MicroRNA in Schizophrenia: Genetic and Expression Analysis of miR-130b (22q11)
O. A. Burmistrova1,2, A. Y. Goltsov2,3, L. I. Abramova2, V. G. Kaleda2, V. A. Orlova2, and E. I. Rogaev1,2,4*
1Research Center of Mental Health, Russian Academy Medical Sciences, Zagorodnoe Shosse 2/2, 113152 Moscow, Russia2Department of Psychiatry, Brudnick Neuropsychiatric Research Institute, University of Massachusetts Medical School, 303 Belmont Street, Worcester, MA 01604, USA; E-mail: Evgeny.Rogaev@umassmed.edu
3Faculty of Bioengineering and Bioinformatics, Lomonosov Moscow State University, 119992 Moscow, Russia
4Vavilov Institute of General Genetics, Russian Academy of Sciences, 119991 Moscow, Russia
* To whom correspondence should be addressed.
Received April 9, 2007
MicroRNAs (miRNAs) are a class of small regulatory RNAs that control a level of expression of protein encoding genes. Their role in brain pathologies is unknown. We made a first attempt to carry out a genetic study coupled with gene expression analysis of microRNA in human neuropsychiatric pathology. Presumably, at least one third of known miRNA genes are expressed in the brain. Mutations disrupting MECP2 protein lead to abnormal development of the brain and resulting behavior. MiR-130b expressed in the brain and potentially targeting MECP2 is located in the susceptibility locus for schizophrenia (22q11). We performed a comparative analysis of the expression of miR-130b in 24 brain neocortex samples from schizophrenic and normal individuals. The stability and effective detection of mature microRNA in postmortem tissues using Real-time PCR have been shown. Screening for mutations has identified a population polymorphism in the 5´-upstream miR-130b gene region containing DNA elements for putative transcription factors. Genetic association analysis of 300 schizophrenia and 316 normal control individuals revealed no statistically significant association of any of the miR-130b allelic variants with schizophrenia. The data demonstrated feasibility and perspective of convergent genetic and expression analysis of human microRNA genes in testing their role in human diseases.
KEY WORDS: microRNA, miR-130b, gene expression, genetic polymorphism, schizophrenia, populations, brainDOI: 10.1134/S0006297907050161
Abbreviations: miRNAs) microRNAs.
MicroRNAs (miRNAs) are a class of 19-22-nt regulatory RNAs that are
products of small non-protein-coding genes found in animals and plants
[1]. MiRNAs are thought to promote degradation of
target mRNAs or suppress translation of corresponding protein by
forming an imperfect duplex with the RNA target. Evolutionary
conservation of many miRNA genes and mutation analysis in invertebrates
and in zebrafish demonstrated their important regulatory role in
development [2, 3]. Using a
hybridization approach with micro-chip technology we have determined
that microRNA is relatively stable in the postmortem-brain and
hypothesized that the role of small non-coding RNA in human brain
development and neuropsychiatric disorders may be tested by genetic and
gene expression analysis [4]. In this study the
genetic and expression analysis of miR-130b gene was conducted in
schizophrenia and normal individuals.
MATERIALS AND METHODS
Selection of miRNA for DNA polymorphism analysis. The choice of miRNA that target 3´-UTR of MECP2 and the miRNA:target duplex prediction were made using the algorithm described in [5]. PCR primers were designed (Primer3; http://www-genome/wi/mit/edu/cgi-bin/primer/primer3www/cgi/) to amplify the precursor miRNA with flanking sequences. For example, for miR-130b we developed PCR with miR130b1.dir 5-tgctctgagaagcagtgcaa-3´ and miR130b1.rev 5´-tcagaagctggggaggtct-3´ primer oligonucleotides resulting in ~600 bp PCR product. The following primers were used for sequencing: miR130b2.dir 5´-tacccaattcgctcccttct-3´, miR130b2.rev 5´-agctctgcacccacctgat-3´. The sequencing reaction was done using ABI BigDye terminators and analyzed on ABI PRISM 310 Genetic Analyzer.
Genotyping. The case-control samples were collected as previously described. The schizophrenia samples from unrelated individuals were mainly from patients with a diagnosis of paranoid schizophrenia. The diagnosis was made in accordance with International Classification of Diseases 10 (ICD-10) criteria. The control group, consisting of age-and gender-matched subjects with no registered psychiatric illnesses, was collected from the same ethnic and population group of Russian origin. DNA was extracted from peripheral venous blood by the phenol-chloroform method.
