Zinc Switch in Pig Heart Lipoamide Dehydrogenase: Steady-State and Transient Kinetic Studies of the Diaphorase Reaction
I. G. Gazaryan1,2,3,4, V. A. Shchedrina3, N. L. Klyachko3,5, A. A. Zakhariants4,6, S. V. Kazakov2, and A. M. Brown1,a*
1Department of Cell Biology and Anatomy, New York Medical College, 15 Dana Road, Valhalla, NY 10605, USA2Department of Chemistry and Physical Sciences, Dyson College of Arts and Sciences, Pace University, 861 Bedford Road, Pleasantville, NY 10570
3Department of Chemical Enzymology, Lomonosov Moscow State University, 119899 Moscow, Russia
4Bach Institute of Biochemistry, Federal Research Centre “Fundamentals of Biotechnology”, Russian Academy of Sciences, 119071 Moscow, Russia
5Derzhavin Tambov State University, 392000 Tambov, Russia
6Shemyakin and Ovchinnikov Institute of Bioorganic Chemistry, Russian Academy of Sciences, 117997 Moscow, Russia
* To whom correspondence should be addressed.
Received May 27, 2020; Revised June 11, 2020; Accepted June 14, 2020
Elevation of intracellular Zn2+ following ischemia contributes to cell death by affecting mitochondrial function. Zn2+ is a differential regulator of the mitochondrial enzyme lipoamide dehydrogenase (LADH) at physiological concentrations (Ka = 0.1 µM free zinc), inhibiting lipoamide and accelerating NADH dehydrogenase activities. These differential effects have been attributed to coordination of Zn2+ by LADH active-site cysteines. A detailed kinetic mechanism has now been developed for the diaphorase (NADH-dehydrogenase) reaction catalyzed by pig heart LADH using 2,6-dichlorophenol-indophenol (DCPIP) as a model quinone electron acceptor. Anaerobic stopped-flow experiments show that two-electron reduced LADH is 15-25-fold less active towards DCPIP reduction than four-electron reduced enzyme, or Zn2+-modified reduced LADH (the corresponding values of the rate constants are (6.5 ± 1.5) × 103 M–1·s–1, (9 ± 2) × 104 M–1·s–1, and (1.6 ± 0.5) × 105 M–1·s–1, respectively). Steady-state kinetic studies with different diaphorase substrates show that Zn2+ accelerates reaction rates exclusively for two-electron acceptors (duroquinone, DCPIP), but not for one-electron acceptors (benzoquinone, ubiquinone, ferricyanide). This implies that the two-electron reduced form of LADH, prevalent at low NADH levels, is a poor two-electron donor compared to the four-electron reduced or Zn2+-modified reduced LADH forms. These data suggest that zinc binding to the active-site thiols switches the enzyme from one- to two-electron donor mode. This zinc-activated switch has the potential to alter the ratio of superoxide and H2O2 generated by the LADH oxidase activity.
KEY WORDS: ubiquinone, 2,6-dichlorophenol indophenol, duroquinone, ferricyanide, enzyme kinetics, dehydrolipoyl dehydrogenaseDOI: 10.1134/S0006297920080064
Abbreviations: DCPIP, 2,6-dichlorophenol-indophenol; LADH, dihydrolipoyl dehydrogenase (=lipoamide dehydrogenase); KGDHC, α-ketoglutarate dehydrogenase complex.
INTRODUCTION
Mounting evidence indicates that mobilization of intracellular Zn2+ following ischemia-reperfusion injury plays a role in cellular toxicity [1-4]. Elevated intracellular Zn2+ has been associated with the loss of mitochondrial membrane potential, production of reactive oxygen species, and cell death [5, 6]. Reduction of zinc accumulation in mitochondria by chelating zinc was shown to decrease the cerebral ischemic injury in a stroke model [7]. We demonstrated that zinc competes with calcium for the entry to mitochondria and proposed the mitochondrial calcium uniporter as an entry port for zinc [8]. This suggestion has been corroborated by the data reported in [9].
Zn2+ is a potent inhibitor of mitochondrial respiration and induces mitochondrial permeability transition [5, 6]. It has also been suggested that Zn2+ can regulate mitochondrial respiration by reversibly inhibiting α-ketoglutarate dehydrogenase complex (KGDHC) [10]. Lipoamide dehydrogenase (LADH) (EC 1.6.4.3), a key component of mitochondrial dehydrogenase complexes including KGDHC, catalyzes transfer of reducing equivalents from the bound dihydrolipoate of the neighboring dihydrolipoyl acyl transferase subunit to NAD+. This reversible reaction involves two reaction centers: a thiol pair, which accepts electrons from dihydrolipoate, and a non-covalently bound FAD/FADH2 moiety, which transfers electrons to NAD+. The enzyme also catalyzes a “diaphorase” reaction, i.e., reduction of quinones (or other electron acceptors) by NADH. This second activity requires only the FAD catalytic center and is greatly enhanced by metal ions or other reagents that bind to [11] or modify [12-17] the two catalytic thiols essential for the lipoamide dehydrogenase activity. Zn2+ increases LADH diaphorase activity towards ubiquinone [18, 19]. Enhancement of diaphorase activity was also reported for the genetically engineered mutants lacking either one of the catalytic dithiols [20]. Although stimulation of the diaphorase activity has been demonstrated [12-17, 20], a kinetic model and precise kinetic parameters have never been reported.
We reported previously that Zn2+ inhibits the reversible lipoamide dehydrogenase reaction catalyzed by the purified pig heart LADH (Ki ca. 0.15 µM) in both directions by blocking the catalytic dithiols [21]. The same zinc-binding interaction also induced a 3-4-fold enhancement in the oxidase activity of LADH [21], which involves only the FAD catalytic center. Objective of the present study is to develop a detailed kinetic mechanism of the diaphorase reaction, with 2,6-dichlorophenol-indophenol (DCPIP) as a model quinone substrate, catalyzed by LADH in the absence and in the presence of zinc. The results of the steady-state and transient kinetic studies suggest that, in addition to switching LADH from thiol reductase to quinone reductase (diaphorase) activity, zinc binding can also switch the enzyme to prefer the two-electron over the one-electron donor mode.
MATERIALS AND METHODS
Reagents/enzymes. NADH, NADPH, Tris, ZnCl2, DCPIP, 1,4-benzoquinone, potassium ferricyanide, duroquinone (2,3,5,6-tetramethyl-1,4-benzoquinone), ubiquinone, were from Sigma-Aldrich (USA), HCl was from J. T. Baker, Inc. (USA). All solutions were prepared using distilled, deionized water with >15 MΩ/cm resistance. All reagents were “SigmaUltra grade”, if available, to reduce the possibility of divalent cation contamination. NADH was freshly made as 10 mM stock solutions in water. ZnCl2 was prepared as a 10 mM stock solution in water.
Lipoamide dehydrogenase from porcine heart (LADH) from Sigma-Aldrich (USA) was shown by SDS-PAGE to contain no detectable (<2%) protein impurities. The enzyme was further purified by FPLC gel-filtration to remove additives such as ammonium sulfate and FAD by application of 0.5 ml portions of the ammonium sulfate slurry diluted 1 : 3 with water onto a Superdex 200 column (Pharmacia Biotech) equilibrated with 0.2 M Tris-HCl buffer, pH 7.5. LADH concentration was determined by its absorbance at 455 nm (ε455 = 11,300 M–1·cm–1 [1]). Thioredoxin reductase from Escherichia coli was purchased from Sigma-Aldrich and used after desalting on Sephadex G-25.
