cells (FGSC) spermidine induces autophagy via inhibition of the PI3K/Akt pathway [134]. Spermidine enhanced the BECN1-dependent autophagy in fibroblasts in the bleomycin-induced idiopathic pulmonary fibrosis in mice [135].
It has been also shown that addition of spermidine affects number and morphology of mitochondria in the heart of old mice by stimulating mitophagy and mitochondrial biogenesis [136]. It was shown in the mouse models that spermidine can cross blood-brain barrier and enhance hypusination of the eukaryotic translation initiation factor 5A (eIF5A) in hippocampus thus improving mitochondrial functions. In the same study it was shown with the model of Drosophila aging that spermidine increases respiratory capacity of mitochondria through the autophagy regulator Atg7 and PINK1/Parkin pathway [131].
Due to the large number of effects of spermidine on cellular metabolism the mechanisms of action of a particular compound differ depending on the cell type, organ, or existence of certain disease. This, from the one hand, could be beneficial for the fight with aging, which is the process with very complex mechanism. However, absence of the unique mechanism could pose serious problems for application of the particular compounds. Despite the fact that safety of spermidine used in the form of plant extract enriched with spermidine has been demonstrated for mice and elderly [132], another study with the rats treated with pure spermidine demonstrated several side effects of this compound [137].
Considering perspectives of spermidine application in different diseases, one of the approaches to overcome limitations of spermidine use is design and synthesis of new derivatives. Structurally related spermidine analogs have been produced, which in the experiments with C. elegans facilitate extension of life expectancy and protection of cells against oxidative stress by activation of mitophagy. In particular, the compound VL-004 (1,8-diaminooctane) has better capability to induce mitophagy than spermidine [138]. Unexpectedly it was discovered during investigation of the mechanism of action of VL-004 that the function of this compound depends on PINK1, but does not depend on PDR-1, Parkin ortholog in C. elegans. The authors of the study assume that another ubiquitin ligase E3 could be involved together with PINK1 inducing mitophagy.
Mitochondrial dysfunction is typical for many neurodegenerative diseases such as Parkinson’s disease, and mitophagy mediated by the PRKN1/Parkin pathway could serve as an approach for treatment of such disorders. It was shown that in addition to individual molecules probiotics could also activate mitophagy thus alleviating symptoms of neurodegenerative diseases. Screening of 49 probiotic strains revealed that two probiotics, Saccharomyces boulardii and Lactococcus lactis, increased recruitment of Parkin to mitochondria, accumulation of phosphoubiquitin, and degradation of mitofusins [139].
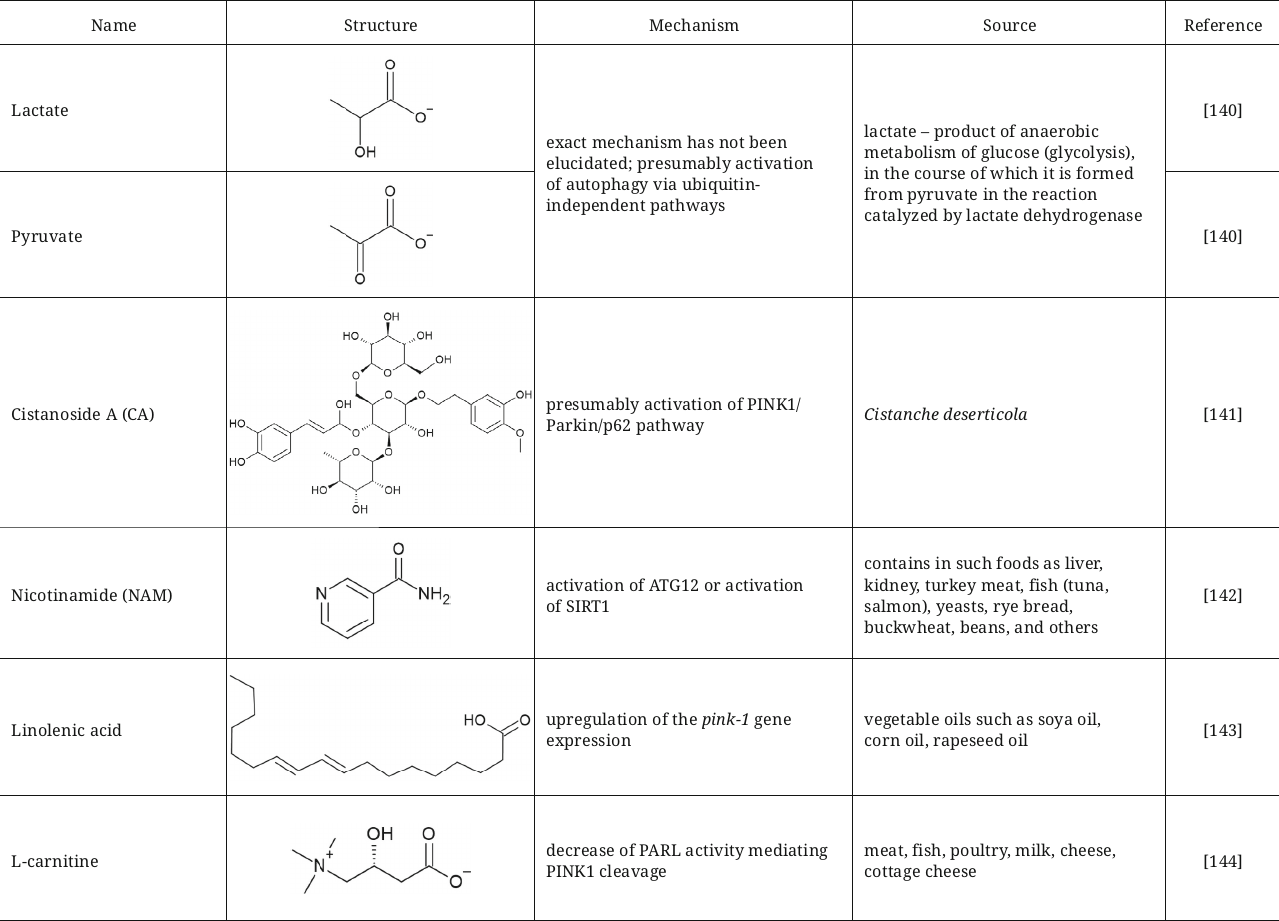
Lactate and pyruvate that are capable of short-term acidification of cytosol were also described as agents activating mitophagy with aim to fight against Parkinson’s disease [140]. Incubation of SH-SY5Y cells or primary neurons and astrocytes with these compounds resulted in activation of mitophagy after treatment with MPP+ (1-methyl-4-phenylpyridinium), which facilitated restoration of mitochondrial functions and protection of these cells from apoptotic and necrotic death. Mechanism of mitophagy activation has not been established in this study, but it was suggested that it could occurred not only via the PINK1/Parkin-dependent pathway, but also via the ubiquitin-independent pathways, such as oxidative stress accompanying Parkinson’s disease.
Another active compound that provides neuroprotective effect in Parkinson’s disease by inducing mitophagy is cistanoside A (CA), phenylethanoid isolated from the desert-broomrape, Cistanche deserticola. Exact mechanism of action has not yet been elucidated; however, it has been suggested that this compound could presumably activate the PINK1/Parkin/p62 pathway leading to degradation of damaged mitochondria and decrease of oxidative stress [141].
Nicotinamide (NAM), the FDA-approved food supplement, also has a potential for improving quality of mitochondria via activation of mitophagy. NAM decreases mitochondrial respiration and ROS production in the human primary fibroblasts and extends their replicative lifespan. NAM in the cells is converted into NAD+ (nicotinamide adenine dinucleotide) with participation of nicotinamide phosphoribosyltransferase (Nampt). The presumed mechanism is associated with either increase of GAPDH activity, which, in turn, stimulates activation of ATG12, or with activation of SIRT1 [142].
