Isoforms of the DHTKD1-Encoded 2-Oxoadipate Dehydrogenase, Identified in Animal Tissues, Are not Observed upon the Human DHTKD1 Expression in Bacterial or Yeast Systems
A. I. Boyko1,a*, A. V. Artiukhov1,2, T. Kaehne3, M. L. di_Salvo4, M. C. Bonaccorsi_di_Patti4, R. Contestabile4, A. Tramonti4,5, and V. I. Bunik1,2,6,b*
1Faculty of Bioengineering and Bioinformatics, Lomonosov Moscow State University, 119234 Moscow, Russia2Belozersky Institute of Physico-Chemical Biology, Lomonosov Moscow State University, 119234 Moscow, Russia
3Institute of Experimental Internal Medicine, Otto-von-Guericke University, 39120 Magdeburg, Germany
4Department of Biological Sciences A. Rossi Fanelli, Sapienza University, 00185 Rome, Italy
5Institute of Molecular Biology and Pathology, Council of National Research, 00185 Rome, Italy
6Department of Biological Chemistry, Sechenov First Moscow State Medical University, 119146 Moscow, Russia
* To whom correspondence should be addressed.
Received June 16, 2020; Revised June 25, 2020; Accepted June 26, 2020
Unlike the OGDH-encoded 2-oxoglutarate dehydrogenase (OGDH), which is an essential enzyme present in all animal tissues, expression of the DHTKD1-encoded isoenzyme, 2-oxoadipate dehydrogenase (OADH), depends on a number of factors, and mutant DHTKD1 phenotypes are rarely manifested. Physiological significance of OADH is also obscured by the fact that both isoenzymes transform 2-oxoglutarate and 2-oxoadipate. By analogy with other members of the 2-oxo acid dehydrogenases family, OADH is assumed to be a component of the multienzyme complex that catalyzes oxidative decarboxylation of 2-oxoadipate. This study aims at molecular characterization of OADH from animal tissues. Phylogenetic analysis of 2-oxo acid dehydrogenases reveals OADH only in animals and Dictyostelium discoideum slime mold, within a common branch with bacterial OGDH. Examination of partially purified animal OADH by immunoblotting and mass spectrometry identifies two OADH isoforms with molecular weights of about 130 and 70 kDa. These isoforms are not observed upon the expression of human DHTKD1 protein in either bacterial or yeast system, where the synthesized OADH is of expected molecular weight (about 100 kDa). Thus, the OADH isoforms present in animal tissues, may result from the animal-specific regulation of the DHTKD1 expression and/or posttranslational modifications of the encoded protein. Mapping of the peptides identified in the OADH preparations, onto the protein structure suggests that the 70-kDa isoform is truncated at the N-terminus, but retains the active site. Since the N-terminal domain of OGDH is required for the formation of the multienzyme complex, it is possible that the 70-kDa isoform catalyzes non-oxidative transformation of dicarboxylic 2-oxo acids that does not require the multienzyme structure. In this case, the ratio of the OADH isoforms in animal tissues may correspond to the ratio between the oxidative and non-oxidative decarboxylation of 2-oxoadipate.
KEY WORDS: DHTKD1, OGDH, carboligase, 2-oxoglutarate dehydrogenase isoenzyme, 2-oxoadipate dehydrogenase isoform, posttranslational modificationsDOI: 10.1134/S0006297920080076
Abbreviations: DHTKD1, 2-oxoadipate dehydrogenase (dehydrogenase E1 and transketolase domain-containing 1) gene; OADH, 2-oxoadipate dehydrogenase; OGDH, 2-oxoglutarate dehydrogenase.