Restriction fragment length polymorphism analysis was developed for genotyping of the case-control groups. MiR130b2.dir and miR130b2.rev primers were used for PCR amplification. The PCR was conducted for 30 cycles each consisting of denaturation at 95°C for 30 sec, annealing at 60°C for 45 sec, and extension at 72°C for 40 sec. The PCR product was digested by FauI restriction enzyme. The resulting restriction fragments were analyzed by electrophoresis in 10% polyacrylamide gel. Statistical analysis was performed using CLUMP software (www.mds.qmw.ac.uk/statgen/dcurtis/software.html).
MiRNA microarray analysis. Total RNA was isolated from parietal cortex (BA7) (Stanley Foundation Medical Research Institute collection) with Trizol Reagent (Invitrogen, USA) and then pooled. The procedures for small RNA enrichment using mirVana miRNA isolation kit (Ambion), human chips containing probes for mature miRNAs (LC Science) and Applied Biosystem (USA) TaqMan MicroRNA assay were used for analysis of miRNAs.
RESULTS
In this pilot study, the miRNAs potentially targeting MECP2 mRNA were selected. The MECP2 gene mutations cause disorders with autistic condition (Rett syndrome) and the contribution of MECP2 gene variation was implicated in the case of schizophrenia [6]. It is intriguing that miR-130b is located in the 22q11 locus. This chromosomal region is among the most consistently reported susceptibility locus for schizophrenia identified via linkage studies. Interstitial deletions of chromosome 22q11 also cause velo-cardio-facial syndrome (VCFS) with a high risk for schizophrenia and depression [7-9]. The 3´UTR of the MECP2 gene was searched for complementarity with 7-nucleotide seed region (bases 2-8) of the miRNAs [5]. The structure of potential duplex was predicted using RNAfold with artificial loop attached to the 3´ end of the seed match in target and 5´ end in miRNA. The putative targets for miR-130b, miR-130a, miR-19a, and miR-19b were detected.
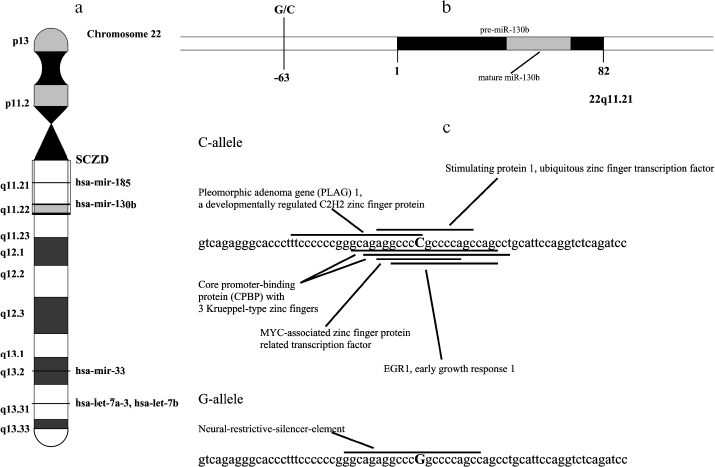
We have screened for population polymorphisms in or nearby these miRNA genes by direct sequencing analysis of five schizophrenia and five normal individuals. The genomic region of about 600 bp that includes the complete sequence of 71-96 bp precursor miRNA was amplified for the sequencing analysis. We found no mutations in the sequence corresponding to mature or precursor miRNA in these genes. However, common single nucleotide polymorphism (C/G) was found at the -63 bp position from the start of the precursor miR-130b (rs861843) (Fig. 1, a and b). Interestingly, the common C allele sequence hits multiple putative regulatory DNA elements for putative transcriptional factors, whereas G mutation disrupts these matches, but generates novel element for neural-restrictive-silencer (Fig. 1c).
