Interaction of HydSL Hydrogenase from the Purple Sulfur Bacterium Thiocapsa roseopersicina BBS with Methyl Viologen and Positively Charged Polypeptides
A. V. Abdullatypov*, N. A. Zorin, and A. A. Tsygankov
Institute of Basic Biological Problems, Russian Academy of Sciences, Institutskaya ul. 2, 142290 Pushchino, Moscow Region, Russia; E-mail: azatik888@yandex.ru* To whom correspondence should be addressed.
Received April 2, 2014; Revision received April 18, 2014
The effect of polypeptides having different charge on the activity of Thiocapsa roseopersicina HydSL hydrogenase was studied. Strong inhibition was shown for poly-L-lysine bearing positive charge. The inhibition was reversible and competitive to methyl viologen, an electron acceptor, in the reaction of hydrogen oxidation catalyzed by the hydrogenase. Peptides carrying less positive charge had weaker inhibiting effect, while neutral and negatively charged peptides did not inhibit the hydrogenase. Molecular docking of poly-L-lysine to T. roseopersicina hydrogenase showed strong affinity of this polypeptide to the acceptor-binding site of the enzyme. The calculated binding constant is close to the experimentally measured value (Ki = 2.1 μM).
KEY WORDS: hydrogenase, methyl viologen, polypeptides, competitive inhibition, molecular dockingDOI: 10.1134/S0006297914080082
Hydrogenase is a key enzyme in hydrogen metabolism of microorganisms catalyzing the reversible reduction of protons to molecular hydrogen:
2H+ + 2e– ↔ H2.
More than 400 genes encoding hydrogenase have been found up to the present time [1, 2], more than thirty hydrogenases have been purified and characterized, and for about ten of these the three-dimensional structure is available [3]. Large studies of the active sites of different types of hydrogenases are being carried out [4]. However, the molecular mechanisms of the interaction of hydrogenases with electron donor/acceptor are poorly described in the literature.
The thermostable HydSL hydrogenase from the purple sulfur bacterium Thiocapsa roseopersicina BBS belongs to the family of Ni-Fe hydrogenases. It has high catalytic activity and resistance to a number of denaturing agents [1]. The high affinity of T. roseopersicina hydrogenase to positively charged metal ions and the positively charged polymer form of the dye neutral red [5] suggests an important role of electrostatic interactions in the functioning of the donor/acceptor-binding site of this enzyme. The full three-dimensional structure of T. roseopersicina hydrogenase has not been established yet. However, its high identity with another hydrogenase with resolved atomic structure allowed us to build its 3D model [6]. In applied aspect, hydrogenases are a subject of interest for development of enzymatic hydrogen electrodes. It was shown that positively charged polypeptides, particularly poly-L-lysine, stabilize monomolecular films of T. roseopersicina hydrogenase, but the mechanism of interaction of such polypeptides has not yet been studied [7].
Besides the experimentally established stabilizing role of poly-L-lysine, it can also serve as an anchoring agent for immobilizing the hydrogenase on electrodes. For its application in this way, we need to find the exact mechanism of interaction of poly-L-lysine with hydrogenase.
The goal of the current work was the experimental investigation of the effect of charged peptides on the reduction of methyl viologen catalyzed by hydrogenase, modeling the spatial interaction of hydrogenase with methyl viologen and polypeptides and comparison of the experimental results with computational data.
MATERIALS AND METHODS
Cells of the purple sulfur bacterium T. roseopersicina BBS were grown under anaerobic photoheterotrophic conditions on modified Pfennig medium [8] in presence of 0.2% sodium acetate.
Cell extract preparation and hydrogenase purification. For extract preparation, cells were harvested at the end of the exponential growth phase. Two hundred grams of cell paste were resuspended in 20 mM potassium phosphate buffer (pH 7.0). The cells were disrupted by acetone treatment and sonication as described earlier [9]. The hydrogenase was purified by fractioning the cell extracts via consecutive ammonium sulfate precipitation and liquid chromatography on CL-4B phenyl-Sepharose columns and DE52 DEAE-cellulose columns [10]. The final step of hydrogenase purification was carried out by preparative electrophoresis in 7% polyacrylamide gel as described earlier [11].