Steady-state kinetics. All spectrophotometric measurements were performed in a 96-well plate reader (SpectraMax Plus, Molecular Dynamics, USA) with a 200 fig reaction volume per well at 20°C. Absorbance of NADH (ε340 = 6.22 mM–1·cm–1 [22]) and DCPIP (E600 = 20.6 mM–1·cm–1 [22]) were recorded. Light path was 0.43 cm. All reactions were carried out in 50 mM Tris-HCl, pH 7.5. In order to correct for non-enzymatic DCPIP reduction, control assays in the absence of added enzyme were performed in parallel wells. These background rates were subtracted from the reaction rate values measured in the presence of enzyme under identical conditions.
The study of DCPIP reduction with NADH in the absence of added zinc was performed by varying NADH concentration in the 10-500 µM range and DCPIP in the 2-50 µM range. The reaction was initiated by adding 400 nM enzyme solution. The reaction rate dependence in double-reciprocal plots on DCPIP concentrations at fixed varied NADH concentrations was fitted to a linear dependence, which was used to calculated rate constants. The final values were calculated as average from three independent experiments.
To study zinc effects, a similar experimental approach was used. Aliquots of NADH and ZnCl2 were placed into the wells first, and reaction was initiated by simultaneous addition of 160 µl enzyme/buffer solution containing different DCPIP concentrations with a multichannel pipette. For assays in which DCPIP reduction by NADH was monitored, final reagent concentrations were in the range of 30-400 nM LADH, 0.002-0.1 mM DCPIP, 0.01-0.5 mM NADH, 0.1-20 µM ZnCl2. Duroquinone, ubiquinone, benzoquinone, and ferricyanide reduction catalyzed by 160, 200, 50, and 150 nM LADH, respectively, was monitored by NADH consumption at 340 nm at a fixed NADH concentration of 0.02 mM and various concentrations of the electron acceptor and Zn2+. In the case of E. coli thioredoxin reductase, the assays were performed at varying concentrations of DCPIP and a fixed concentration of NADPH (0.02 mM) in the absence and presence of 5 µM Zn2+. Benzoquinone reduction with LADH and DCPIP reduction with E. coli TRR was performed in duplicate and triplicate, respectively, in a single run.
Zn2+ activation data were analyzed using Lineweaver–Burk plots and fitted by the method of least squares using Sigma Plot 12.5 software (SPSS, Chicago). Each experiment was performed in triplicate. All data are reported as a mean ± S.E. from three independent experiments. Significance of the changes in the slopes of the double-reciprocal plots were tested by performing an unpaired t-test (Sigmastat software, SPSS, Chicago), using the estimated slopes and standard errors from the regression lines of the highest Zn2+ dose and control series in each panel.
Transient kinetic studies. Anaerobic stopped-flow measurements were performed in quadruplicate with a High-Tech SF-61 stopped-flow rapid scan spectrophotometer (High-Tech Scientific, U.K.) installed in an anaerobic glove box operating under N2 with less than 1 p.p.m. O2. The dead time was 0.8 ms. Temperature was maintained at 22°C with a Techne-400 circulator [Techne (Cambridge) Ltd., Duxford, Cambs., U.K.] with an external cooler. The enzyme solution and substrate powder were placed into the sealed serum vials and deoxygenated before being placed into the glove box. Buffer solution (0.05 M Tris-HCl, pH 7.5) used in all experiments was deoxygenated overnight in the glove box. NADH (0.75 mM) and ZnCl2 (1.875 mM) stock solutions were prepared anaerobically under N2 in the same buffer.
Changes in the protein-bound flavin absorbance were determined in the rapid-scan mode with a xenon lamp. The enzyme stock solution (15 µM) was shot against increasing concentrations of NADH (7.5-60 µM) to determine the NADH concentration that gave the highest absorbance at 530 nm, which was an indicator of two-electron reduced “charge-transfer complex” form of LADH [10]. The charge-transfer complex has one reduced thiol and the other sharing an electron with FAD [10]. The highest concentration of the charge-transfer complex was observed with a 1.5 molar NADH excess over the enzyme.
Changes in DCPIP absorbance were determined using a single-wavelength regime at 600 nm. To determine the rate constant for oxidation of the two-electron reduced enzyme by DCPIP, the enzyme (2 µM) was pre-reduced with a 1.5-molar excess of NADH and then shot against increasing concentrations of DCPIP in the range 2-60 µM. To determine rate constants for the Zn2+-modified enzyme, 2 µM LADH was pre-reduced with 1.5 equivalents of NADH in the presence of 100 µM Zn2+. The LADH-Zn complex was then reduced by further addition of 1.5 equivalents of NADH to generate a fully reduced FAD and shot against increasing DCPIP concentrations (2-60 µM).
Experiments with the two-electron reduced enzyme and DCPIP satisfied the condition of a single turnover experiment under pseudo first-order conditions with [S]o >> [E]o. Therefore, the absorbance vs time curves were fitted to the exponential function A = Aoexp(-k[S]ot) using a least-squares minimization program supplied by High-Tech Scientific. The second order rate constant was calculated from the slope of a plot of apparent first order rate constants against DCPIP concentration.
To determine the rate constant for oxidation of the four-electron reduced enzyme by DCPIP, the enzyme was pre-reduced with 50 equivalents of NADH to ensure complete reduction of the FAD and then shot against rising concentrations of DCPIP. Since the enzyme reduction with NADH is known to be very rapid [23] under these conditions, essentially all the enzyme is present in the four-electron reduced state over the time-course of DCPIP reduction, i.e., the experiment can be considered as a steady-state one monitoring decline of DCPIP absorption at the millisecond scale. Therefore, the rate equation is [S] = [S]oexp(-k[E]ot), and as t → 0, the exponential function can be presented as a linear one [S] = [S]o (1-k[E]ot), allowing the rate constant to be calculated from the initial slope of the kinetic curve.