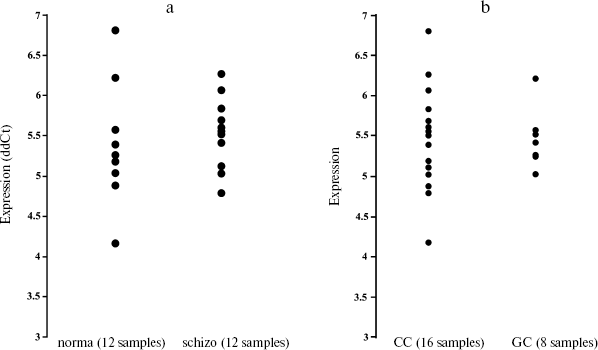
To test whether the variation in 5´-region in miR-130b may directly contribute to a genetic risk for schizophrenia, we analyzed the age- and gender-matched groups of schizophrenia and normal individuals. Three hundreds schizophrenic patients and 316 control individuals from Moscow (Russia) were collected and used for the case-control association study as described previously [10]. All patients in this study met criteria for paranoid schizophrenia (F20.0x) according to the International Classification of Diseases 10 (ICD-10). DNA was extracted from peripheral venous blood samples using the phenol-chloroform method. Direct and reverse primer oligonucleotides were designed to amplify the polymorphic region (Fig. 1c) and 450 bp PCR fragment was digested by FauI endonuclease for the genotyping. Exact tests and chi2-test were used to perform association analysis between the SNP allele or genotype and schizophrenia (Table 1). No statistically significant prevalence of the C allele was found in the total group of paranoid schizophrenia (Ms cohort) (P = 0.960, OR = 1.012, C. I. = 0.737-1.391). Comparison of CC and GG genotype frequencies in schizophrenia versus control did not produce a statistically significant difference (P = 0.738, OR = 1.176, C. I. = 0.456-3.032). Real-time PCR analysis was undertaken using TaqMan MicroRNA Assays (Applied Biosystems) to elucidate whether expression of miR-130b in the adult human neocortex is modified in individuals with schizophrenia and whether its expression correlates with genotype. The total RNA was isolated from specimens of human parietal neocortex (Brodman area 7) from different individuals using mirVana miRNA isolation kit (Ambion). The frozen tissue specimens were kindly provided by the Stanley Medical Research Institute (Brain Collection). The control PCR experiments with original RNA (prior generation of cDNA) and with genomic DNA revealed no effective PCR in TaqMan MicroRNA Assay (Applied Biosystems). The data indicate that potential genomic contamination in RNA extracts cannot contribute to quantitative analysis of miRNAs in this assay. PCR of each specimen was replicated and the average mean was used for comparison. Normalization was performed using expression of let-7a abundant in most tissues including brain. On average, there was no strong difference in expression of miR-130b between schizophrenia and norm groups (12 normal individuals and 12 individuals with schizophrenia, p > 0.5) (Fig. 2a). No correlation of expression of miR-130b and the -63 C/G genotypes was observed (Fig. 2b). Using Human MicroChip hybridization array we determined what miRNAs are expressed in the adult human brain, which may be subjects of further investigation (Table 2). Comparative expression analysis of multiple miRNA in collection of brain specimens from schizophrenia and normal individuals using hybridization or direct RT-PCR profiles (our unpublished data) and other hybridization assay [11] detected putative variations in expression of some miRNAs. More detailed validation of the expression of the miRNA and the role of genetic factors in variations in miRNA gene expression is required to determine their role in schizophrenia. It must be noted that variability in level of miR-130b RNA between individual specimens regardless the disease status was clearly observed (Fig. 2). Theoretically, a variety of factors may contribute to the individual variability in miRNA gene expression, including cellular heterogeneity of brain tissues used in analysis, partial degradation of selected miRNA in some post-mortem specimens or, indeed, inter-individual difference in expression of miRNA caused by variable genetic or epigenetic factors. Further studies are required to address this question. To our knowledge, the data present the first genetic study coupled with expression analysis of miRNA gene in schizophrenia and implicate the feasibility of convergent miRNA analysis in neuropsychiatric disorders.Fig. 1. Human gene for miR-130b: a) chromosomal location; b) precursor gene structure and polymorphism found in 5´-upstream region; c) putative regulatory regions for two allelic variants.
Table 1. Genotype and allele frequency in
case and control groups
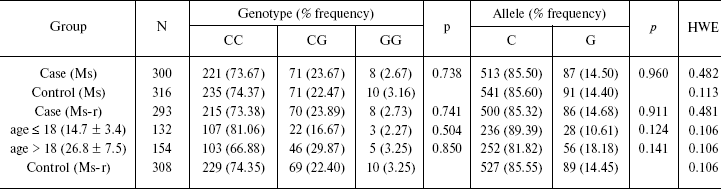
Note: Case-control group (Ms) represents the total sample of the
paranoid schizophrenia patients and control individuals from the Moscow
population; case-control group (Ms-r) excludes the individuals of
non-Russian ethnic background documented in medical records; Ms-r group
was stratified by gender and age of onset of schizophrenia (patients
with unknown age-of onset were excluded); p values for allelic and
genotype distributions are shown. HWE, p values for deviation
from Hardy-Weinberg equilibrium.