INTRODUCTION
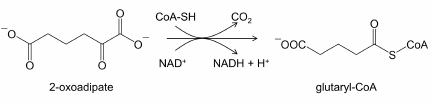
2-Oxoadipate dehydrogenase (OADH), a product of the DHTKD1 (dehydrogenase E1 and transketolase domain-containing 1) gene, was discovered by the automatic annotation of animal genomes. Based on the sum of identical amino acids and conservative amino acid substitutions, the OADH sequence has 56% similarity with 2-oxoglutarate dehydrogenase (OGDH) [1], a key enzyme of the tricarboxylic acid cycle encoded by the OGDH gene. Both enzymes catalyze transformation of dicarboxylic 2-oxo acids (2-oxoglutarate and 2-oxoadipate), albeit with different efficiencies [2-4]. The structural and functional similarities allow to consider OADH and OGDH as isoenzymes. Enzymological studies of the recombinant OADH [5] have demonstrated that within the 2-oxoadipate dehydrogenase multienzyme complex, assembled in vitro from the enzymatic components produced in bacterial expression system, OADH catalyzes the reaction of oxidative decarboxylation of 2-oxoadipate, analogous to that of OGDH (reaction 1):
 (1)
(1)However, manifestation of mutations in the human DHTKD1 gene, as well as accumulation of 2-oxoadipate and 2-aminoadipate (intermediates of lysine and tryptophan catabolism) in the cases of such mutations [6, 7], are fundamentally different from the outcomes of mutations in OGDH, that are incompatible with life [8] due to severely impaired CNS development and function [9]. The studies of such mutants suggest that OADH and OGDH are not redundant at the organism level, despite the similarity of their catalytic functions. Thus, the functions of OADH and OGDH differ more significantly in vivo than in vitro.
Investigation of physiological significance of OADH-catalyzed oxidative decarboxylation of 2-oxoadipate (a common intermediate of lysine and tryptophan catabolism) is hindered by the limited understanding and cell-specific expression of metabolic pathways employing the reaction. Moreover, the absence of a clearly pronounced phenotype associated with the DHTKD1 mutations, may be also due to a significant variability in the DHTKD1 expression in different organs and tissues of the same organism and at the population level. Down-regulation of OADH synthesis in cells perturbs mitochondrial biogenesis and energy metabolism [10]. At the organism level, changes in the DHTKD1 expression are associated with the development of systemic pathologies of glucose metabolism, such as diabetes and obesity [11-13]. Accumulation of 2-oxo- and 2-aminoadipate due to mutations of essential OADH residues accompanies neuromuscular disorders [6, 7, 14]. On the other hand, 2-aminoadipate demonstrates a neuroprotective effect in a model of Parkinson disease [15], which indicates complexity of (patho)physiological significance of the OADH-catalyzed reaction. Therefore, investigation of OADH is important from both fundamental and medical points of view.
In this study, the structural features of OADH that underlie physiologically relevant regulation of this enzyme in animal tissues, are characterized. We demonstrate that the molecular weight of OADH detected in rat tissues, is significantly different from that calculated from the enzyme primary structure and inherent in the recombinant protein synthesized in either bacterial or yeast expression systems. These results imply alternative protein products of the DHTKD1 gene and/or their posttranslational modifications in animals, which do not occur when the recombinant OADH is produced.
MATERIALS AND METHODS
Materials. All used reagents were of the highest purity grade available. Polyethylene glycol-6000, EDTA, and PMSF were purchased from Serva (Germany); protease inhibitor cocktail – from Roche (Switzerland); methanol – from Merck (Germany); Triton-X 100, KH2PO4, and NaCl – from Panreac (Spain); glycerol and sucrose – from Biomedicals, LLC (USA); MgCl2 and CaCl2 – from DIA-M (Russia); MOPS – from Helicon (Russia); yeast extract – from Fluka (USA); Tripton – from GeneSpin (Italy); and glucose – from VWR (USA). All other reagents were from Sigma (USA).
Animals. Male Wistar rats (body weight 300-350 g) were kept at the Animal Facility of the Faculty of Biology, Lomonosov Moscow State University, and fed with a standard diet. The animals had free access to water and food and were kept at a 12/12 h light/dark cycle (light phase from 9:00 am to 9:00 pm local time) at a temperature of 21 ± 2°C and relative humidity of 53 ± 5%. The animals were sacrificed by decapitation with a guillotine.
Preparation of the tissue homogenates and OADH-enriched fractions. Homogenization of rat tissues was performed according to the previously published protocol [16]. OADH-enriched preparations from the heart, liver, and brain were obtained using the procedure described earlier [3].