Restoration of mitochondrial functions by activation of mitophagy is also considered as an approach for treatment of sarcopenia, syndrome of age-related muscle and strength loss. Linolenic acid was suggested as a compound with such potential, which significantly reduced sarcopenia in C. elegans by restoring mitochondrial functions, stimulating mitophagy, and fighting against oxidative stress. The mechanism of action is associated with upregulation of the pink1 gene expression [143].
Another activator of the PINK1/Parkin-dependent mitophagy is L-carnitine, which maintains mitochondrial functions and normal state of heart microvessels. The described mechanism of L-carnitine action involves decrease of protease activity of the mitochondrial intramembrane cleaving protease PARL (presenilin-associated rhomboid-like protein), which mediates PINK1 cleavage by increasing interaction between PHB2 (Prohibitin-2) and PARL [144].
Hence, numerous activators of autophagy and mitophagy have been identified among the natural compounds, which exhibit a wide range of the mechanisms of action that involve several targets in the majority of cases (Fig. 8) including many proteins autophagy regulators. In addition to the effects on the PINK/Parkin pathway observed for the majority of the described molecules activators of mitophagy, they enhance autophagy via activation of AMPK and SIRT1 or by inhibiting mTOR.
Fig. 8. Mechanism of action of natural activators of mitophagy. Normal mitochondria are shown in green, and mitochondria with disrupted function – in red. Ub, ubiquitination; P, activating phosphorylation. Green arrows show activating interaction. Red line shows interaction inhibiting the target. Black arrows show the sequence of stages in the process of mitophagy, as well as the targets of the compounds.
Summarized data on activators of autophagy and mitophagy including their structures, brief description of mechanism, and sources are presented in table.
Natural activators of auto- and mitophagy


COMMERICIAL DEVELOPMENTS OF NATURAL AUTOPHAGY ACTIVATORS
Research and development in the area of autophagy became more and more popular in recent years as an approach for search of the compounds capable to slow down aging, improve health of elderly, and to delay onset of diverse age-related diseases. The companies Amazentis, Longevity labs, Mimio Health, Nestlé Health Science produce various biologically active supplements that aim to activate autophagy in cells and slow down aging and improve health status of an organism.
Some companies do not emphasize the fact that their product is capable of modulating autophagy, however, they use ingredients described in the literature as autophagy activators. In particular, the Napa Hills company produces drinks with resveratrol and other antioxidants, which help to reduce inflammation, facilitate improvement of cardiovascular system functioning, and slow down aging. Resveratrol also is in composition of food supplements produced by the Vinomis and Life Extension companies. The Taiyo company is a manufacturer of isoquercetin, precursor of quercetin, which provides defense against oxidative stress and also could initiate autophagy. Jarrow Formulas uses quercetin and other ingredients in their products to maintain mitochondrial functions, and Zeus Hygia uses bioavailable form of curcumin to restore muscle and joint functions. The list of companies using natural autophagy activators in compositions of their products does not end here, because natural compounds exhibit a wide spectrum of effects, and potential of their application is not limited to their use for autophagy activation.
Let us begin our detailed description with the companies that use natural autophagy activators in their products. The company Amazentis, founded in 2007 in Lausanne, Switzerland, uses modern science and clinical research for discovery and development of supplements aiming at extension of healthy life. The company targets age-related mitochondrial dysfunctions. History of the company starts with the Mitopure™ ingredient (patented highly purified form of urolithin A). Despite the fact that urolithin could be produced in an organism by the gut microflora, microflora composition in individuals could be very different and synthesis of urolithin could occur with varying efficiency. That is why the Amazentis team created the Mitopure preparation protected by the patents (EP-3393467-B1, US-9994542-B2, US-10906883-B2, and others), efficiency of which was verified in several clinical trials [145, 146]. The company patented the method of urolithin synthesis and well as use of the compositions containing this ingredient in food supplements, food products, and drinks. Mitopure induces mitophagy and supports improved mitochondrial functions and muscle strength. By using the Mitopure ingredient, the Timeline company (subsidiary of Amazentis) produces food supplements and well as anti-aging cosmetics.
Commercialization of urolithin A attracted attention of the Nestlé Health Science company, which took an equity stake in Amazentis and received global rights for using the patented ingredient Mitopure. That is why urolithin A in the form of Mitopure ingredient is also in the composition of the Celltrient Cellular Nutrition line of the dietary supplements produced by the Nestlé Health Science company.
Among the companies producing biologically active supplements for modulation of autophagy the Austrian company Longevity Labs+ should be mentioned. This company was established in 2016, although its history begins six years earlier in 2010. Professor Frank Madeo, discoverer of spermidine, was at that time the head of research group investigating polyamine spermidine in the Graz university. This research attracted attention of Doctor Gerald Sitte and professional engineer Herbert Pock, founders of the company. Immediately they started collaboration with the researchers from Graz university in the area of development of special extraction process, which eventually led to establishment of the Longevity Labs+ company. The company produces supplements Spermidine LIFE, which contain 100% natural and clinically tested spermidine from wheat germ. The company patented the use of spermidine in dietary supplements and pharmaceutical preparations for treating diseases or disorders associated with mitochondrial dysfunction.
Despite the fact that the Longevity Labs+ company was the first company to introduce supplements with spermidine on the market in 2019, at present it is not the only one using this biologically active substance. The USA company Mimio Health also uses spermidine as an ingredient of the patented composition mimicking fasting effects and intended for activation of the natural capability of cells for self-cleaning, restoration, and defence. Clinical study conducted by the company revealed that intermittent long-term fasting activates effects of several biologically active metabolites (spermidine, 1-methylnicotinamide, palmitoylethanolamide, and oleoylethanolamide), which were capable to increase average lifespan of C. elegans by as much as 96% [5]. The compounds mentioned above are in composition of the food supplements produced by the company. With regards of the mechanism of action, this composition helps to activate autophagy via regulation of mTOR, Akt, and histone acetyl transfer, as well as stimulation of AMPK and SIRT1.
Despite the fact that spermidine is recognized as a promising autophagy activator, its mechanism of action could vary depending on the cell type and type of disease. A complicated and poorly understood mechanism of action in combination with high activity with regard to autophagy regulation promoted the idea of development of more specific analogues of spermidine. At present the Israeli startup Vitalunga actively develops the methods for production of clinical preparations and conducts investigation of the properties of spermidine analogues that enhance mitophagy and autophagy. The startup’s founding researchers already tested a family of compounds structurally related to spermidine and demonstrated that the 1,8-diaminooctane (VL-004) molecule induced mitophagy and protected from oxidative stress more effectively than spermidine, which facilitated extension of the C. elegans lifespan [138].
Research in the Vitalunga startup has a goal of developing preparations for prophylactics of numerous diseases associated with aging, such as Alzheimer’s and Parkinson’s diseases, heart failure, sarcopenia, and others.
CONCLUSIONS
Autophagy plays an important role among the cellular processes, and its dysfunction is associated with many human diseases. Disruption of the autophagy process could lead to the development of cardiovascular, neurodegenerative diseases, as well as diabetes, cancer, and other disorders. However, the role of natural compounds in prevention of these diseases is quite complicated due to pleiotropic mechanism of their action. Moreover, depending on concentration, cell type, and type of the disease natural compounds could exert not only activating effect on autophagy, but also an inhibitory effect. In particular, the well-known and described in this review natural compound resveratrol activates autophagy via several mechanisms, and also initiates apoptosis of cancer cells. Another good example is the compound Kazinol C, which was shown to induce autophagy at low concentrations and apoptosis – at high concentrations. Use of many natural compounds in food supplements and pharmaceutical preparations is limited, because mechanisms of their action are not fully understood, and there are quite a few of side effects.