Determination of hydrogenase activity. Hydrogenase activity was determined by spectrophotometric measurement of the rate of reduction of oxidized methyl viologen in a Thunberg cuvette. The reaction mixture (total volume 2 ml) contained 50 mM Tris-HCl buffer (pH 9.0), 4 mM methyl viologen, and 1-10 μg of the hydrogenase. A trace (5-10 μl) of 20 mM sodium dithionite solution prepared under anaerobic conditions was added for initializing the reaction [9]. All the measurements of enzymatic activity were carried out at 30°C. The activity was calculated using the molar extinction coefficient of reduced methyl viologen (ε600 = 13.0 mM–1·cm–1) [12]. The enzymatic activity was expressed in μmol H2 per minute per mg of protein.
Modeling the interaction of hydrogenase with methyl viologen and polypeptides. Molecular docking was carried out for mapping the binding sites for methyl viologen and the polypeptides in the hydrogenase enzyme. The full model of the HydSL hydrogenase from T. roseopersicina built and described previously [6] was taken as the receptor.
Molecular docking was carried out using AutoDock Vina [13]. During docking, the following assumptions were made: 1) methyl viologen is a rigid planar molecule and carries a total charge +2 distributed equally on the atoms of the pyridine rings (see Fig. 1a); 2) molecules of polypeptides K20 (poly-L-lysine) and K(KLK)6K (poly-L-lysine-L-leucine) are rigid α-helices (rigidity of the molecules was assumed due to low accuracy of calculation in case of large flexible ligands); 3) the hydrogenase molecule is rigid; 4) interaction of methyl viologen with the large subunit of HydSL hydrogenase does not lead to electron transfer, thus interaction of polypeptides with the large subunit is not inhibiting; 5) the size of the grid box used for docking exceeded the size of hydrogenase molecule (needed to cover both enzyme and ligand molecules) and was 90 × 90 × 90 Å.
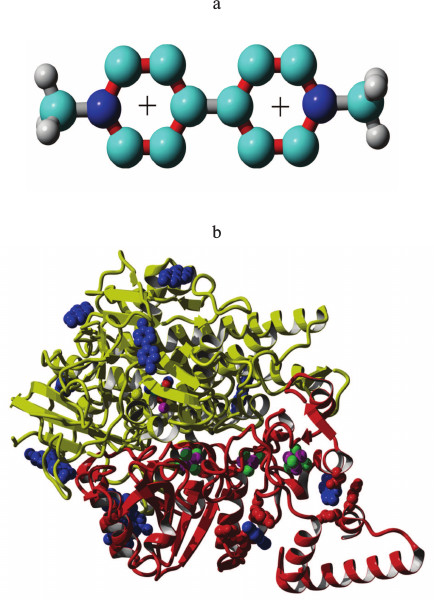
Fig. 1. a) Methyl viologen molecule. Atom colors: gray, hydrogen; cyan, carbon; blue, nitrogen. Bond colors: gray, single bonds; red, resonance bonds (bond order 1.5). b) Interaction of methyl viologen with hydrogenase. Molecule colors: red, small hydrogenase subunit; yellow, large hydrogenase subunit; blue, methyl viologen molecules. Atom colors (atoms shown in balls): magenta, iron; green, sulfur; cyan, carbon; blue, nitrogen; red, oxygen (in the active site).
Ligands (methyl viologen and polypeptides K20 and K(KLK)6K) were built in the YASARA model [14] and AutoDock Tools [15] programs. The charges for methyl viologen were corrected manually in the AkelPad text editor according to the following: charges of methyl groups equal zero, charges of other groups equal 0.167 (this assumption is explained by the fact that methyl viologen has a total charge +2 distributed equally on 12 atoms of the aromatic pyridine rings).
The binding constants for complexes of ligands with hydrogenase were calculated using the formula:
Ki = eΔG/RT,
where ΔG is the Gibbs energy of formation of the enzyme–ligand complex expressed in J/mol, R = 8.314 J·mol–1·K–1 is the universal gas constant, and T is the temperature (303 K) [16].