OADH production in bacterial expression system. The human DHTKD1 cDNA was inserted into the pET28 vector and expressed in E. coli BL21(DE3) cells according to a standard protocol [17]. The cells were pelleted by centrifugation for 5 min at 2550g and frozen at –20°C. Bacterial lysates were produced by incubation of the bacterial cells with lysozyme followed by ultrasonic disintegration. The recombinant protein with the His6-tag at the C-terminus was purified from the bacterial lysate on a Ni-NTA-agarose resin in 50 mM KH2PO4, pH 8.0, containing 300 mM KCl and 300 mM imidazole.
OADH production in yeast expression system. The pIB2 vector with the human DHTKD1 cDNA under the GAP promoter was linearized with the SalI restriction endonuclease and used for the transformation of competent Pichia pastoris GS115 (his4) cells by electroporation according to the published procedure [18]. The yeast was cultivated in a sterile YPD medium containing 2% glucose for ~48 h at 28°C with shaking, followed by centrifugation and freezing. The cells were disrupted using glass beads in the presence of DNase 1 and RNase. OADH with the His6-tag at the C-terminus was purified on a Ni-NTA Superflow column (1-ml Cartridge, Qiagen, Germany) equilibrated with 50 mM KH2PO4 buffer (pH 8.0) containing 300 mM KCl. OADH was eluted from the column with an increasing gradient of imidazole concentration (30 mM to 300 mM) in the same buffer using an ÄKTA Prime chromatographic system (GE Healthcare, USA).
SDS-PAGE and immunoblotting. Protein samples were mixed with the solubilizing buffer (250 mM Tris-HCl, 8% SDS, 40% (v/v) glycerol, 20% β-mercaptoethanol, and 0.04% bromophenol blue, pH 6.8) at a ratio 3 : 1. The samples were boiled for 5 min and cooled to room temperature prior to application onto a gel.
OADH was detected by a standard immunoblotting procedure using mouse antibodies against OADH (H00055526-B01P, 1 : 500, Abnova, Taiwan). Horseradish peroxidase-conjugated secondary antibodies were from Boehringer Mannheim GmbH (1520709, 1 : 6000, Germany) or Imtek (P-GAM Iss, 1 : 1000, Russia). In the former case, freshly prepared chemiluminescent substrate was used: 10 mg of 3-amino-9-ethyl-carbazole dissolved in 25 ml of 50 mM CH3COONa, pH 5.0, containing 5% dimethyl formamide, was mixed with 30% hydrogen peroxide at a ratio 1 : 2000. In the latter case, the ECL substrate kit (Bio-Rad, USA) was used. Band images were acquired with a ChemiDoc MP imaging system (Bio-Rad).
The molecular weights of the protein bands stained by the OADH antibodies, were determined from the linear dependence of the logarithm of the protein molecular weight on its electrophoretic mobility. The known weights of proteins of Unstained Protein Standards kit (Bio-Rad) were used for the calibration.
Identification of OADH peptides in rat tissues. After SDS-PAGE, the gel area corresponding to proteins with the molecular weights of 130 ± 10 kDa and 70 ± 5 kDa were analyzed by tandem mass spectroscopy (LC-MS/MS) employing trypsinolysis as described previously [19]. Rat proteins in these bands were identified using the Rat Swiss-Prot database.
Construction of phylogenetic tree for the 2-oxo acid dehydrogenase family. To construct the phylogenetic tree, we searched for rat OADH (UniProt identifier Q4KLP0) homologues in Swiss-Prot and RefSeq databases, using the BLASTP tool [20]. Except for E-value threshold of 10–3 and word size of 2, default search parameters were applied. Sequences produced by automatic translation of DNA (XM_/XP_ prefix) and non-unique sequences (WP_ prefix) were excluded. The search results from the two databases were downloaded as sequences in the fasta format excluding duplicate hits. Multiple sequence alignment was conducted with the Muscle algorithm in the MEGA X program [21]. Phylogenetic tree visualizing the results of the alignment, was constructed using the neighbor-joining method.
Modeling. The amino acid sequence of the rat protein with the UniProt identifier Q4KLP0 (DHTK1_RAT) was used for predicting the 3D structure of mammalian OADH monomer. Considering that phylogenetic analysis indicated the similarity of OADH to the bacterial OGDH, the resolved 3D structure of the mycobacterial OGDH holoenzyme (RSCB PDB identifier 2XT6) was selected as a template for OADH modeling. The model was built with the I-TASSER server [22]. The PyMOL 2.0.4. program (Schrödinger, Inc.) was used for protein visualization.