Despite the large number of studies devoted to natural autophagy activators, it is necessary in future studies to address the issue of elucidation of exact targets of the plant-derived autophagy activators, as well as to search for new compounds with more targeted action. Deeper investigation of novel plant-derived sources seems very promising for the discovery of new natural activators of autophagy, which should involve establishing of exact formulas of the molecules in the plant composition, as well as investigation of the metabolites of these compounds produced by microbiota of the animals resistant to long-term starvation, because starvation is one of the well-known conditions facilitating activation of autophagy in an organism.
It should be noted that autophagy attracts attention not only from the medical point of view, but also as a process, which could be instrumental in extension of human longevity and delay onset of the age-related diseases. In this regard, the compounds activating mitophagy are the most interesting. It is remarkable that several companies already produce biologically active supplements based on natural compounds activating autophagy such as urolithin A (Amazentis, Nestlé Health Science) and spermidine (Longevity labs, Mimio Health) that are intended for improving the state of cells and slow down aging. It must be emphasized that at present there are no companies in Russia that produce food supplements based on autophagy activators. Considering the fact that investigation of autophagy activators becomes more and more popular worldwide, the number of dietary supplements or clinical preparations on their bases introduced to the market would only grow in future. That is why it is very important to develop this research area in Russia in order to establish our own industry of functional food supplements targeting extension of healthy life and, as a consequence, quality of life of humans.
Acknowledgments. The authors are grateful to all research team and to academicians A. G. Gabibov and A. A. Makarov for fruitful discussions.
Contributions. J.A.P. and E.A.G. writing text of the paper, O.A.D. discussion of the material, editing text of the paper, P.V.S. concept and supervision of the study, editing text of the paper.
Funding. This work was financially supported by the company EFKO.
Ethics declarations. This work does not describe any studies involving human participants and animals performed by and of the authors. The authors of this work declare that they have no conflicts of interest.
REFERENCES
1.Shintani, T., and Klionsky, D. J. (2004) Autophagy
in health and disease: a double-edged sword, Science,
306, 990-995, doi: 10.1126/science.1099993.
2.Yang, Q., Wang, R., and Zhu, L. (2019)
Chaperone-mediated autophagy, Adv. Exp. Med. Biol., 1206,
435-452, doi: 10.1007/978-981-15-0602-4_20.
3.Khaminets, A., Behl, C., and Dikic, I. (2016)
Ubiquitin-dependent and independent signals in selective autophagy,
Trends Cell Biol., 26, 6-16, doi:
10.1016/j.tcb.2015.08.010.
4.Kaizuka, T., Morishita, H., Hama, Y., Tsukamoto,
S., Matsui, T., et al. (2016) An autophagic flux probe that releases an
internal control, Mol. Cell, 64, 835-849, doi:
10.1016/j.molcel.2016.09.037.
5.Rhodes, C. H., Zhu, C., Agus, J., Tang, X., Li, Q.,
et al. (2023) Human fasting modulates macrophage function and
upregulates multiple bioactive metabolites that extend lifespan in
Caenorhabditis elegans: a pilot clinical study, Am. J. Clin.
Nutr., 117, 286-297, doi: 10.1016/j.ajcnut.2022.10.015.
6.Madeo, F., Carmona-Gutierrez, D., Hofer, S. J., and
Kroemer, G. (2019) Caloric restriction mimetics against age-associated
disease: targets, mechanisms, and therapeutic potential, Cell
Metab., 29, 592-610, doi: 10.1016/j.cmet.2019.01.018.
7.Mizushima, N., Yoshimori, T., and Levine, B. (2010)
Methods in mammalian autophagy research, Cell, 140,
313-326, doi: 10.1016/j.cell.2010.01.028.
8.Bravo-San Pedro, J. M., Kroemer, G., and Galluzzi,
L. (2017) Autophagy and mitophagy in cardiovascular disease, Circ.
Res., 120, 1812-1824, doi:
10.1161/CIRCRESAHA.117.311082.
9.Georgakopoulos, N. D., Wells, G., and Campanella,
M. (2017) The pharmacological regulation of cellular mitophagy, Nat.
Chem. Biol., 13, 136-146, doi: 10.1038/nchembio.2287.
10.Vaduganathan, M., Mensah, G. A., Turco, J. V.,
Fuster, V., and Roth, G. A. (2022) The global burden of cardiovascular
diseases and risk, J. Am. Coll. Cardiol., 80, 2361-2371,
doi: 10.1016/j.jacc.2022.11.005.
11.Ryu, D., Mouchiroud, L., Andreux, P. A.,
Katsyuba, E., Moullan, N., et al. (2016) Urolithin A induces mitophagy
and prolongs lifespan in C. elegans and increases muscle
function in rodents, Nat. Med., 22, 879-888, doi:
10.1038/nm.4132.
12.Andreux, P. A., Blanco-Bose, W., Ryu, D., Burdet,
F., Ibberson, M., et al. (2019) The mitophagy activator urolithin A is
safe and induces a molecular signature of improved mitochondrial and
cellular health in humans, Nat. Metab., 1, 595-603, doi:
10.1038/s42255-019-0073-4.
13.Yang, S., Liu, T., Hu, C., Li, W., Meng, Y., et
al. (2022) Ginsenoside compound K protects against obesity through
pharmacological targeting of glucocorticoid receptor to activate
lipophagy and lipid metabolism, Pharmaceutics, 14, 1192,
doi: 10.3390/pharmaceutics14061192.
14.Lim, H., and Lee, M.-S. (2018) Amelioration of
obesity-induced diabetes by a novel autophagy enhancer, CST,
2, 181-183, doi: 10.15698/cst2018.07.146.
15.Minami, Y., Hoshino, A., Higuchi, Y., Hamaguchi,
M., Kaneko, Y., et al. (2023) Liver lipophagy ameliorates nonalcoholic
steatohepatitis through extracellular lipid secretion, Nat.
Commun., 14, 4084, doi: 10.1038/s41467-023-39404-6.
16.Jung, J., Park, J., Kim, M., Ha, J., Cho, H., et
al. (2023) SB2301-mediated perturbation of membrane composition in
lipid droplets induces lipophagy and lipid droplets ubiquitination,
Commun. Biol., 6, 300, doi:
10.1038/s42003-023-04682-9.
17.Yerra, V. G., Kalvala, A. K., and Kumar, A.
(2017) Isoliquiritigenin reduces oxidative damage and alleviates
mitochondrial impairment by SIRT1 activation in experimental diabetic
neuropathy, J. Nutr. Biochem., 47, 41-52, doi:
10.1016/j.jnutbio.2017.05.001.
18.Campbell, G. R., and Spector, S. A. (2012)
Vitamin D inhibits human immunodeficiency virus type 1 and
Mycobacterium tuberculosis infection in macrophages through the
induction of autophagy, PLoS Pathog., 8, e1002689, doi:
10.1371/journal.ppat.1002689.
19.Liang, X. H., Jackson, S., Seaman, M., Brown, K.,
Kempkes, B., et al. (1999) Induction of autophagy and inhibition of
tumorigenesis by beclin 1, Nature, 402, 672-676, doi:
10.1038/45257.
20.Marinković, M., Šprung, M.,
Buljubašić, M., and Novak, I. (2018) Autophagy modulation
in cancer: current knowledge on action and therapy, Oxid. Med. Cell.
Longev., 2018, 8023821, doi: 10.1155/2018/8023821.