The following reagents were used in the work: polypeptides K20, K(KLK)6K, L20, and E20 (Sigma, Germany), reagents for PAGE (Sigma), phenyl-Sepharose CL-4B (Pharmacia, Sweden), DEAE-cellulose DE52 (Whatman, England), and methyl viologen and sodium dithionite (Fluka, Switzerland). Other reagents used were made in Russia (grades “chemically pure” and “high purity”).
RESULTS AND DISCUSSION
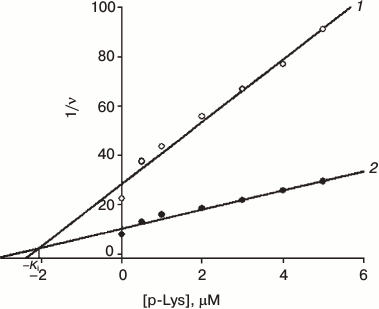
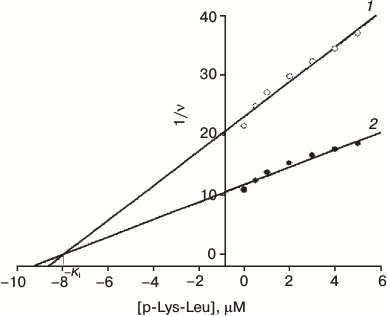
Effect of polypeptides on hydrogenase activity. The electron acceptors participating in the reaction of hydrogen oxidation catalyzed by hydrogenase are positively charged. Along with natural electron acceptors, different viologen dyes (charge in oxidized state, +2) and nickel and cadmium cations (which can be reduced to metal state by hydrogenase under a hydrogen atmosphere) can also serve as electron acceptors [17]. Thus, electrostatic interactions play a significant role in the interaction between the enzyme and its substrate (acceptor). As shown earlier [9], a number of metal cations have high affinity to T. roseopersicina hydrogenase, being reversible inhibitors of the enzyme competitive to methyl viologen. The increase in the affinity of hydrogenase to cations on increase in pH, when the negative charge of the enzyme molecule is increases [9], gives evidence for the important contribution of electrostatic interactions to inhibition of T. roseopersicina hydrogenase by these metal cations. We supposed that other positively charged substances could have high affinity to the hydrogenase, being competitive inhibitors to the electron acceptor. To check this supposition, we used polypeptides with different charge as model polyions. The most powerful inhibitor was poly-L-lysine (K20) bearing a significant positive charge (Fig. 2). The inhibiting effect of poly-L-lysine was reversible and competitive with regard to the electron acceptor, oxidized methyl viologen, in the hydrogen oxidation reaction catalyzed by the hydrogenase. The inhibiting constant reflecting the affinity of this peptide to the hydrogenase was Ki = 2.1 μM. This constant is two orders of magnitude lower than the Km = 182 μM for methyl viologen [18]. Polypeptides carrying less positive charge had less inhibitory effect. For example, poly-L-lysine-L-leucine (K(KLK)6K), which contains 14 lysine residues and six neutral leucine residues, had significantly lower inhibitory effect (also competitive to oxidized methyl viologen), the inhibition constant being Ki = 8.0 μM (Fig. 3). Neutral and negatively charged polypeptides, i.e. poly-L-leucine (L20) and poly-L-glutamate (E20), showed no inhibitory effect on the hydrogenase (data not shown). Thus, it was shown experimentally that polypeptides with positive charge are effective inhibitors of T. roseopersicina hydrogenase competitive to methyl viologen.
Fig. 2. Dependence of reverse H2 oxidation rate in the presence of hydrogenase on poly-L-lysine (K20) concentration at certain methyl viologen concentrations: 1) 1 mM; 2) 4 mM at pH 9.0 (Ki = 2.1 μM).
Fig. 3. Dependence of reverse H2 oxidation rate in presence of hydrogenase from poly-L-lysine-leucine (K(KLK)6K) concentration at certain methyl viologen concentrations: 1) 1 mM; 2) 4 mM at pH 9.0 (Ki = 8 μM).