RESULTS AND DISCUSSION
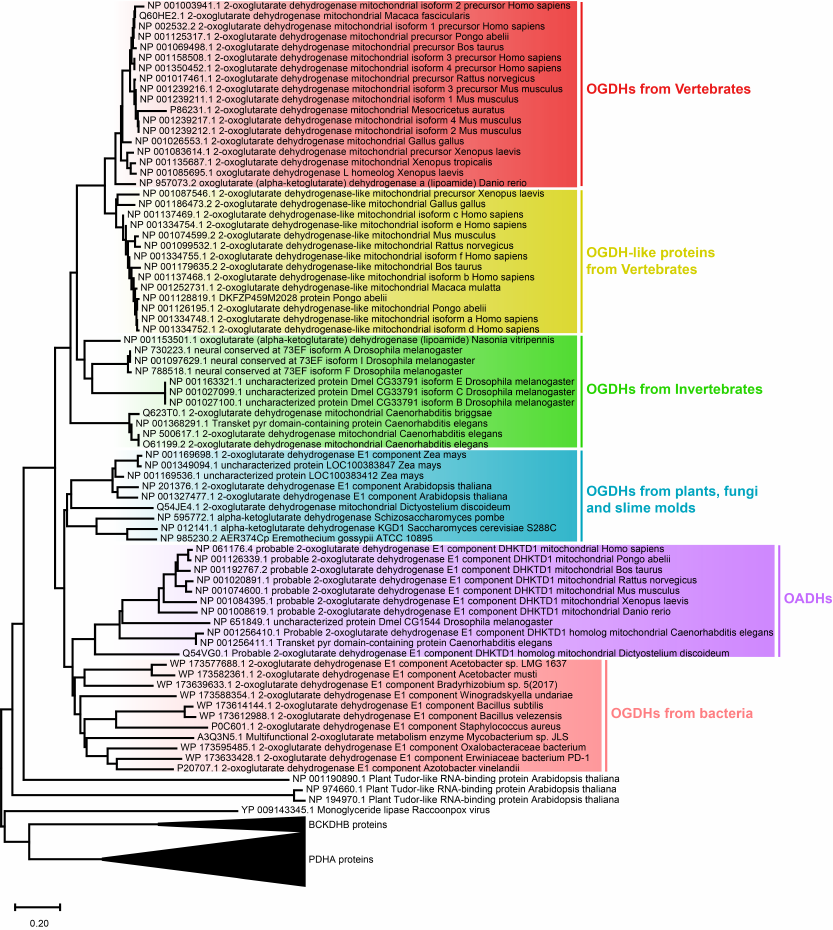
Phylogenetic analysis of the OADH origin. The phylogenetic tree of the 2-oxo acid dehydrogenase family presented in Fig. 1, shows that the DHTKD1-encoded OADH diverged from OGDH during early evolution of multicellular eukaryotes. Currently, the DHTKD1-encoded OADH has been annotated only in animals and slime mold Dictyostelium discoideum (Fig. 1). Based on this finding, it may be suggested that OADH evolved in the organisms that started feeding on other organisms and, as a result, lost their ability for the biosynthesis of some metabolically essential compounds. In particular, the ability to synthesize lysine and tryptophan which are catabolized by OADH, had been independently lost in animals and Dictyostelium in the course of evolution [23].
Fig. 1. Phylogenetic tree of 2-oxo acid dehydrogenases. For the optimal visualization of results, α-subunits of pyruvate dehydrogenases (PDHA proteins) are presented as one common branch; β-subunits of the branched-chain 2-oxo acid dehydrogenases (BCKDHB proteins) are presented in a similar manner. Scale bar: 0.20 amino acid substitutions per residue.
Another interesting finding of the phylogenetic analysis is the formation of a common cluster by eukaryotic OADHs and bacterial OGDHs (Fig. 1), pointing to their close relationship. Such structural similarity suggests an evolutionary need to preserve certain properties and functions of bacterial OGDHs that have been lost by eukaryotic OGDHs due to their increased specialization. In particular, in comparison with the bacterial complexes, those of eukaryotes employ additional protein–protein interactions during self-assembly and display increased selectivity of the 2-oxo substrates transformation [24].