RESULTS
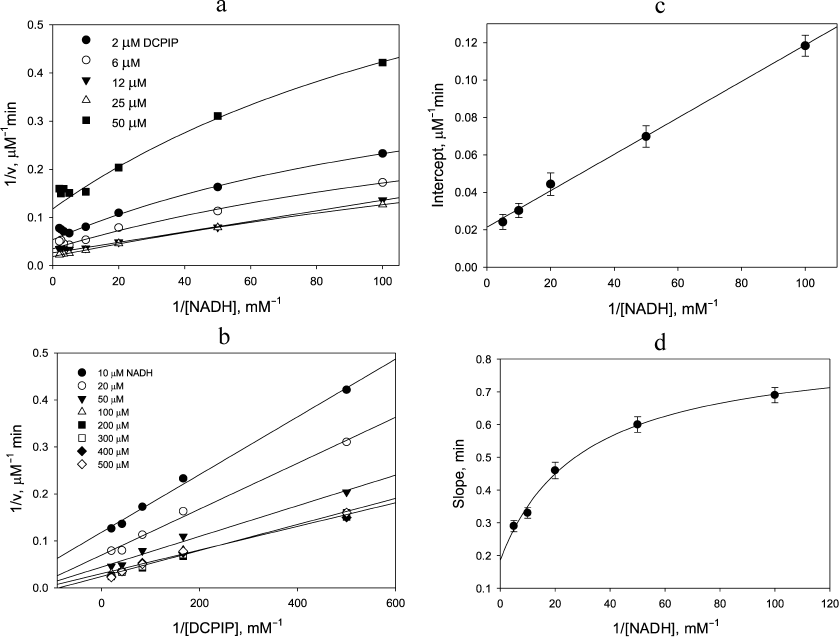
Steady-state kinetics of the diaphorase reaction. The dependence of the initial reaction rate on NADH concentration in double-reciprocal plots exhibits hyperbolic behavior up to concentrations of 0.2 mM followed by the onset of NADH inhibition (Fig. 1a). The latter effect is due to the NADH-induced enzyme inactivation via a dissociation process, which is known to occur for the four-electron reduced form of LADH from different sources [24]. It seems that complex nature of the rate dependence has not been recognized in the previous publications, and therefore the authors concluded that ternary complex formation [25, 26] occurred during the DCPIP reduction by LADH. The double reciprocal plots in Fig. 1a at non-saturating DCPIP concentrations can be only poorly approximated by the plots corresponding to the mechanism involving ternary complex formation that requires the data to be forced into a linear dependence. Moreover, the straight-line fits would not result in a single intersection point, which would be indicative of the mechanism involving ternary complex formation.
Fig. 1. Dependence of the rate of DCPIP reduction by NADH catalyzed by 400 nM LADH on the concentration of one substrate at the various fixed concentrations of the second substrate. a) Dependence on [NADH]; b) dependence on [DCPIP]; c and d) secondary plots showing dependence of the intercepts and slopes from (b), respectively, on NADH concentration (enzyme-inactivating high NADH concentrations omitted). 50 mM Tris-HCl buffer, pH 7.5, 22°C. The curves in panels a and d are least square fits to hyperbolic curves. The curves in panels b and c are linear regression lines. The data were generated in a single representative experimental run (see experimental details under “Materials and Methods” section).
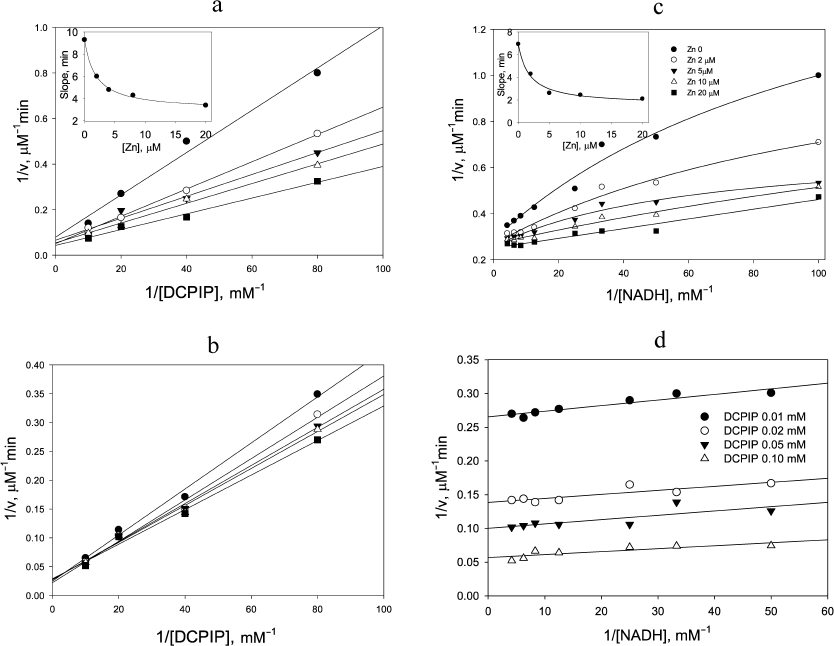
Replotting the experimental data in double reciprocal coordinates for the reaction rate vs DCPIP concentration gives a linear dependence for non-inactivating NADH concentrations, i.e., below 0.2 mM (Fig. 1b), easily permitting further analysis (Fig. 1, c and d). A similar trend is observed, when the zinc effects are examined: linear dependence of the reaction rate on DCPIP concentrations with rising zinc concentrations both at low (Fig. 2a) and high (Fig. 2b) NADH concentrations, and hyperbolic character of the reaction rate dependence on NADH concentration at low zinc concentrations, which transforms into a linear dependence with increasing zinc concentrations (Fig. 2c).
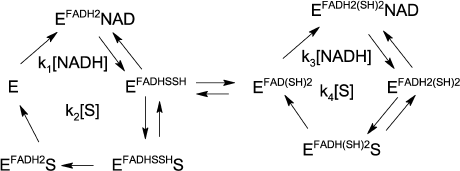
By analogy to our work on the effect of zinc ions on LADH oxidase activity [21], we suggest a two-cycle scheme to explain hyperbolic dependence observed for the LADH-catalyzed DCPIP reduction by NADH. Since DCPIP has been shown to be an exclusively two-electron acceptor [26], we assume that only enzyme forms with fully reduced FAD are capable of reducing the substrate. Thus, the charge-transfer complex is likely to be a poor catalyst of DCPIP reduction, and we must assume enzyme isomerization into a form with fully reduced FAD either preceding DCPIP binding or within the enzyme-substrate complex. We favor the latter option since our earlier transient-kinetic studies demonstrated direct formation of the charge-transfer complex when oxidized LADH was reduced by NADH [21]. Moreover, partitioning between the charge-transfer complex and the form with both thiols reduced was found to be 1 : 1 upon enzyme reduction with equimolar NADH under anaerobic conditions [21].
To simplify the rate equation, we assign the composite apparent rate constants k1 and k3 for the steps related to NADH binding and enzyme reduction, and composite apparent rate constants k2 and k4 for DCPIP binding and release in the left and right cycles, respectively, as shown in Scheme 1. Equilibrium constant between the charge-transfer complex and the form with both thiols reduced is taken as 1 [21]. The main differences between the two cycles are in the relative rates of the enzyme reduction step by NADH, with k1 >> k3, and DCPIP reduction, with k4 >> k2.
Scheme 1
The rate equation in this case is a combination of two hyperbolic functions:
The above equation can be rearranged (Eq. 2) to show contributions of the rate constant ratios to the overall dependence:
Thus, the double-reciprocal plots should have a hyperbolic character for both substrates, which has been observed in our experiments. The fact that this hyperbolic character is hardly pronounced with respect to DCPIP, especially at high concentrations of NADH (see Fig. 1b and Fig. 2b) is to be expected, since k1 is at least 5 times higher than k2 and k3 [21]. With rising NADH and assuming that k3 ≈ k4 >> k2, equation 2 is reduced to equation 3 and predicts a linear dependence on DCPIP at high NADH concentrations:
Thus, the value of the apparent rate constant for DCPIP reduction calculated from the slope of the plot 1/v vs. 1/[DCPIP] at high NADH concentrations will be close to the rate constant of its reduction by the four-electron reduced enzyme form, k4. The apparent rate constant for NADH oxidation calculated from the intercept of the above plot will be equal to k3/2.