21.Xie, Z., Lau, K., Eby, B., Lozano, P., He, C., et
al. (2011) Improvement of cardiac functions by chronic metformin
treatment is associated with enhanced cardiac autophagy in diabetic
OVE26 mice, Diabetes, 60, 1770-1778, doi:
10.2337/db10-0351.
22.Wang, B., Yang, Q., Sun, Y., Xing, Y., Wang, Y.,
et al. (2014) Resveratrol-enhanced autophagic flux ameliorates
myocardial oxidative stress injury in diabetic mice, J. Cell. Mol.
Med., 18, 1599-1611, doi: 10.1111/jcmm.12312.
23.Pierzynowska, K., Gaffke, L., Cyske, Z.,
Puchalski, M., Rintz, E., et al. (2018) Autophagy stimulation as a
promising approach in treatment of neurodegenerative diseases,
Metab. Brain. Dis., 33, 989-1008, doi:
10.1007/s11011-018-0214-6.
24.Guseva, E. A., Pavlova, J. A., Dontsova, O. A.,
and Sergiev, P. V. (2024) Synthetic activators of autophagy,
Biochemistry (Moscow), 89, 27-52, doi:
10.1134/S0006297924010024.
25.Li, X., Feng, Y., Wang, X.-X., Truong, D., and
Wu, Y.-C. (2020) The critical role of SIRT1 in Parkinson’s
disease: mechanism and therapeutic considerations, Aging Dis.,
11, 1608-1622, doi: 10.14336/AD.2020.0216.
26.Palmieri, M., Pal, R., Nelvagal, H. R., Lotfi,
P., Stinnett, G. R., et al. (2017) mTORC1-independent TFEB activation
via Akt inhibition promotes cellular clearance in neurodegenerative
storage diseases, Nat. Commun., 8, 14338, doi:
10.1038/ncomms14338.
27.Feldman, M. E., Apsel, B., Uotila, A., Loewith,
R., Knight, Z. A., et al. (2009) Active-site inhibitors of mTOR target
rapamycin-resistant outputs of mTORC1 and mTORC2, PLoS Biol.,
7, e38, doi: 10.1371/journal.pbio.1000038.
28.He, C., and Klionsky, D. J. (2009) Regulation
mechanisms and signaling pathways of autophagy, Annu. Rev.
Genet., 43, 67-93, doi:
10.1146/annurev-genet-102808-114910.
29.Kimura, S., Noda, T., and Yoshimori, T. (2007)
Dissection of the autophagosome maturation process by a novel reporter
protein, tandem fluorescent-tagged LC3, Autophagy, 3,
452-460, doi: 10.4161/auto.4451.
30.Bjørkøy, G., Lamark, T., Brech, A.,
Outzen, H., Perander, M., et al. (2005) p62/SQSTM1 forms protein
aggregates degraded by autophagy and has a protective effect on
huntingtin-induced cell death, J. Cell. Biol., 171,
603-614, doi: 10.1083/jcb.200507002.
31.Guan, Y., Wang, Y., Li, B., Shen, K., Li, Q., et
al. (2021) Mitophagy in carcinogenesis, drug resistance and anticancer
therapeutics, Cancer Cell Int., 21, 350, doi:
10.1186/s12935-021-02065-w.
32.Yang, Y., Li, T., Li, Z., Liu, N., Yan, Y., et
al. (2020) Role of mitophagy in cardiovascular disease, Aging
dis., 11, 419, doi: 10.14336/AD.2019.0518.
33.Jetto, C. T., Nambiar, A., and Manjithaya, R.
(2022) Mitophagy and neurodegeneration: between the knowns and the
unknowns, Front. Cell Dev. Biol., 10, 837337, doi:
10.3389/fcell.2022.837337.
34.Ma, X., McKeen, T., Zhang, J., and Ding, W.-X.
(2020) Role and mechanisms of mitophagy in liver diseases,
Cells, 9, 837, doi: 10.3390/cells9040837.
35.Ke, P.-Y. (2020) Mitophagy in the pathogenesis of
liver diseases, Cells, 9, 831, doi:
10.3390/cells9040831.
36.Liu, H., Dai, C., Fan, Y., Guo, B., Ren, K.,
Tangna Sun, T., and Wang W. (2017) From autophagy to mitophagy: the
roles of P62 in neurodegenerative diseases, J. Bioenerg.
Biomembr., 49, 413-422, doi: 10.1007/s10863-017-9727-7.
37.Zheng, M., Bai, Y., Sun, X., Fu, R., Liu, L., et
al. (2022) Resveratrol reestablishes mitochondrial quality control in
myocardial ischemia/reperfusion injury through
Sirt1/Sirt3-Mfn2-Parkin-PGC-1α pathway, Molecules,
27, 5545, doi: 10.3390/molecules27175545.
38.McLelland, G.-L., Goiran, T., Yi, W., Dorval, G.,
Chen, C. X., et al. (2018) Mfn2 ubiquitination by PINK1/parkin gates
the p97-dependent release of ER from mitochondria to drive mitophagy,
eLife, 7, e32866, doi: 10.7554/eLife.32866.
39.Kopustinskiene, D. M., Jakstas, V., Savickas, A.,
and Bernatoniene, J. (2020) Flavonoids as anticancer agents,
Nutrients, 12, E457, doi: 10.3390/nu12020457.
40.Rodríguez-García, C.,
Sánchez-Quesada, C., and Gaforio, J. J. (2019) Dietary
flavonoids as cancer chemopreventive agents: an updated review of human
studies, Antioxidants (Basel), 8, E137, doi:
10.3390/antiox8050137.
41.Serra, D., Almeida, L. M., and Dinis, T. C. P.
(2020) Polyphenols in the management of brain disorders: Modulation of
the microbiota-gut-brain axis, Adv. Food. Nutr. Res., 91,
1-27, doi: 10.1016/bs.afnr.2019.08.001.
42.Shabbir, U., Rubab, M., Daliri, E. B.-M.,
Chelliah, R., Javed, A., et al. (2021) Curcumin, quercetin, catechins
and metabolic diseases: the role of gut microbiota, Nutrients,
13, 206, doi: 10.3390/nu13010206.
43.Benvenuto, M., Albonici, L., Focaccetti, C.,
Ciuffa, S., Fazi, S., et al. (2020) Polyphenol-mediated autophagy in
cancer: evidence of in vitro and in vivo studies, Int.
J. Mol. Sci., 21, E6635, doi: 10.3390/ijms21186635.
44.Deng, S., Shanmugam, M. K., Kumar, A. P., Yap, C.
T., Sethi, G., et al. (2019) Targeting autophagy using natural
compounds for cancer prevention and therapy, Cancer, 125,
1228-1246, doi: 10.1002/cncr.31978.
45.García-Aguilar, A., Palomino, O., Benito,
M., and Guillén, C. (2021) Dietary polyphenols in metabolic and
neurodegenerative diseases: molecular targets in autophagy and
biological effects, Antioxidants (Basel), 10, 142, doi:
10.3390/antiox10020142.
46.Lewandowska, H., Kalinowska, M., Lewandowski, W.,
Stępkowski, T. M., and Brzóska, K. (2016) The role of
natural polyphenols in cell signaling and cytoprotection against cancer
development, J. Nutr. Biochem., 32, 1-19, doi:
10.1016/j.jnutbio.2015.11.006.
47.Moosavi, M. A., Haghi, A., Rahmati, M.,
Taniguchi, H., Mocan, A., et al. (2018) Phytochemicals as potent
modulators of autophagy for cancer therapy, Cancer Lett.,
424, 46-69, doi: 10.1016/j.canlet.2018.02.030.
48.Pang, X., Zhang, X., Jiang, Y., Su, Q., Li, Q.,
et al. (2021) Autophagy: mechanisms and therapeutic potential of
flavonoids in cancer, Biomolecules, 11, 135, doi:
10.3390/biom11020135.