Thus, the evolvement of OADH from its homologue OGDH likely reflects divergence of the OGDH and OADH functions due to natural selection, with the associated necessity of OADH regulation independent of OGDH.
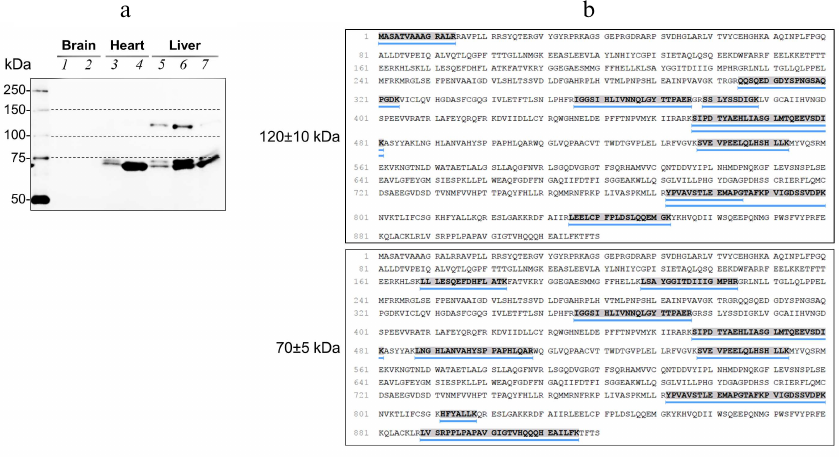
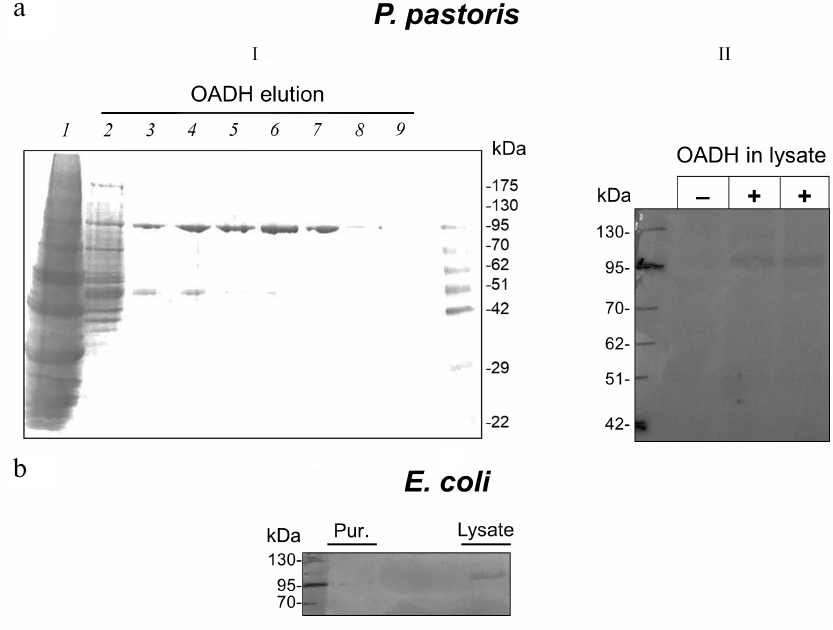
OADH isoforms in rat tissues. Analysis of rat tissue homogenates by immunoblotting demonstrates that OADH-specific antibodies react with two bands of proteins (Fig. 2a). The apparent molecular weights of these bands determined by averaging the results of analysis of 25 samples in 5 independent experiments, are 127 ± 2 and 69 ± 1 kDa. In some cases, the low-molecular-weight band is represented by two closely located bands, typical for the proteins forming intramolecular disulfide bonds [25]. Considering the standard deviations of the molecular weight estimation, the weights of the isoforms are further mentioned as 130 and 70 kDa to simplify discussion. None of these bands has the molecular weight of 100 kDa calculated for the native OADH based on its amino acid sequence excluding the 2.5-kDa peptide targeting the protein to mitochondria. The 130-kDa band is mainly detected in the liver (Fig. 2a), where the levels of DHTKD1 mRNA and OADH are the highest in comparison with other tissues [3]. The presence of OADH in both protein bands reacting with the antibodies, is confirmed by mass spectrometry identification of the OADH-specific peptides (Fig. 2b). The obtained data indicate that the difference between the molecular weights of OADH isoforms identified by immunoblotting, and the calculated weight of OADH, is due to changes in the OADH structure.