The apparent linearity of the plots shown in Fig. 1b (r2 = 0.991 ± 0.005) allows the dependence of the intercept and slope on NADH concentration to be analyzed (Fig. 1, c and d) as discussed above. The intercept depends linearly on 1/[NADH], while the slope depends hyperbolically on 1/[NADH]. At very high NADH concentrations the slope approaches the value of the apparent rate constant for the reaction of the enzyme with DCPIP (k4 = (2.1 ± 0.1) × 105M–1·s–1, see Fig. 1, b and d). The apparent rate constant for NADH (k3/2) can be calculated as a slope from the secondary plot (Fig. 1c, kNADH = (4.6 ± 0.4) × 104 M–1·s–1). The steady-state data on DCPIP reduction were obtained using comparatively low DCPIP concentrations to minimize contribution of the non-enzymatic reaction and, therefore, do not allow us to make an accurate estimate of k1 and k2. However, the value of k1 has been reported for benzoquinone reduction, which is reduced within a single catalytic cycle, i.e., by the two-electron reduced enzyme (see Table 1) [26].
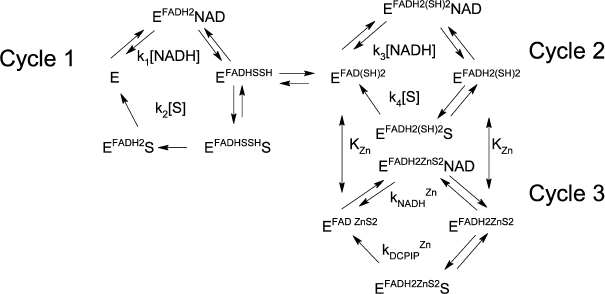
In the presence of Zn2+ strong activation is observed both for DCPIP reduction (Fig. 2, a and b) and NADH oxidation (Fig. 2c). As can be seen from Fig. 2c, at low non-saturating DCPIP concentrations, Zn2+ accelerates the reaction by increasing the apparent rate constant for NADH oxidation up to 4 × 105 M–1·s–1. The half-activation concentration of Zn2+ is ~1.8 µM (see insets to Figs. 2, a and c) in 50 mM Tris-HCl, pH 7.5. After correcting for sequestration of Zn2+ by Tris buffer this corresponds to ca. 0.1 µM free Zn2+ [21, appendix]. This value is similar to the one obtained from our previous experiments on Zn2+ inhibition of lipoamide dehydrogenase reaction and activation of NADH-oxidase activity [21] and supports the hypothesis that the same molecular mechanism accounts for both effects, i.e., Zn2+ binding to the catalytic dithiols (Scheme 2).
Scheme 2
Fig. 2. Zinc effects on steady-state kinetics of DCPIP reduction with NADH. The effect of 0-20 µM Zn2+ on the rate of NADH oxidation at varied [DCPIP] and fixed [ NADH] equal to: a) 20 µM and b) 240 µM. The inset in (a) shows Zn2+dependence of apparent rate constant for DCPIP reduction. c) Effect of Zn2+ on the rate of 12.5 µM DCPIP reduction at various concentrations of NADH. The inset shows Zn2+dependence of the apparent rate constant for NADH oxidation. d) Dependence of the rate of DCPIP reduction by NADH at various fixed concentrations of both substrates in the presence of 20 µM Zn2+. Each panel presents an independent representative experiment. Reaction conditions as for Fig. 1, except 30 nM LADH is used.
Zn2+ ion also accelerates the DCPIP reduction step. This effect is most evident at low NADH concentrations (Fig. 2a), when the enzyme functions predominantly within the left cycle in Scheme 1. However, at high NADH concentrations acceleration in the presence of Zn2+ is still detectable (Fig. 2b). The estimated apparent rate constant for DCPIP reduction in the presence of saturating Zn2+ is 1.85 × 105 M–1·s–1 (Fig. 2a).
Double-reciprocal plots at saturating Zn2+ concentration display parallel lines (Fig. 2d), which are characteristic for a ping-pong mechanism for DCPIP reduction in the presence of zinc and give the values for the rate constants for NADH as (8.9 ± 2.0) × 105 M–1·s–1 and DCPIP as (2.2 ± 0.4) × 105 M–1·s–1. The value for the DCPIP rate constant is the same as the extrapolated value of k4 in the absence of zinc derived from Fig. 1d, and the value for the NADH rate constant is close to the value determined in the benzoquinone reduction studies [26].
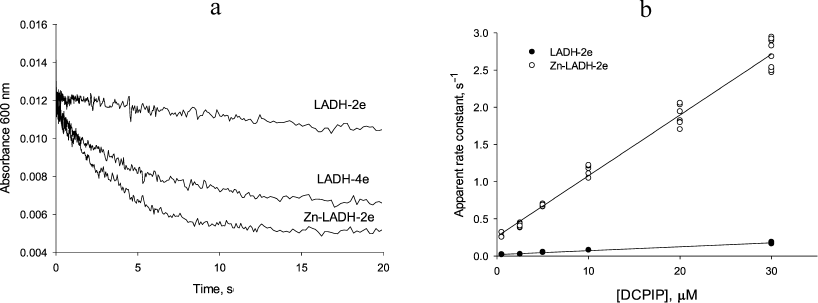
Transient kinetic studies of DCPIP reduction in the absence and presence of Zn2+. Acceleration in the presence of zinc ions is clearly seen in comparison of the time-courses for different reduced forms of LADH with DCPIP (Fig. 3a). In the case of DCPIP reduction, Zn-modified enzyme behaves similarly to the four-electron reduced LADH, while the two-electron reduced LADH is ca. 20-fold less active with respect to DCPIP (Fig. 3b, Table 1). The much lower activity of the two-electron reduced enzyme form with respect to DCPIP (i.e., k2 << k4, kDCPIPZn), likely originates from the additional intramolecular electron transfer step found in Cycle 1 that is absent in Cycles 2 and 3 in Scheme 2.
Fig. 3. a) Time courses for the reduction of 5 µM DCPIP by three different LADH forms. Preparation of these reduced LADH forms is described in “Materials and Methods” section. b) Determination of rate constants for DCPIP reduction by the two-electron reduced unmodified (closed circles) and Zn-modified (open circles) forms of LADH. Each point corresponds to a single run. Rate constants are calculated from the slopes. Reaction conditions are: 1 µM LADH and 1.5 µM NADH in 50 mM Tris-HCl buffer, pH 7.5, 22°C.
The Y-intercept of the plot of the apparent rate constants against the substrate concentration corresponds to the rate of DCPIP dissociation from the enzyme (Fig. 3b), which combined with the rate constant for DCPIP binding determined from the slope of the above dependence allows estimating a binding constant for DCPIP. Binding constants for the Zn2+-modified and unmodified two-electron reduced forms of the enzyme (3.5 ± 0.6 µM) are identical within experimental error. These values are in agreement with the previously reported [25], although the author postulated formation of a ternary complex in the case of DCPIP reduction. The value of the rate constant obtained from anaerobic stopped-flow experiments for the DCPIP reduction step by Zn-modified enzyme (Table 1) is in agreement with the one determined from the steady-state kinetics and supports the reaction mechanism presented in Schemes 1 and 2.