Molecular docking of methyl viologen and polypeptides to HydSL hydrogenase. The results of molecular docking showed that the Gibbs energy of methyl viologen binding varies from –5.6 to –4.7 kcal/mol (from –23.43 to –19.66 kJ/mol). The binding constants calculated from these energies vary in the range 91.3-407.2 μM, which is quite close to the experimentally determined binding constant of 182 μM. Also, two methyl viologen–hydrogenase complexes obtained via docking had methyl viologen bound to areas of the small subunits very close to the distal Fe-S cluster (distances 8.5 and 15 Å, binding constants 127 and 292 μM, respectively). This makes theoretical confirmation for the experimentally known possibility of electron transfer from the hydrogenase to methyl viologen (Fig. 1b). For comparison, the distance between the FeS clusters is 9 Å and the distance between the active site and the proximal FeS cluster is 13 Å.
Binding the methyl viologen molecule in the closest site to the distal FeS cluster (8.5 Å distance between the cluster and methyl viologen) is presumably due to electrostatic interactions: the methyl viologen molecule interacts with negatively charged residues D247, E363, and E366 (distances from charged groups to methyl viologen are 3.7, 4.8, and 3.8 Å, respectively; here and below the distance means minimal distance from centers of atoms of charged groups to centers of atoms of methyl viologen pyridine rings; numbering of the residues is according to numbering of the sequence from the UniProt database (ID o51820)).
Binding of methyl viologen with the site more distant from the FeS cluster occurs due to both electrostatic interaction (with aspartic acid residue D223, distance to charged group 3.8 Å) and stacking interaction with tyrosine residues (Y225, Y277, distances to aromatic groups being 3.8 and 4.6 Å, respectively).
We proposed that K20 and K(KLK)6K are α-helical molecules. To check the reasonability of this assumption, we predicted the secondary structure of these peptides by the hierarchical neural network method [19] on the Pole BioInformatique Lyonnais server [20]. The results showed that K20 consists of 50% α-helical conformation and 50% random coiled conformation, whereas K(KLK)6K is 80% α-helical and 20% (four terminal lysine residues on N- and C-termini) random coiled.
The results of rigid polypeptide docking showed that the Gibbs energy of K20 binding varies from –9.2 to –8.1 kcal/mol, which corresponds to the binding constant range 0.131-1.44 μM; for K(KLK)6K the range of Gibbs energies was from –9.9 to –8.1 kcal/mol, which corresponds to binding constant range from 0.072 to 1.44 μM.
According to our experimental data, the binding constant for poly-L-lysine (K20) was 2.1 μM, which corresponds to Gibbs energy of –33 kJ/mol (–7.9 kcal/mol). The binding constant for poly-L-lysine-leucine (K(KLK)6K) was 8 μM, which corresponds to Gibbs energy of –29.6 kJ/mol (–7.1 kcal/mol).
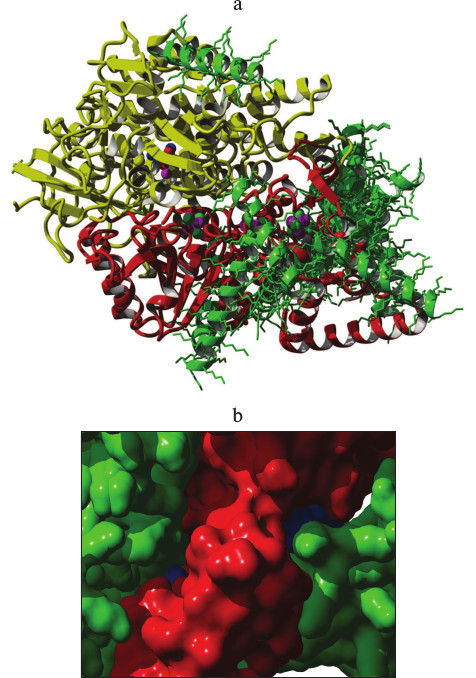
Thus, the docking results showed the following. 1) There are two possible sites for binding methyl viologen near the distal FeS-cluster. The most possible electron transfer process from the hydrogenase could occur in the negatively charged site closest to the distal FeS-cluster (residues D247, E363, and E366); another site (residues D223, Y225, and Y277) is situated farther from the FeS-cluster, thus the electron transfer to the methyl viologen at this site is less possible. 2) Peptides K20 and K(KLK)6K are also capable of binding to these sites, which proves the competitive mechanism of inhibition of methyl viologen reduction by these polycations (Fig. 4, a and b). 3) The binding constants for methyl viologen, K20, and K(KLK)6K are close to the experimentally determined values, but no difference between the binding of the two positively charged peptides was found.