Fig. 2. OADH isoforms in the rat tissues. a) Proteins reactive to antibodies to OADH in the preparations from different tissues: 1 and 2) cerebral cortex homogenates; 3) heart homogenate; 4) heart sample enriched with OADH; 5) liver homogenate; 6 and 7) two samples from liver, with different degree of enrichment with OADH. An aliquot of the protein samples (~35 µg) was loaded in each lane. Dashed lines indicate the protein markers with the molecular weights of 75, 100, and 150 kDa. b) OADH peptides identified by LC-MS/MS in the rat liver preparation are highlighted and underlined. The mass spectrometry analysis was carried out for the gel regions corresponding to the molecular weights of 120-140 and 65-75 kDa.
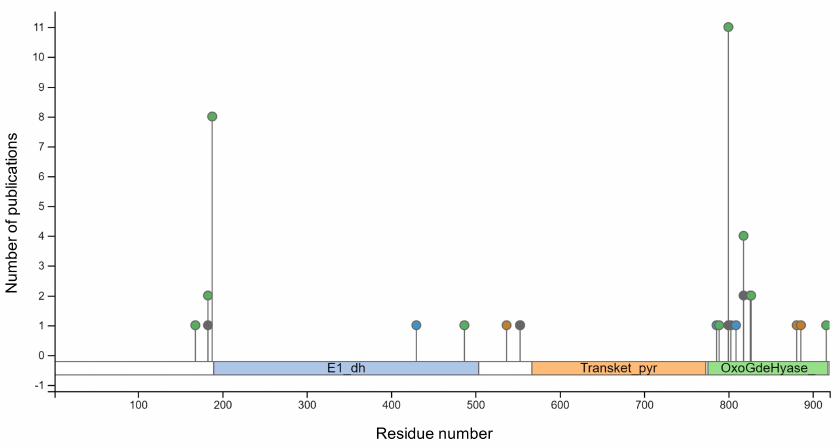
It is known from high-throughput studies that OADH interacts with ubiquitin and ubiquitin-like proteins (NEDD8, SUMO1) [26, 27]. Ubiquitination of OADH at Lys537, Lys881, and Lys886 residues is demonstrated in one of these studies (Fig. 3). Unlike other modification found in OADH (Fig. 3), ubiquitination could increase the enzyme molecular weight from 100 kDa to 130 kDa, as observed in this work, and even further.
Fig. 3. Modifications of rat OADH, identified by high-throughput screening. The data are obtained from PhosphoSitePlus v.6.5.9.1. database [28]. Domains of the OADH sequence are shown in different colors: E1_dh, dehydrogenase domain (blue); Transket_pyr, transketolase pyrimidine-binding domain (orange); OxoGdeHyase, C-terminal 2-oxo acid dehydrogenase domain (green). Modifications of the indicated amino acid residues include acetylation (green circles); phosphorylation (blue circles); succinylation (grey circles), ubiquitination (brown circles).
Peptides containing characteristic Lys-ε-Gly-Gly triplets are found during identification of ubiquitinated proteins by mass spectrometry after trypsinolysis [29]. However, in the examined rat liver samples such peptides could not have been identified; hence, the possibility of OADH modification with ubiquitin or ubiquitin-like proteins requires further investigation. Nevertheless, an OADH-specific peptide is revealed also during mass-spectrometric analysis of the gel region above the protein band of 130 kDa. This finding suggests the existence of OADH isoforms with molecular weight(s) higher than 130 kDa. Obviously, the amount of these high-molecular-weight forms detectable by mass spectrometry, is low compared to that of the main OADH isoforms, which are detectable by immunoblotting (Fig. 2a).