Table 1. Rate constants for the diaphorase
reaction depicted in Scheme 2
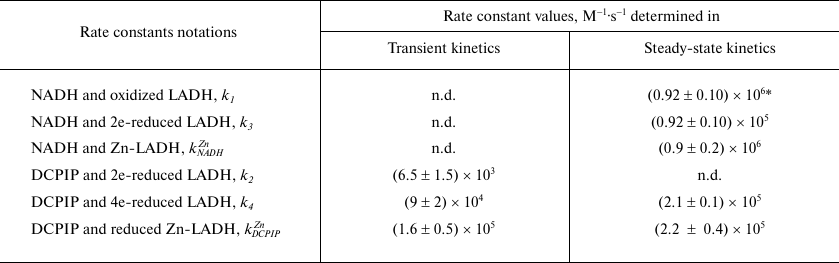
* k1 with benzoquinone as electron acceptor [26]; n.d. – not determined.
The degree of experimentally observed stimulation of the steady-state reaction in the presence of saturating Zn2+ concentrations varies from 2-2.5-fold at low DCPIP concentrations (because the rate-limiting step changes from k3 to kDCPIPZn ≈ k4) to 4-5-fold at low NADH concentrations (because the rate-limiting step changes from k2 to kDCPIPZn ≈ k4). Assuming equal values of the rate constants for DCPIP reduction by four-electron reduced and Zn-modified enzyme, k4 ≈ kDCPIPZn, and for NADH oxidation by the oxidized native and Zn-modified forms, k1 ≈ kNADHZn, the equation for the steady-state reaction rate in the presence of Zn2+ can be written as follows:
Thus, the extent to which Zn2+ accelerates the reaction under steady-state conditions is determined by the ratio of the composite rate constants k1, k2, k3, k4 and the range of substrate concentrations used in the particular experiment (Eq. 4). The consequences of this for catalytic behavior of LADH and related enzymes are discussed below.
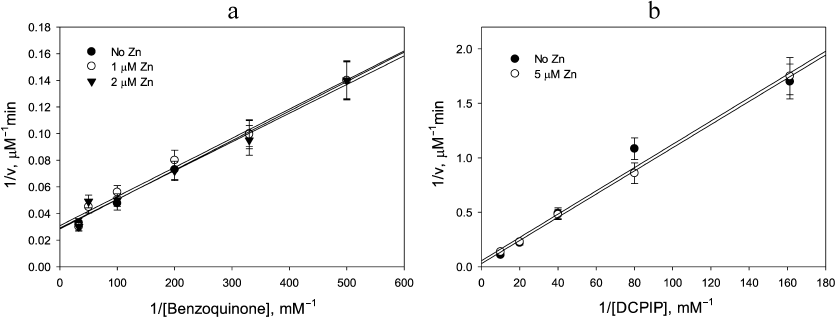
Lack of Zn2+ stimulatory effect in the benzoquinone reduction by LADH and in reactions catalyzed by E. coli thioredoxin reductase. The catalytic disulfide present in all FAD thiol oxidoreductases, including LADH, glutathione reductase, and thioredoxin reductase, complicates the mechanism of diaphorase reaction due to the existence of two- and four-electron reduced forms of the enzyme. As demonstrated above, Zn2+switches the enzyme mechanism from a two-cycle scheme to a single cycle by preventing electron exchange between the two reaction centers. Stimulation by Zn2+ would not be observed if the enzyme functions only within a single cycle, i.e., either the left or right cycles in Scheme 1, in the absence of Zn2+. If k2 ≥ k1 > k3 (i.e., the rate limiting step is 2-electron reduction of enzyme by NADH), then the enzyme will cycle exclusively between the oxidized and two-electron reduced forms. This condition is met for benzoquinone reduction, where kNADH = 9.2 × 105 M–1·s–1 < kBQ = 1.3 × 106M–1·s–1[26]. Therefore, Zn2+ binding to the catalytic dithiols should not produce any measurable effect on the reaction rate. The experimental data presented in Fig. 4a corroborate this prediction by showing no effect of Zn2+ addition on the rate of benzoquinone reduction (kBQ ≈ kBQZn ≈ 1.5 × 106M–1·s–1).
If k3 ≥ k1 > k2, the enzyme would cycle exclusively between the two- and four-electron reduced forms, i.e., with fully reduced FAD. This situation is never realized in mammalian LADH, glutathione reductase, or thioredoxin reductase, which form stable charge transfer complexes where an electron is shared between FAD and catalytic thiol [21, 27]. In contrast, E. coli thioredoxin reductase does not form a charge transfer complex and therefore cycles in the reaction course with fully reduced FAD [28-30]. As expected, no Zn2+ effect is observed for the E. coli thioredoxin reductase (Fig. 4b).
Fig. 4. Demonstration of lack of Zn2+ effect on (a) benzoquinone reduction by 20 µM NADH catalyzed by 50 nM LADH; and (b) DCPIP reduction by 20 µM NADPH catalyzed by 0.2 µM E. coli thioredoxin reductase. a) Closed circles, no Zn2+; open circles, 1 µM Zn2+; triangles, 2 µM Zn2+; b) Closed circles, no Zn2+; open circles, 5 µM Zn2+. The apparent rate constants determined from the slopes are: 1.5 × 106 M–1·s–1 (a), and 1.0 × 104 M–1·s–1 (b).
Dissimilar effects of Zn2+ on one- and two-electron substrate reduction reactions. It was suggested above that lower activity of the two-electron reduced enzyme form with respect to DCPIP (i.e., k2 << k4, kDCPIPZn), likely originates from the rate-limiting intramolecular electron transfer (see Cycle 1, Scheme 2) that supplies second electron for the obligatory two-electron reduction of DCPIP [31]. If this statement is correct, then replacement of DCPIP with a one-electron acceptor would be expected to eliminate the observed difference between k2 and kDCPIPZn.
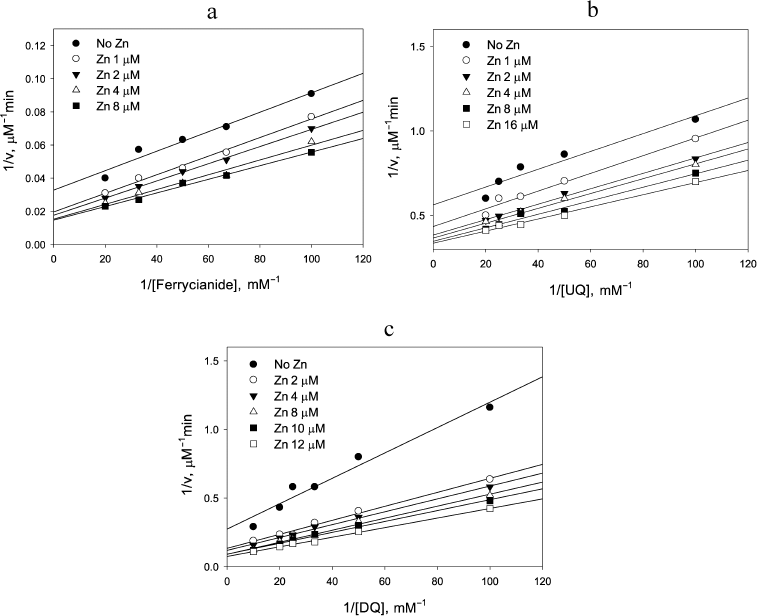
This was found to be the case for reduction of the one-electron acceptor, ferricyanide, by NADH: Zn2+ clearly affected the NADH oxidation step as can be judged from the decrease in the intercept in Fig. 5a with increasing Zn2+, but had essentially no effect on the ferricyanide reduction step because the slope did not change significantly (p = 0.063). The apparent rate constant observed for the ferricyanide reduction, (2.5 ± 0.2) × 105 M–1·s–1, was the same as reported earlier by Vienozinskis et al [26].