Fig. 4. a) Interaction of hydrogenase with poly-L-lysine (K20). Molecule colors: red, small hydrogenase subunit; yellow, large hydrogenase subunit; blue, methyl viologen molecules; green, poly-L-lysine molecules. Atom colors: green, sulfur; magenta, iron. b) Overlapping of poly-L-lysine and methyl viologen atoms in two acceptor-binding sites. Molecular surface colors: red, hydrogenase; blue, methyl viologen; green, poly-L-lysine. The figure was made by superposition of two complexes: hydrogenase–poly-L-lysine and hydrogenase–methyl viologen.
Although hydrogenase activates the simplest molecule, i.e. molecular hydrogen, the catalytic mechanism of this enzyme is quite sophisticated. Progress in the investigation of hydrogenase led to determination of spatial structure of these enzymes and their hydrogen-activating center containing a unique nickel–iron cluster [3]. The structure and functional mechanism of acceptor/donor binding site is less well described. According to the spatial structure, this site is close to the distal FeS-cluster [21]. In hydrogenase from Desulfovibrio desulfuricans, the distal FeS cluster is surrounded by several amino acid residues carrying negative charge. This increases the efficiency of interaction with the in vivo redox partner of the hydrogenase, namely cytochrome c3, which has positive charge on its surface [21].
It could seem that our data on inhibition of the interaction of hydrogenase with methyl viologen by positively charged poly-L-lysine are contradictory to the data on its hydrogenase-stabilizing action [7]. However, we should take into account the differences in the measurements that have been carried out. Poly-L-lysine prevents binding of methyl viologen by the hydrogenase region close to distal FeS-cluster, which was shown in the current work. But if we propose that on an electrode surface poly-L-lysine binds to the acceptor-binding site of the hydrogenase, then the hydrogenase would be anchored on the electrode surface by its region close to the FeS cluster. This allows direct electron transferring from the hydrogenase to the electrode. Apparently, immobilization of hydrogenase onto the electrode surface with a positively charged polypeptide, particularly K20, increases the stability of the hydrogenase, as shown earlier [7], and allows direct electron transfer to the electrode. Thus, our data could be helpful in choosing polypeptides for co-immobilization with the hydrogenase on electrodes.
This work was supported by the Russian Foundation for Basic Research (grant No. 14-04-01676).
REFERENCES
1.Gogotov, I. N., Zadvorny, O. A., Zorin, N. A., and
Serebryakova, L. T. (2010) Bacterial hydrogenases, in Proc.
Winogradsky Institute of Microbiology, issue 15: Photosynthetic
Microorganisms (Galchenko, V. F., ed.) [in Russian], MAKS Press,
Moscow, pp. 260-289.
2.Vignais, P., and Billoud, B. (2007) Occurrence,
classification, and biological function of hydrogenases: an overview,
Chem. Rev., 107, 4206-4272.
3.Yagi, T., and Higuchi, Y. (2013) Studies on
hydrogenase, Proc. Jpn. Acad. Ser. B, Phys. Biol.
Sci., 89, 16-33.
4.Armstrong, F., and Albracht, S. (2005)
[NiFe]-hydrogenases: spectroscopic and electrochemical definition of
reactions and intermediates, Phil. Trans. R. Soc.,
363, 937-954.
5.Voronin, O. G., Konischeva, E. V., Zorin, N. A.,
Fedotenkov, F. A., Karyakina, E. E., Karpachova, G. P., Orlov, A. V.,
Kiseleva, S. G., and Karyakin, A. A. (2013) Modification of the
electrode surface with analogues of hydrogenase substrates for highly
active fuel bioelectrocatalysts development, Nano- Microsystems
Techn., 5, 15-19.