49.Patra, S., Pradhan, B., Nayak, R., Behera, C.,
Panda, K. C., et al. (2021) Apoptosis and autophagy modulating dietary
phytochemicals in cancer therapeutics: Current evidences and future
perspectives, Phytother Res., 35, 4194-4214, doi:
10.1002/ptr.7082.
50.Zhao, Y., Hu, X., Zuo, X., and Wang, M. (2018)
Chemopreventive effects of some popular phytochemicals on human colon
cancer: a review, Food Funct., 9, 4548-4568, doi:
10.1039/C8FO00850G.
51.Espín, J. C., Larrosa, M.,
García-Conesa, M. T., and Tomás-Barberán, F.
(2013) Biological significance of urolithins, the gut microbial ellagic
acid-derived metabolites: the evidence so far, Evid. Based
Complement Alternat. Med., 2013, 270418, doi:
10.1155/2013/270418.
52.Sallam, I. E., Abdelwareth, A., Attia, H., Aziz,
R. K., Homsi, M. N., et al. (2021) Effect of gut microbiota
biotransformation on dietary tannins and human health implications,
Microorganisms, 9, 965, doi:
10.3390/microorganisms9050965.
53.Beltrán, D., Romo-Vaquero, M.,
Espín, J. C., Tomás-Barberán, F. A., and Selma, M.
V. (2018) Ellagibacter isourolithinifaciens gen. nov., sp. nov.,
a new member of the family Eggerthellaceae, isolated from human gut,
Int. J. Syst. Evol. Microbiol., 68, 1707-1712, doi:
10.1099/ijsem.0.002735.
54.Selma, M. V., Beltrán, D.,
García-Villalba, R., Espín, J. C., and
Tomás-Barberán, F. A. (2014) Description of urolithin
production capacity from ellagic acid of two human intestinal
Gordonibacter species, Food Funct., 5, 1779-1784,
doi: 10.1039/C4FO00092G.
55.Tomás-Barberán, F. A.,
García-Villalba, R., González-Sarrías, A., Selma,
M. V., and Espín, J. C. (2014) Ellagic acid metabolism by human
gut microbiota: consistent observation of three urolithin phenotypes in
intervention trials, independent of food source, age, and health
status, J. Agric. Food. Chem., 62, 6535-6538, doi:
10.1021/jf5024615.
56.Rafii, F. (2015) The role of colonic bacteria in
the metabolism of the natural isoflavone daidzin to equol,
Metabolites, 5, 56-73, doi: 10.3390/metabo5010056.
57.Stevens, J. F., and Maier, C. S. (2016) The
chemistry of gut microbial metabolism of polyphenols, Phytochem.
Rev., 15, 425-444, doi: 10.1007/s11101-016-9459-z.
58.Kurtz, C. C., Otis, J. P., Regan, M. D., and
Carey, H. V. (2021) How the gut and liver hibernate, Comp. Biochem.
Physiol. A Mol. Integr. Physiol., 253, 110875, doi:
10.1016/j.cbpa.2020.110875.
59.Martens, E. C., Chiang, H. C., and Gordon, J. I.
(2008) Mucosal glycan foraging enhances fitness and transmission of a
saccharolytic human gut bacterial symbiont, Cell Host Microbe,
4, 447-457, doi: 10.1016/j.chom.2008.09.007.
60.Sonnenburg, J. L., Xu, J., Leip, D. D., Chen,
C.-H., Westover, B. P., et al. (2005) Glycan foraging in vivo by an
intestine-adapted bacterial symbiont, Science, 307,
1955-1959, doi: 10.1126/science.1109051.
61.Bergman, E. N. (1990) Energy contributions of
volatile fatty acids from the gastrointestinal tract in various
species, Physiol. Rev., 70, 567-590, doi:
10.1152/physrev.1990.70.2.567.
62.Iannucci, L. F., Sun, J., Singh, B. K., Zhou, J.,
Kaddai, V. A., et al. (2016) Short chain fatty acids induce
UCP2-mediated autophagy in hepatic cells, Biochem. Biophys. Res.
Commun., 480, 461-467, doi: 10.1016/j.bbrc.2016.10.072.
63.Russo, M., and Russo, G. L. (2018) Autophagy
inducers in cancer, Biochem. Pharmacol., 153, 51-61, doi:
10.1016/j.bcp.2018.02.007.
64.Scarlatti, F., Maffei, R., Beau, I., Codogno, P.,
and Ghidoni, R. (2008) Role of non-canonical Beclin 1-independent
autophagy in cell death induced by resveratrol in human breast cancer
cells, Cell Death Differ., 15, 1318-1329, doi:
10.1038/cdd.2008.51.
65.Cai, H., Scott, E., Kholghi, A., Andreadi, C.,
Rufini, A., et al. (2015) Cancer chemoprevention: evidence of a
nonlinear dose response for the protective effects of resveratrol in
humans and mice, Sci. Transl. Med., 7, 298ra117, doi:
10.1126/scitranslmed.aaa7619.
66.Ferraresi, A., Titone, R., Follo, C.,
Castiglioni, A., Chiorino, G., et al. (2017) The protein restriction
mimetic Resveratrol is an autophagy inducer stronger than amino acid
starvation in ovarian cancer cells, Mol. Carcinog., 56,
2681-2691, doi: 10.1002/mc.22711.
67.Tang, Q., Li, G., Wei, X., Zhang, J., Chiu,
J.-F., et al. (2013) Resveratrol-induced apoptosis is enhanced by
inhibition of autophagy in esophageal squamous cell carcinoma,
Cancer. Lett., 336, 325-337, doi:
10.1016/j.canlet.2013.03.023.
68.Fukuda, T., Oda, K., Wada-Hiraike, O., Sone, K.,
Inaba, K., et al. (2016) Autophagy inhibition augments
resveratrol-induced apoptosis in Ishikawa endometrial cancer cells,
Oncol. Lett., 12, 2560-2566, doi:
10.3892/ol.2016.4978.
69.Wang, J., Li, J., Cao, N., Li, Z., Han, J., et
al. (2018) Resveratrol, an activator of SIRT1, induces protective
autophagy in non-small-cell lung cancer via inhibiting Akt/mTOR and
activating p38-MAPK, Onco Targets Ther., 11, 7777-7786,
doi: 10.2147/OTT.S159095.
70.Bode, L. M., Bunzel, D., Huch, M., Cho, G.-S.,
Ruhland, D., et al. (2013) In vivo and in vitro
metabolism of trans-resveratrol by human gut microbiota, Am. J.
Clin. Nutr., 97, 295-309, doi: 10.3945/ajcn.112.049379.
71.Jarosova, V., Vesely, O., Marsik, P., Jaimes, J.
D., Smejkal, K., et al. (2019) Metabolism of stilbenoids by human
faecal microbiota, Molecules, 24, E1155, doi:
10.3390/molecules24061155.
72.Luca, S. V., Macovei, I., Bujor, A., Miron, A.,
Skalicka-Woźniak, K., et al. (2020) Bioactivity of dietary
polyphenols: the role of metabolites, Crit. Rev. Food. Sci.
Nutr., 60, 626-659, doi: 10.1080/10408398.2018.1546669.
73.Kasi, P. D., Tamilselvam, R.,
Skalicka-Woźniak, K., Nabavi, S. F., Daglia, M., et al. (2016)
Molecular targets of curcumin for cancer therapy: an updated review,
Tumour Biol., 37, 13017-13028, doi:
10.1007/s13277-016-5183-y.