Fig. 5. Effect of Zn2+ on reduction of different electron acceptors by 20 µM NADH catalyzed by LADH. a) Ferricyanide, 0.15 µM LADH; b) ubiquinone, 0.20 µM LADH; c) duroquinone, 0.18 µM LADH.
Since k1 >> k3 in the case of pig heart LADH, Eq. 4 can be simplified to:
In the case of equal rate constants for reduction of an electron acceptor by two- and four-electron reduced forms of LADH (k2 ≈ k4 << k1) and under the condition of comparable NADH and substrate concentrations, only the rate of NADH oxidation will be enhanced by Zn2+:
Our model predicts that Zn2+ would increase the apparent rate constant for the substrate reduction step if there is a significant difference in the rate constants for substrate reduction by the two- and four-electron reduced forms of LADH (i.e., k2 << k4 in Scheme 1). The maximum effect of Zn2+ addition on the substrate reduction step would be observed at low NADH concentrations satisfying the condition k3[NADH] ≤ k2[S] where, in the absence of zinc, the enzyme steady-state form is mainly represented by the two-electron reduced form:
In this case, zinc addition at a concentration equal to KZn would increase the apparent rate constant for the electron acceptor from (1/k4 + 1/k2) to 1/k4. To visualize most clearly the impact of Zn2+ on the substrate reduction steps, the slopes that are estimated from Figure 5 are considered only for the cases of moderate NADH concentrations.
Two other substrates were investigated to further test the suggestion that Zn2+ acceleration would only be observed for the two-electron substrates. The effect of Zn2+ on ubiquinone reduction step (Fig. 5b) resembles that for ferricyanide (Fig. 5a), while the effect in the case of duroquinone (Fig. 5c) resembles the one for DCPIP (Fig. 2a). Rate constants for the reduction step of both duroquinone and ubiquinone by Zn2+-modified two-electron reduced enzyme are the same within the experimental error, (2.4 ± 0.2) × 104 M–1·s–1. However, acceleration under the steady-state conditions in the case of ubiquinone is low and the difference in slopes is not statistically significant (p = 0.065). In contrast, the Zn2+-induced acceleration for duroquinone is 2-3-fold, similar to that observed for DCPIP, and is highly significant (p = 0.0003). The observed difference between substrates is consistent with our model. Ubiquinone can serve as either a one or two-electron acceptor, while duroquinone acts exclusively as a two-electron acceptor (standard reduction potential for duroquinone/semiquinone is –240 mV, whereas that for duro-semiquinone/hydroquinone is +350 mV, making two-electron reduction more beneficial). Thus, zinc increases the rate constant for reduction of two-electron acceptors because the two-electron reduced Zn2+-modified enzyme carries both electrons on the FAD center.
DISCUSSION
This paper, together with the previous publication [21], clearly demonstrates that Zn2+ acts as a differential effector of the dehydrogenase and diaphorase activities of LADH. By coordinating to the reduced active site cysteines, Zn2+ switches the enzyme from dehydrogenase to diaphorase mode and selectively accelerates the reduction of exclusively two-electron acceptor substrates. This switch operates within the physiological range of Zn2+ concentrations.
The magnitude of Zn2+ effect on the substrate reduction step is determined by the k2/k4 ratio, which appeared to be affected by whether the substrate is a one- or two-electron acceptor (see Table 2). Ferricyanide is known to be exclusively a one-electron acceptor, while most quinones can act both as one and two-electron acceptors [26]. The data presented here are consistent with DCPIP and duroquinone acting as two-electron acceptors, while ferricyanide and ubiquinone act as one-electron acceptors. Benzoquinone can also act as a one-electron acceptor, which likely accounts for the absence of Zn2+ induced acceleration. However, the absence of apparent acceleration of NADH kinetics (i.e., the Y-intercept) suggests that the predominant difference for benzoquinone is that k2 ≥ k1 > k3 (i.e., the rate limiting step is 2-electron reduction of enzyme by NADH).
The conclusion that the two-electron reduced enzyme in the absence of Zn2+ is a very poor two-electron donor compared to the four-electron reduced enzyme or its Zn-modified form is further supported by the rate constants determined from anaerobic stopped-flow experiments (Fig. 3 and Table 1). Thus, we argue that Zn2+ switches the enzyme from a one-electron to a two-electron reductase. Within the class of FAD-dithiol oxido-reductases the switching effect will be most pronounced for those enzymes that have a large difference in redox potentials between the two and four-electron reduced states.
Table 2. Zinc effects on diaphorase reaction
steps for different acceptor substrates
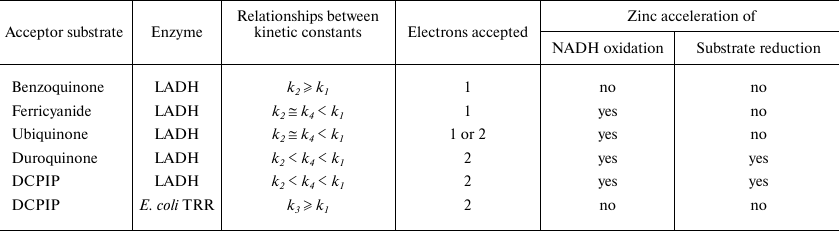
The nature of Zn2+effects on kinetics of ferricyanide and ubiquinone reduction catalyzed by LADH resembles that for the LADH-catalyzed reaction with oxygen [21]. In particular, the unchanged slope for the enzyme saturated with zinc in double-reciprocal plots of 1/v versus 1/[O2] caused us to conclude that the rate constants for interaction of the two- and four electron reduced enzyme with oxygen (k2 and k4) were equal [21]. However, this does not prove that the reaction products are the same. Oxygen could serve both as one- and two-electron acceptor yielding superoxide and hydrogen peroxide, respectively. It can be suggested that reduction of oxygen by the charge-transfer complex could yield substantially greater amounts of superoxide radical than initially reported [21, 32]. Type of the products formed in the course of the oxidase reaction and effect of zinc on their ratio should be reinvestigated. Zn2+ binding to LADH could produce the physiologically significant effect of switching enzyme from generating superoxide radical to generating hydrogen peroxide.
It has been suggested that the intracellular Zn2+ concentrations could be important for modulating mitochondrial interchange between pools of reduced nucleotides (NAD(P)H) and reduced thiols (e.g., LADH, thioredoxin and glutathione reductase activities) [21, 33]. In addition, varying intracellular Zn2+ could alter distribution of the one- and two-electron reduction products generated. Zinc strongly stimulates reduction of ubiquinone to ubiquinol catalyzed by lipoamide dehydrogenase and glutathione reductase, but inhibits thioredoxin reductase, a seleno-enzyme [34]. One-electron reduction products of LADH, such as semiquinones or superoxide radicals, are highly reactive and potentially damaging. Zn2+ could play a protective role against radical damage by promoting production of the two-electron products, such as quinones or H2O2, which are less damaging to cells.