Table 2. miRNA genes expressed in adult human brainFig. 2. Expression of miR-130b (ddCt, normalized by miR-let-7a as endogenous control) in human neocortex brain according to Real-Time PCR data. The experiment was undertaken in two replicates on 7900HT Fast Real-Time PCR System using 386-well plates and according to ABI protocol for TaqMan miRNA Assays. a) Comparative expression analysis of miR-130b in schizophrenia and normal individuals; b) comparative expression analysis of individuals with miR130b CC or GC genotypes.
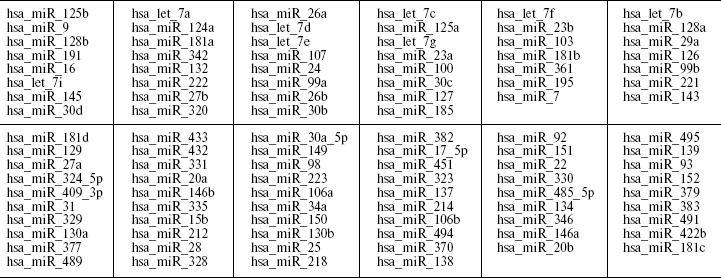
Note: MiRNA transcripts detected by hybridization of Human miRNA Array (probes for 312 mature miRNAs spotted in 4-5 replications) with a Cy3- or Cy5-labeled fraction of small RNA isolated from the human neocortex from adult individuals. The miRNAs in two sections for relatively high/medium and low expression: order from the highest (at the top) to the lowest (at the bottom) level of detectable signal (read from left to the right).
This work was supported by the Stanley Research Medical Institute and, in part, by the Presidium of the Russian Academy of Sciences grant program “Biodiversity and Gene Pools”. The frozen tissue specimens were donated by the Stanley Medical Research Institute Brain Collection courtesy of Drs. Michael B. Knable, E. Fuller Torrey, Maree J. Webster, and Robert H. Yolken.
REFERENCES
1.Ambros, V. (2001) Cell, 107,
823-826.
2.Giraldez, A. J., Cinalli, R. M., Glasner, M. E.,
Enright, A. J., Thomson, J. M., Baskerville, S., Hammond, S. M.,
Bartel, D. P., and Schier, A. F. (2005) Science, 308,
833-838.
3.Johnston, R. J., and Hobert, O. (2003)
Nature, 426, 845-849.
4.Rogaev, E. I. (2005) Biochemistry (Moscow),
70, 1404-1407.
5.Lewis, B. P., Shih, I. H., Jones-Rhoades, M. W.,
Bartel, D. P., and Burge, C. B. (2003) Cell, 115,
787-798.
6.Cohen, D., Lazar, G., Couvert, P., Desportes, V.,
Lippe, D., Mazet, P., and Heron, D. (2002) Am. J. Psychiatry,
159, 148-149.
7.Bassett, A. S., Hodgkinson, K., Chow, E. W.,
Correia, S., Scutt, L. E., and Weksberg, R. (1998) Am. J. Med.
Genet., 81, 328-337.
8.Karayiorgou, M., Morris, M. A., Morrow, B.,
Shprintzen, R. J., Goldberg, R., Borrow, J., Gos, A., Nestadt, G.,
Wolyniec, P. S., and Lasseter, V. K. (1995) Proc. Natl. Acad. Sci.
USA, 92, 7612-7616.
9.Murphy, K. C., Jones, L. A., and Owen, M. J. (1999)
Arch. Gen. Psychiatry, 56, 940-945.
10.Goltsov, A. Y., Loseva, J. G., Andreeva, T. V.,
Grigorenko, A. P., Abramova, L. I., Kaleda, V. G., Orlova, V. A.,
Moliaka, Y. K., and Rogaev, E. I. (2006) Mol. Psychiatry,
11, 325-326.
11.Perkins, D. O., Jeffries, C. D., Jarskog, L. F.,
Thomson, J. M., Woods, K., Newman, M. A., Parker, J. S., Jin, J., and
Hammond, S. M. (2007) Genome Biol., 8, R27 [Epub ahead of
print].