74.Fu, H., Wang, C., Yang, D., Wei, Z., Xu, J., et
al. (2018) Curcumin regulates proliferation, autophagy, and apoptosis
in gastric cancer cells by affecting PI3K and P53 signaling, J.
Cell. Physiol., 233, 4634-4642, doi: 10.1002/jcp.26190.
75.Yang, C., Ma, X., Wang, Z., Zeng, X., Hu, Z., et
al. (2017) Curcumin induces apoptosis and protective autophagy in
castration-resistant prostate cancer cells through iron chelation,
Drug Des. Devel. Ther., 11, 431-439, doi:
10.2147/DDDT.S126964.
76.Xu, Y., Wu, Y., Wang, L., Qian, C., Wang, Q., et
al. (2020) Identification of curcumin as a novel natural inhibitor of
rDNA transcription, Cell Cycle, 19, 3362-3374, doi:
10.1080/15384101.2020.1843817.
77.Xiao, K., Jiang, J., Guan, C., Dong, C., Wang,
G., et al. (2013) Curcumin induces autophagy via activating the AMPK
signaling pathway in lung adenocarcinoma cells, J. Pharmacol.
Sci., 123, 102-109, doi: 10.1254/jphs.13085FP.
78.Bi, Y., Shen, C., Li, C., Liu, Y., Gao, D., et
al. (2016) Inhibition of autophagy induced by quercetin at a late stage
enhances cytotoxic effects on glioma cells, Tumour. Biol.,
37, 3549-3560, doi: 10.1007/s13277-015-4125-4.
79.Heger, M., van Golen, R. F., Broekgaarden, M.,
and Michel, M. C. (2014) The molecular basis for the pharmacokinetics
and pharmacodynamics of curcumin and its metabolites in relation to
cancer, Pharmacol. Rev., 66, 222-307, doi:
10.1124/pr.110.004044.
80.Jabczyk, M., Nowak, J., Hudzik, B., and
Zubelewicz-Szkodzińska, B. (2021) Curcumin and its potential
impact on microbiota, Nutrients, 13, 2004, doi:
10.3390/nu13062004.
81.Scazzocchio, B., Minghetti, L., and
D’Archivio, M. (2020) Interaction between gut microbiota and
curcumin: a new key of understanding for the health effects of
curcumin, Nutrients, 12, 2499, doi:
10.3390/nu12092499.
82.Shen, L., and Ji, H.-F. (2019) Bidirectional
interactions between dietary curcumin and gut microbiota, Crit. Rev.
Food Sci. Nutr., 59, 2896-2902, doi:
10.1080/10408398.2018.1478388.
83.Zam, W. (2018) Gut microbiota as a prospective
therapeutic target for curcumin: a review of mutual influence, J.
Nutr. Metab., 2018, 1367984, doi: 10.1155/2018/1367984.
84.Song, G., Lu, H., Chen, F., Wang, Y., Fan, W., et
al. (2018) Tetrahydrocurcumin-induced autophagy via suppression of
PI3K/Akt/mTOR in nonsmall cell lung carcinoma cells, Mol. Med.
Rep., 17, 5964-5969, doi: 10.3892/mmr.2018.8600.
85.Wu, J.-C., Lai, C.-S., Badmaev, V.,
Nagabhushanam, K., Ho, C.-T., et al. (2011) Tetrahydrocurcumin, a major
metabolite of curcumin, induced autophagic cell death through
coordinative modulation of PI3K/Akt-mTOR and MAPK signaling pathways in
human leukemia HL-60 cells, Mol. Nutr. Food Res., 55,
1646-1654, doi: 10.1002/mnfr.201100454.
86.Russo, M., Milito, A., Spagnuolo, C., Carbone,
V., Rosén, A., et al. (2017) CK2 and PI3K are direct molecular
targets of quercetin in chronic lymphocytic leukaemia,
Oncotarget, 8, 42571-42587, doi:
10.18632/oncotarget.17246.
87.Wang, K., Liu, R., Li, J., Mao, J., Lei, Y., et
al. (2011) Quercetin induces protective autophagy in gastric cancer
cells: involvement of Akt-mTOR- and hypoxia-induced factor
1α-mediated signaling, Autophagy, 7, 966-978, doi:
10.4161/auto.7.9.15863.
88.Moon, J.-H., Eo, S. K., Lee, J. H., and Park,
S.-Y. (2015) Quercetin-induced autophagy flux enhances TRAIL-mediated
tumor cell death, Oncol. Rep., 34, 375-381, doi:
10.3892/or.2015.3991.
89.Guo, H., Ding, H., Tang, X., Liang, M., Li, S.,
et al. (2021) Quercetin induces pro-apoptotic autophagy via SIRT1/AMPK
signaling pathway in human lung cancer cell lines A549 and H1299 in
vitro, Thoracic. Cancer, 12, 1415-1422, doi:
10.1111/1759-7714.13925.
90.Wu, Q., Tian, A.-L., Durand, S., Aprahamian, F.,
Nirmalathasan, N., et al. (2020) Isobacachalcone induces autophagy and
improves the outcome of immunogenic chemotherapy, Cell Death
Dis., 11, 1015, doi: 10.1038/s41419-020-03226-x.
91.Xu, P., Chen, Q., Chen, X., Qi, H., Yang, Y., et
al. (2023) Morusin and mulberrin extend the lifespans of yeast and
C. elegans via suppressing nutrient-sensing pathways,
GeroScience, 45, 949-964, doi:
10.1007/s11357-022-00693-2.
92.Zhou, X., Yue, G. G.-L., Chan, A. M.-L., Tsui, S.
K.-W., Fung, K.-P., et al. (2017) Eriocalyxin B, a novel autophagy
inducer, exerts anti-tumor activity through the suppression of
Akt/mTOR/p70S6K signaling pathway in breast cancer, Biochem.
Pharmacol., 142, 58-70, doi: 10.1016/j.bcp.2017.06.133.
93.Sun, J., Feng, Y., Wang, Y., Ji, Q., Cai, G., et
al. (2019) α-Hederin induces autophagic cell death in colorectal
cancer cells through reactive oxygen species dependent AMPK/mTOR
signaling pathway activation, Int. J. Oncol., 54,
1601-1612, doi: 10.3892/ijo.2019.4757.
94.Shin, S., Jing, K., Jeong, S., Kim, N., Song,
K.-S., et al. (2013) The omega-3 polyunsaturated fatty acid DHA induces
simultaneous apoptosis and autophagy via mitochondrial ROS-mediated
Akt-mTOR signaling in prostate cancer cells expressing mutant p53,
Biomed. Res. Int., 2013, 568671, doi:
10.1155/2013/568671.
95.Prajapat, S. K., Subramani, C., Sharma, P.,
Vrati, S., and Kalia, M. (2022) Avobenzone, Guaiazulene and Tioxolone
identified as potent autophagy inducers in a high-throughput image
based screen for autophagy flux, Autophagy Rep., 1,
523-536, doi: 10.1080/27694127.2022.2132075.
96.Ortiz, M. I., Fernández-Martínez,
E., Soria-Jasso, L. E., Lucas-Gómez, I.,
Villagómez-Ibarra, R., et al. (2016) Isolation, identification
and molecular docking as cyclooxygenase (COX) inhibitors of the main
constituents of Matricaria chamomilla L. extract and its
synergistic interaction with diclofenac on nociception and gastric
damage in rats, Biomed. Pharmacother., 78, 248-256, doi:
10.1016/j.biopha.2016.01.029.
97.Ye, Q., Zhou, L., Jin, P., Li, L., Zheng, S., et
al. (2021) Guaiazulene triggers ROS-induced apoptosis and protective
autophagy in non-small cell lung cancer, Front. Pharmacol.,
12, 621181, doi: 10.3389/fphar.2021.621181.