A dual role of zinc in ischemia/reperfusion injury has been widely discussed [35-37], since benefits of zinc administration have been recently reported [38, 39]. Obviously, protecting effects of zinc supplementation could be associated with activation of antioxidant (Nrf2-driven) and antihypoxic genetic programs (HIF stabilization by Zn-induced inhibition [40] of HIF prolyl hydroxylase, a negative regulator of HIF stability), however, these mechanisms cannot explain the reported positive effects of post-injury administration of high zinc concentrations. The bound zinc concentration in mammalian cells is estimated as 200-300 µM [41], whereas the free zinc concentration in PC12 cells’ mitochondria matrix determined using a ratiometric fluorescent sensor is only 0.2 pM [42]. The switch described in this work operates with an apparent inhibition constant for zinc equal to ca. 2 µM in Tris buffer. This value corresponds to 100 nM free zinc concentration as determined in [21] for the same conditions as we used in this work. Elevation in the free zinc level upon oxidative stress, which results in chemical oxidation of protein thiols including those in metallothioneines, zinc storage proteins, can easily reach submicromolar concentrations sufficient to convert LADH into diaphorase. LADH is a model enzyme for the flavo-dithiol NAD(P)H-dependent oxidoreductases such as glutathione reductase and thioredoxin reductase. And observations made in this work could point to a possible pro-survival mechanism upon zinc coordination by active site thiols of these enzymes, namely, switching the enzyme diaphorase activity from one-electron to two-electron reduction mode.
Funding. The work in part was supported by the Russian Foundation for Basic Research (projects Nos. 20-04-00921, 17-54-33027, and 18-29-09154).
Acknowledgements. The authors thank Mrs. Gill A. Ashby and Prof. Roger N. F. Thorneley (UK) for their invaluable help with transient kinetics experiments.
Ethics declarations. The authors declare no conflict of interest in financial or any other sphere. This article does not contain any studies with human participants or animals performed by any of the authors.
REFERENCES
1.Koh, J.-Y., Suh, S. W., Gwag, B. J., He, Y. Y.,
Hsu, C. Y., and Choi, D. W. (1996) The role of zinc in selective
neuronal death after transient global cerebral ischemia,
Science, 272, 1013-1016,
doi: 10.1126/science.272.5264.1013.
2.Choi, D. W., and Koh, J. Y. (1998) Zinc and brain
injury, Ann. Rev. Neurosci., 21, 347-375,
doi: 10.1146/annurev.neuro.21.1.347.
3.Sensi, S. L., Paoletti, P., Bush, A. I., and
Sekler, I. (2009) Zinc in the physiology and pathology of the CNS,
Nat. Rev. Neurosci., 10, 780-791,
doi: 10.1038/nrn2734.
4.Shuttleworth, C. W., and Weiss, J. H. (2011)
Zinc: new clues to diverse roles in brain ischemia, Trends
Pharmacol. Sci., 32, 480-486,
doi: 10.1016/j.tips.2011.04.001.
5.Skulachev, V. P., Christyakov, V. V., Jasaitis, A.
A., and Smirnova, E. G. (1967) Inhibition of the respiratory chain by
zinc ions, Biochem. Biophys. Res. Commun., 26, 1-6,
doi: 10.1016/0006-291x(67)90242-2.
6.Sensi, S. L., Yin, H. Z., Carriedo, S. G., Rao, S.
S., and Weiss, J. H. (1999) Preferential Zn2+ influx through
Ca2+-permeable AMPA/kainate channels triggers prolonged
mitochondrial superoxide production, Proc. Natl. Acad. Sci. USA,
96, 2414-2419, doi: 10.1073/pnas.96.5.2414.
7.Dong, W., Qi, Z., Liang, J., Shi, W., Zhao, Y.,
Luo, Y., Ji, X., and Liu, K. J. (2015) Reduction of zinc accumulation
in mitochondria contributes to decreased cerebral ischemic injury by
normobaric hyperoxia treatment in an experimental stroke model, Exp.
Neurol., 272, 181-189,
doi: 10.1016/j.expneurol.2015.04.005.
8.Gazaryan, I. G., Krasinskaya, I. P., Kristal, B.
S., and Brown, A. M. (2007) Zinc irreversibly damages major enzymes of
energy production and antioxidant defense prior to mitochondrial
permeability transition, J Biol. Chem., 282, 24373-24380,
doi: 10.1074/jbc.M611376200.
9.Ji, S. G., Medvedeva, Yu. V., and Weiss, J. H.
(2020) Zn2+ entry through the mitochondrial calcium
uniporter is a critical contributor to mitochondrial dysfunction and
neurodegeneration, Exp. Neurol., 325, 113161,
doi: 10.1016/j.expneurol.2019.113161.
10.Brown, A. M., Kristal, B. S., Effron, M. S.,
Shestopalov, A. I., Ullucci, P. A., Sheu, K.-F. R., Blass, J. P., and
Cooper, A. J. L. (2000) Zn2+ inhibits
alpha-ketoglutarate-stimulated mitochondrial respiration and the
isolated alpha-ketoglutarate dehydrogenase complex, J. Biol.
Chem., 275, 13441-13447,
doi: 10.1074/jbc.275.18.13441.
11.Casola, L., Brumby, P. E., and Massey, V. (1966)
The reversible conversion of lipoyl dehydrogenase to an artifactual
enzyme by oxidation of sulfhydryl groups, J. Biol. Chem.,
241, 4977-4984.
12.Veeger, C., and Massey, V. (1962) Inhibition of
lipoyl dehydrogenase by trace metals, Biochim. Biophys. Acta,
64, 83-100, doi: 10.1016/0006-3002(60)90108-6.
13.Casola, L., and Massey, V. (1966) Differential
effects of mercurial on the lipoyl reducatase and diaphorase activities
of lipoyl dehydrogenase, J. Biol. Chem., 241,
4985-4993.
14.Nakamura, M., and Yamazaki, I. (1972)
One-electron transfer reactions in biochemical systems. VI. Changes in
electron transfer mechanism of lipoamide dehydrogenase by modification
of sulfhydryl groups, Biochim. Biophys. Acta, 267,
249-257, doi: 10.1016/0005-2728(72)90113-2.
15.Thorpe, C., and Williams, C. H., Jr. (1975)
Modification of pig heart lipoamide dehydrogenase by cupric ions,
Biochemistry, 14, 2419-2424,
doi: 10.1021/bi00682a023.
16.Lowe, C. R. (1977) Immobilised lipoamide
dehydrogenase. 2. Properties of the enzyme immobilised to agarose
through spacer molecules of various lengths, Eur. J. Biochem.,
76, 401-409, doi: 10.1111/j.1432-1033.1977.tb11608.x.
17.Gutierrez Correa, J., and Stoppani, A. O. M.
(1993) Inactivation of lipoamide dehydrogenase by cobalt(II) and
iron(II) Fenton systems: effect of metal chelators, thiol compounds and
adenine nucleotides, Free Radic. Res. Commun., 19,
303-314, doi: 10.3109/10715769309056519.
18.Olsson, J. M., Xia, L., Eriksson, L. C., and
Bjornstedt, M. (1999) Ubiquinone is reduced by lipoamide dehydrogenase
and this reaction is potently stimulated by zinc, FEBS Lett.,
448, 190-192, doi: 10.1016/s0014-5793(99)00363-4.