6.Abdullatypov, A. V., and Tsygankov, A. A. (2013)
Homology modeling of the spatial structure of HydSL hydrogenase from
purple sulfur bacterium Thiocapsa roseopersicina
BBS, Comput. Res. Model. (Moscow), 5,
737-747.
7.Noda, K., Zorin, N. A., Nakamura, C., Miyake, M.,
Gogotov, I. N., Asada, Y., Akutsu, H., and Miyake, J. (1998)
Langmuir–Blodgett film of hydrogenase for electrochemical
hydrogen production, Thin Solid Films, 327-329,
639-642.
8.Bogorov, L. V. (1974) The properties of
Thiocapsa roseopersicina, strain BBS, isolated from an estuary
of the White Sea, Mikrobiologiya, 43,
326-333.
9.Zadvorny, O. A., Zorin, N. A., and Gogotov, I. N.
(2000) Influence of metal ions on hydrogenase from the purple sulfur
bacterium Thiocapsa roseopersicina, Biochemistry
(Moscow), 65, 1287-1291.
10.Sherman, M. B., Orlova, E. V., Zorin, N. A.,
Smirnova, E. A., Kuranova, I. P., and Gogotov, I. N. (1987) The
structure of hydrogenase microcrystals from Thiocaspa
roseopersicina, Doklady Akad. Nauk SSSR, 295,
509-512.
11.Gogotov, I. N., Zorin, N. A., and Kondratieva, E.
N. (1976) Purification and properties of Thiocapsa roseopersicina
hydrogenase, Biokhimiya, 41, 836-842.
12.Thorneley, R. N. F. (1974) A convenient
electrochemical preparation of reduced methyl viologen and a kinetic
study of the reaction with oxygen using an anaerobic stopped-flow
apparatus, Biochim. Biophys. Acta, 333,
487-496.
13.Trott, O., and Olson, A. J. (2010) AutoDock Vina:
improving the speed and accuracy of docking with a new scoring
function, efficient optimization, and multithreading, J. Comput.
Chem., 31, 455-461.
14.Krieger, E., Dunbrack, R. L., Jr., Hooft, R. W.,
and Krieger, B. (2012) Assignment of protonation states in proteins and
ligands: combining pKa prediction with hydrogen
bonding network optimization, Methods Mol. Biol., 819,
405-421.
15.Sanner, F. (1999) Python: a programming language
for software integration and development, J. Mol. Graphics
Model., 17, 57-61.
16.Krasnov, K. S., Vorobyov, N. K., Godnev, I. N.,
Vasilieva, V. N., Vasiliev, V. P., Kiseleva, V. L., Belonogov, K. N.,
and Gostikin, V. N. (2001) Physical Chemistry (Krasnov, K. S.,
ed.) [in Russian], Vol. 1, Vysshaya Shkola, Moscow.
17.Zadvorny, O. A., Zorin, N. A., and Gogotov, I. N.
(2006) Transformation of metals and metal ions by hydrogenases from
phototrophic bacteria, Arch. Microbiol., 184,
279-285.
18.Kondratieva, E. N., and Gogotov, I. N. (1981)
Molecular Hydrogen in Microbial Metabolism [in Russian], Nauka,
Moscow.
19.Guermuer, Y. (1999) Improved performance in
protein secondary structure prediction by inhomogeneous score
combination, Bioinformatics, 15, 413-421.
20.Combet, C., Blanchet, C., Geourjon, C., and
Deleage, G. (2000) NPS: network protein sequence analysis, Trends
Biochem. Sci., 25, 147-150.
21.Matias, P. M., Soares, C. M., Saraiva, L. M.,
Coelho, R., Morais, J., Le Gall, J., and Carrondo, M. A. (2001) [NiFe]
hydrogenase from Desulfovibrio desulfuricans ATCC 27774: gene
sequencing, three-dimensional structure determination and refinement at
1.8 Å and modeling studies of its interaction with the tetraheme
cytochrome c3, J. Biol. Inorg. Chem.,
6, 63-81.