98.Reinke, A., Chen, J. C.-Y., Aronova, S., and
Powers, T. (2006) Caffeine targets TOR complex I and provides evidence
for a regulatory link between the FRB and kinase domains of Tor1p,
J. Biol. Chem., 281, 31616-31626, doi:
10.1074/jbc.M603107200.
99.Saiki, S., Sasazawa, Y., Imamichi, Y., Kawajiri,
S., Fujimaki, T., et al. (2011) Caffeine induces apoptosis by
enhancement of autophagy via PI3K/Akt/mTOR/p70S6K inhibition,
Autophagy, 7, 176-187, doi: 10.4161/auto.7.2.14074.
100.Wanke, V., Cameroni, E., Uotila, A., Piccolis,
M., Urban, J., et al. (2008) Caffeine extends yeast lifespan by
targeting TORC1, Mol. Microbiol., 69, 277-285, doi:
10.1111/j.1365-2958.2008.06292.x.
101.Sinha, R. A., Farah, B. L., Singh, B. K.,
Siddique, M. M., Li, Y., et al. (2014) Caffeine stimulates hepatic
lipid metabolism by the autophagy-lysosomal pathway in mice,
Hepatology, 59, 1366-1380, doi: 10.1002/hep.26667.
102.Moon, J.-H., Lee, J.-H., Park, J.-Y., Kim,
S.-W., Lee, Y.-J., et al. (2014) Caffeine prevents human prion
protein-mediated neurotoxicity through the induction of autophagy,
Int. J. Mol. Med., 34, 553-558, doi:
10.3892/ijmm.2014.1814.
103.Pietrocola, F., Malik, S. A., Mariño,
G., Vacchelli, E., Senovilla, L., et al. (2014) Coffee induces
autophagy in vivo, Cell Cycle, 13, 1987-1994, doi:
10.4161/cc.28929.
104.Freedman, N. D., Park, Y., Abnet, C. C.,
Hollenbeck, A. R., and Sinha, R. (2012) Association of coffee drinking
with total and cause-specific mortality, N. Engl. J. Med.,
366, 1891-1904, doi: 10.1056/NEJMoa1112010.
105.Sun, C., Zhang, J., Hou, J., Hui, M., Qi, H.,
et al. (2023) Induction of autophagy via the PI3K/Akt/mTOR signaling
pathway by Pueraria flavonoids improves non-alcoholic fatty liver
disease in obese mice, Biomed. Pharmacother., 157,
114005, doi: 10.1016/j.biopha.2022.114005.
106.Kang, A.-W., Sun, C., Li, H.-T., Zhong, K.,
Zeng, X.-H., et al. (2023) Puerarin extends the lifespan of
Drosophila melanogaster by activating autophagy, Food
Funct., 14, 2149-2161, doi: 10.1039/D2FO02800J.
107.Wu, Q., Tian, A.-L., Li, B., Leduc, M.,
Forveille, S., et al. (2021) IGF1 receptor inhibition amplifies the
effects of cancer drugs by autophagy and immune-dependent mechanisms,
J. Immunother. Cancer, 9, e002722, doi:
10.1136/jitc-2021-002722.
108.Honma, Y., Sato-Morita, M., Katsuki, Y.,
Mihara, H., Baba, R., et al. (2018) Trehalose activates autophagy and
decreases proteasome inhibitor-induced endoplasmic reticulum stress and
oxidative stress-mediated cytotoxicity in hepatocytes, Hepatol.
Res., 48, 94-105, doi: 10.1111/hepr.12892.
109.Meier, J. L., and Grose, C. (2017) Variable
effects of autophagy induction by trehalose on herpesviruses depending
on conditions of infection, Yale J. Biol. Med., 90,
25-33.
110.Chen, X., Li, M., Li, L., Xu, S., Huang, D., et
al. (2016) Trehalose, sucrose and raffinose are novel activators of
autophagy in human keratinocytes through an mTOR-independent pathway,
Sci. Rep., 6, 28423, doi: 10.1038/srep28423.
111.He, Q., Koprich, J. B., Wang, Y., Yu, W., Xiao,
B., et al. (2016) Treatment with trehalose prevents behavioral and
neurochemical deficits produced in an AAV α-synuclein rat model
of Parkinson’s disease, Mol. Neurobiol., 53,
2258-2268, doi: 10.1007/s12035-015-9173-7.
112.Krüger, U., Wang, Y., Kumar, S., and
Mandelkow, E.-M. (2012) Autophagic degradation of tau in primary
neurons and its enhancement by trehalose, Neurobiol. Aging,
33, 2291-2305, doi: 10.1016/j.neurobiolaging.2011.11.009.
113.Aguib, Y., Heiseke, A., Gilch, S., Riemer, C.,
Baier, M., et al. (2009) Autophagy induction by trehalose counteracts
cellular prion infection, Autophagy, 5, 361-369, doi:
10.4161/auto.5.3.7662.
114.Wada, S., Kubota, Y., Sawa, R., Umekita, M.,
Hatano, M., et al. (2015) Novel autophagy inducers lentztrehaloses A, B
and C, J. Antibiot. (Tokyo), 68, 521-529, doi:
10.1038/ja.2015.23.
115.Campbell, G. R., and Spector, S. A. (2011)
Hormonally active vitamin D3 (1alpha,25-dihydroxycholecalciferol)
triggers autophagy in human macrophages that inhibits HIV-1 infection,
J. Biol. Chem., 286, 18890-18902, doi:
10.1074/jbc.M110.206110.
116.Høyer-Hansen, M., Bastholm, L.,
Mathiasen, I. S., Elling, F., and Jäättelä, M. (2005)
Vitamin D analog EB1089 triggers dramatic lysosomal changes and
Beclin-1 mediated autophagic cell death, Cell Death Differ.,
12, 1297-1309, doi: 10.1038/sj.cdd.4401651.
117.Høyer-Hansen, M., Nordbrandt, S. P. S.,
and Jäättelä, M. (2010) Autophagy as a basis for the
health-promoting effects of vitamin D, Trends Mol. Med.,
16, 295-302, doi: 10.1016/j.molmed.2010.04.005.
118.Lee, Y., Kwon, J., Jeong, J. H., Ryu, J.-H.,
and Kim, K. I. (2022) Kazinol C from Broussonetia kazinoki
stimulates autophagy via endoplasmic reticulum stress-mediated
signaling, Anim. Cells Syst., 26, 28-36, doi:
10.1080/19768354.2021.2023628.
119.Lan, F., Cacicedo, J. M., Ruderman, N., and
Ido, Y. (2008) SIRT1 modulation of the acetylation status, cytosolic
localization, and activity of LKB1, J. Biol. Chem., 283,
27628-27635, doi: 10.1074/jbc.M805711200.
120.DiNicolantonio, J. J., McCarty, M. F., Assanga,
S. I., Lujan, L. L., and O’Keefe, J. H. (2022) Ferulic acid and
berberine, via Sirt1 and AMPK, may act as cell cleansing promoters of
healthy longevity, Open Heart, 9, e001801, doi:
10.1136/openhrt-2021-001801.
121.Guo, P., Wang, P., Liu, L., Wang, P., Lin, G.,
et al. (2023) Naringin alleviates glucose-induced aging by reducing fat
accumulation and promoting autophagy in Caenorhabditis elegans,
Nutrients, 15, 907, doi: 10.3390/nu15040907.
122.Salama, A. A. A., Yassen, N. N., and Mansour,
H. M. (2023) Naringin protects mice from D-galactose-induced lung aging
and mitochondrial dysfunction: implication of SIRT1 pathways, Life
Sci., 324, 121471, doi: 10.1016/j.lfs.2023.121471.