19.Xia, L., Bjornstedt, M., Nordman, T., Eriksson,
L. C., and Olsson, J. M. (2001) Reduction of ubiquinone by lipoamide
dehydrogenase. An antioxidant regenerating pathway, Eur. J.
Biochem., 268, 1486-1490,
doi: 10.1046/j.1432-1327.2001.02013.x.
20.Hopkins, N., and Williams, C. H., Jr. (1995)
Characterization of lipoamide dehydrogenase from Escherichia
coli lacking the redox active disulfide: C44S and C49S,
Biochemistry, 34, 11757-11765,
doi: 10.1021/bi00037a013.
21.Gazaryan, I. G., Krasnikov, B. F., Ashby, G. A.,
Thorneley, R. N., Kristal, B. S., and Brown, A. M. (2002) Zinc is a
potent inhibitor of thiol oxidoreductase activity and stimulates
reactive oxygen species production by lipoamide dehydrogenase, J.
Biol. Chem., 277, 10064-10072,
doi: 10.1074/jbc.M108264200.
22.Popov, V. O., Gazarian, I. G., Egorov, A. M., and
Berezin, I. V. (1985) NAD-dependent hydrogenase from the
hydrogen-oxidizing bacterium Alcaligenes eutrophus Z-1. Kinetic
studies of the NADH-dehydrogenase activity, Biochim. Biophys.
Acta, 827, 466-471.
23.Maeda-Yorita, K., Russell, G. C., Guest, J. R.,
Massey, V., and Williams, C. H., Jr. (1991) Properties of lipoamide
dehydrogenase altered by site-directed mutagenesis at a key residue
(I184Y) in the pyridine nucleotide binding domain, Biochemistry,
30, 11788-11795, doi: 10.1021/bi00115a008.
24.Van Berkel, W. J. H., Regelink, A. G., Beintema,
J. J., and de Kok, A. (1991) The conformational stability of the redox
states of lipoamide dehydrogenase from Azotobacter vinelandii,
Eur. J. Biochem., 202, 1049-1055,
doi: 10.1111/j.1432-1033.1991.tb16469.x.
25.Tsai, C. S. (1980) Kinetic studies of
multifunctional reactions catalysed by lipoamide dehydrogenase, Int.
J. Biochem., 11, 407-413,
doi: 10.1016/0020-711x(80)90311-0.
26.Vienozinskis, J., Butkus, A., Cenas, N., and
Kulys, J. (1990) The mechanism of the quinone reductase reaction of pig
heart lipoamide dehydrogenase, Biochem. J., 269, 101-105,
doi: 10.1042/bj2690101.
27.Williams, C. H., Arscott, L. D., Muller, S.,
Lennon, B. W., Ludwig, M. L., Wang, P. F., Veine, D. M., Becker, K.,
and Schirmer, R. H. (2000) Thioredoxin reductase two modes of catalysis
have evolved, Eur. J. Biochem., 267, 6110-6117,
doi: 10.1046/j.1432-1327.2000.01702.x.
28.Lennon, B. W., and Williams, C. H., Jr. (1996)
Enzyme-monitored turnover of Escherichia coli thioredoxin
reductase: insights for catalysis, Biochemistry, 35,
4704-4712, doi: 10.1021/bi952521i.
29.Lennon, B. W., Williams, C. H., Jr., and Ludwig,
M. L. (2000) Twists in catalysis: alternating conformations of
Escherichia coli thioredoxin reductase, Science,
289, 1190-1194, doi: 10.1126/science.289.5482.1190.
30.Williams, C. H., Jr. (1995) Mechanism and
structure of thioredoxin reductase from Escherichia coli,
FASEB J., 9, 1267-1276,
doi: 10.1096/fasebj.9.13.7557016.
31.Dooijewaard, G., and Slater, E. C. (1976)
Steady-state kinetics of high molecular weight (type-I) NADH
dehydrogenase, Biochim. Biophys. Acta, 440, 1-15,
doi: 10.1016/0005-2728(76)90109-2.
32.Bando, Y., and Aki, K. (1991) Mechanisms of
generation of oxygen radicals and reductive mobilization of ferritin
iron by lipoamide dehydrogenase, J. Biochem., 109,
450-454, doi: 10.1093/oxfordjournals.jbchem.a123402.
33.Gaballa, A., and Helmann, J. D. (2002) A
Peroxide-induced zinc uptake system plays an important role in
protection against oxidative stress in Bacillus subtilis,
Mol. Microbiol., 45, 997-1005,
doi: 10.1046/j.1365-2958.2002.03068.x.
34.Nordman, T., Xia, L., Björkhem-Bergman, L.,
Damdimopoulos, A., Nalvarte, I., Arnér, E. S., Spyrou, G.,
Eriksson, L. C., Björnstedt, M., and Olsson, J. M. (2003)
Regeneration of the antioxidant ubiquinol by lipoamide dehydrogenase,
thioredoxin reductase and glutathione reductase, Biofactors,
18, 45-50, doi: 10.1002/biof.5520180206.
35.Galasso, S. L., and Dyck, R. H. (2007) The role
of zinc in cerebral ischemia, Mol. Med., 13, 380-387,
doi: 10.2119/2007-00044.Galasso.
36.Levenson, C. W. (2005) Zinc supplementation:
neuroprotective or neurotoxic? Nutr. Rev., 63, 122-125,
doi: 10.1111/j.1753-4887.2005.tb00130.x.
37.Ischia, J., Bolton, D. M., and Patel, O. (2019)
Why is it worth testing the ability of zinc to protect against
ischaemia reperfusion injury for human application, Metallomics,
11, 1330-1343, doi: 10.1039/c9mt00079h.
38.Aquilani, R., Baiardi, P., Scocchi, M., Iadarola,
P., Verri, M., Sessarego, P., Boschi, F., Pasini, E., Pastoris, O., and
Viglio, S. (2009) Normalization of zinc intake enhances neurological
retrieval of patients suffering from ischemic strokes, Nutr.
Neurosci., 12, 219-225,
doi: 10.1179/147683009X423445.
39.Kitamura, Y., Iida, Y., Abe, J., Ueda, M.,
Mifune, M., Kasuya, F., Ohta, M., Igarashi, K., Saito, Y., and Saji, H.
(2006) Protective effect of zinc against ischemic neuronal injury in a
middle cerebral artery occlusion model, J. Pharmacol. Sci.,
100, 142-148, doi: 10.1254/jphs.fp0050805.
40.Osipyants, A. I., Smirnova, N. A., Khristichenko,
A. Yu., Nikulin, S. V., Zakhariants, A. A., Tishkov, V. I., Gazaryan,
I. G., and Poloznikov, A. A. (2018) Metal ions as activators of hypoxia
inducible factor, Moscow University Chem. Bull., 73,
13-18.
41.Maret, W. (2015) Analyzing free zinc(II) ion
concentrations in cell biology with fluorescent chelating molecules,
Metallomics, 7, 202-211,
doi: 10.1039/c4mt00230j.
42.McCranor, B. J., Bozym, R. A., Vitolo, M. I.,
Fierke, C. A., Bambrick, L., Polster, B. M., Fiskum, G., and Thompson,
R. B. (2012) Quantitative imaging of mitochondrial and cytosolic free
zinc levels in an in vitro model of ischemia/reperfusion, J.
Bioenerg. Biomembr., 44, 253-263,
doi: 10.1007/s10863-012-9427-2.