123.Kepp, O., Chen, G., Carmona-Gutierrez, D.,
Madeo, F., and Kroemer, G. (2020) A discovery platform for the
identification of caloric restriction mimetics with broad
health-improving effects, Autophagy, 16, 188-189, doi:
10.1080/15548627.2019.1688984.
124.Liu, M., Li, N., Lu, X., Shan, S., Gao, X., et
al. (2022) Sweet tea (Rubus Suavissmus S. Lee) polysaccharides
promote the longevity of Caenorhabditis elegans through
autophagy-dependent insulin and mitochondrial pathways, Int. J.
Biol. Macromol., 207, 883-892, doi:
10.1016/j.ijbiomac.2022.03.138.
125.Espín, J. C.,
González-Sarrías, A., and Tomás-Barberán,
F. A. (2017) The gut microbiota: a key factor in the therapeutic
effects of (poly)phenols, Biochem. Pharmacol., 139,
82-93, doi: 10.1016/j.bcp.2017.04.033.
126.Palikaras, K., Daskalaki, I., Markaki, M., and
Tavernarakis, N. (2017) Mitophagy and age-related pathologies:
Development of new therapeutics by targeting mitochondrial turnover,
Pharmacol. Ther., 178, 157-174, doi:
10.1016/j.pharmthera.2017.04.005.
127.Heilman, J., Andreux, P., Tran, N., Rinsch, C.,
and Blanco-Bose, W. (2017) Safety assessment of Urolithin A, a
metabolite produced by the human gut microbiota upon dietary intake of
plant derived ellagitannins and ellagic acid, Food Chem.
Toxicol., 108, 289-297, doi: 10.1016/j.fct.2017.07.050.
128.Singh, A., Andreux, P., Blanco-Bose, W., Ryu,
D., Aebischer, P., et al. (2017) Orally administered urolithin a is
safe and modulates muscle and mitochondrial biomarkers in elderly,
Innov. Aging, 1, 1223-1224, doi:
10.1093/geroni/igx004.4446.
129.Muñoz-Esparza, N. C., Latorre-Moratalla,
M. L., Comas-Basté, O., Toro-Funes, N., Veciana-Nogués,
M. T., et al. (2019) Polyamines in food, Front. Nutr., 6,
108, doi: 10.3389/fnut.2019.00108.
130.Eisenberg, T., Abdellatif, M., Schroeder, S.,
Primessnig, U., Stekovic, S., et al. (2016) Cardioprotection and
lifespan extension by the natural polyamine spermidine, Nat.
Med., 22, 1428-1438, doi: 10.1038/nm.4222.
131.Schroeder, S., Hofer, S. J., Zimmermann, A.,
Pechlaner, R., Dammbrueck, C., et al. (2021) Dietary spermidine
improves cognitive function, Cell Rep., 35, 108985, doi:
10.1016/j.celrep.2021.108985.
132.Schwarz, C., Stekovic, S., Wirth, M., Benson,
G., Royer, P., et al. (2018) Safety and tolerability of spermidine
supplementation in mice and older adults with subjective cognitive
decline, Aging, 10, 19-33, doi:
10.18632/aging.101354.
133.Pietrocola, F., Lachkar, S., Enot, D. P.,
Niso-Santano, M., Bravo-San Pedro, J. M., et al. (2015) Spermidine
induces autophagy by inhibiting the acetyltransferase EP300, Cell
Death Differ., 22, 509-516, doi: 10.1038/cdd.2014.215.
134.Yuan, X., Tian, G. G., Pei, X., Hu, X., and Wu,
J. (2021) Spermidine induces cytoprotective autophagy of female
germline stem cells in vitro and ameliorates aging caused by oxidative
stress through upregulated sequestosome-1/p62 expression, Cell
Biosci., 11, 107, doi: 10.1186/s13578-021-00614-4.
135.Baek, A. R., Hong, J., Song, K. S., Jang, A.
S., Kim, D. J., et al. (2020) Spermidine attenuates bleomycin-induced
lung fibrosis by inducing autophagy and inhibiting endoplasmic
reticulum stress (ERS)-induced cell death in mice, Exp. Mol.
Med., 52, 2034-2045, doi: 10.1038/s12276-020-00545-z.
136.Messerer, J., Wrede, C., Schipke, J.,
Brandenberger, C., Abdellatif, M., et al. (2023) Spermidine
supplementation influences mitochondrial number and morphology in the
heart of aged mice, J. Anat., 242, 91-101, doi:
10.1111/joa.13618.
137.Til, H. P., Falke, H. E., Prinsen, M. K., and
Willems, M. I. (1997) Acute and subacute toxicity of tyramine,
spermidine, spermine, putrescine and cadaverine in rats, Food Chem.
Toxicol., 35, 337-348, doi:
10.1016/S0278-6915(97)00121-X.
138.Srivastava, V., Zelmanovich, V., Shukla, V.,
Abergel, R., Cohen, I., et al. (2023) Distinct designer diamines
promote mitophagy, and thereby enhance healthspan in C. elegans
and protect human cells against oxidative damage, Autophagy,
19, 474-504, doi: 10.1080/15548627.2022.2078069.
139.Hawrysh, P. J., Gao, J., Tan, S., Oh, A.,
Nodwell, J., et al. (2023) PRKN/parkin-mediated mitophagy is induced by
the probiotics Saccharomyces boulardii and Lactococcus
lactis, Autophagy, 19, 2094-2110, doi:
10.1080/15548627.2023.2172873.
140.Fedotova, E. I., Dolgacheva, L. P., Abramov, A.
Y., and Berezhnov, A. V. (2022) Lactate and pyruvate activate autophagy
and mitophagy that protect cells in toxic model of Parkinson’s
disease, Mol. Neurobiol., 59, 177-190, doi:
10.1007/s12035-021-02583-8.
141.Xu, C., Wu, Y., Tang, L., Liang, Y., and Zhao,
Y. (2023) Protective effect of cistanoside A on dopaminergic neurons in
Parkinson’s disease via mitophagy, Biotechnol. Appl.
Biochem., 70, 268-280, doi: 10.1002/bab.2350.
142.Kang, H. T., and Hwang, E. S. (2009)
Nicotinamide enhances mitochondria quality through autophagy activation
in human cells, Aging Cell, 8, 426-438, doi:
10.1111/j.1474-9726.2009.00487.x.
143.Zhang, L., Zhang, X., Zhang, T., Guo, Y., Pei,
W., et al. (2023) Linolenic acid ameliorates sarcopenia in C.
elegans by promoting mitophagy and fighting oxidative stress,
Food Funct., 14, 1498-1509, doi: 10.1039/D2FO02974J.
144.Li, S., Liu, M., Chen, J., Chen, Y., Yin, M.,
et al. (2023) L‐carnitine alleviates cardiac microvascular
dysfunction in diabetic cardiomyopathy by enhancing PINK1‐Parkin
‐dependent mitophagy through the CPT1a‐PHB2‐PARL
pathways, Acta Physiol., 238, e13975, doi:
10.1111/apha.13975.
145.D’Amico, D., Olmer, M., Fouassier, A. M.,
Valdés, P., Andreux, P. A., et al. (2022) Urolithin A improves
mitochondrial health, reduces cartilage degeneration, and alleviates
pain in osteoarthritis, Aging Cell, 21, e13662, doi:
10.1111/acel.13662.
146.Singh, A., D’Amico, D., Andreux, P. A.,
Dunngalvin, G., Kern, T., et al. (2022) Direct supplementation with
Urolithin A overcomes limitations of dietary exposure and gut
microbiome variability in healthy adults to achieve consistent levels
across the population, Eur. J. Clin. Nutr., 76, 297-308,
doi: 10.1038/s41430-021-00950-1.