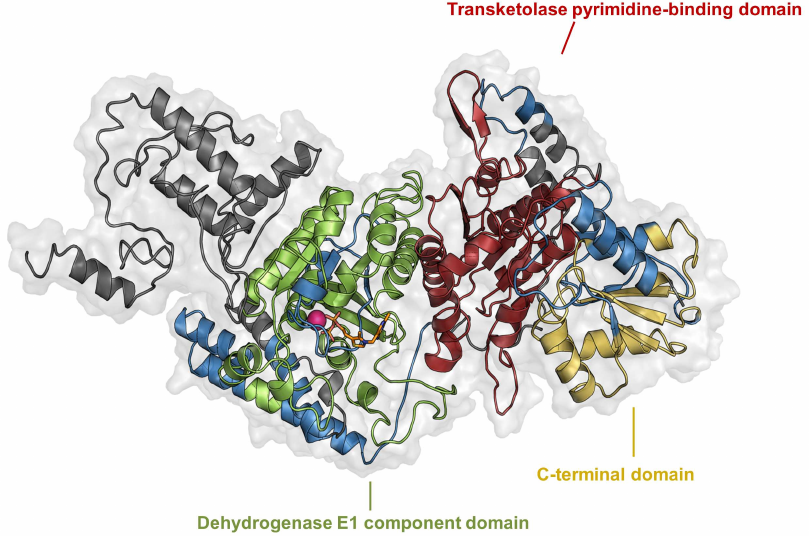
OADH peptides identified by mass spectrometry of the gel region corresponding to proteins with molecular weight of 65-75 kDa (Fig. 2b), are marked on the protein monomer model (Fig. 4), based on the structure of Mycobacterium smegmatis OGDH. As can be seen in Fig. 4, the identified peptides cover the dehydrogenase, transketolase, and C-terminal domains of OADH, but are absent at the N-terminus. The obtained results suggest that the 70-kDa OADH isoform may arise due to alternative site of transcription start of DHTKD1 gene or alternative splicing of mRNA. Besides, it could result from specific cleavage of the N-terminal fragment.
Fig. 4. Model of rat OADH monomer with thiamine diphosphate and Mg2+. Magnesium ion is shown in pink, thiamine diphosphate molecule – in orange, dehydrogenase domain of E1-components of 2-oxo acid dehydrogenases – in green, transketolase pyrimidine-binding domain – in red, C-terminal domain – in yellow. OADH peptides identified in the fraction of proteins with molecular weight of 70 kDa (Fig. 2b), are marked in blue.
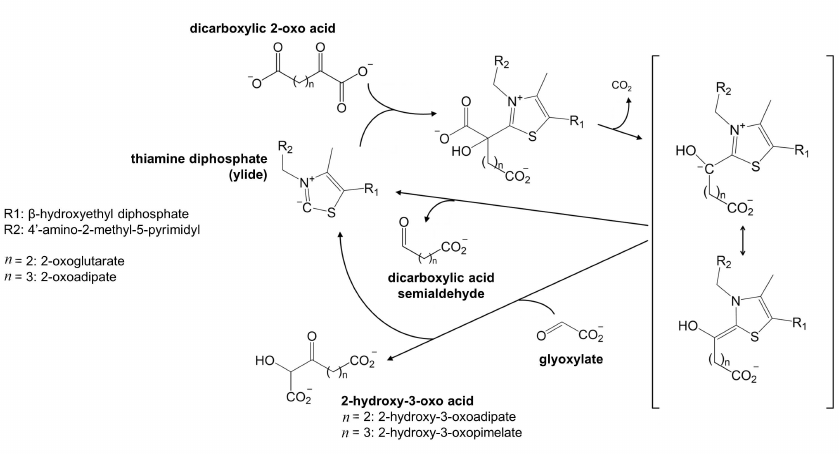
Catalytic competence of 70-kDa isoform is unclear. In eukaryotic OGDH, the N-terminal fragment participates in the formation of the multienzyme complex, and proteolytic removal of this fragment inactivates the complex-catalyzed reaction [30]. The existence of the N-terminus-truncated 70-kDa isoform may therefore testify that OADH has functions that do not require formation of the multienzyme complex. Non-oxidative decarboxylation of 2-oxo acids, including the carboligase reactions predicted for OADH [1], could be among these functions. It is worth noting in this regard that mycobacterial OGDH, which has significant structural similarity with OADH according to our phylogenetic analysis (Fig. 1), is known to efficiently catalyze non-oxidative decarboxylation of 2-oxoglutarate. In the non-oxidative reaction, the decarboxylation product is either released as succinic semialdehyde or condensed with glyoxylate in the so-called carboligase reaction (Fig. 5). The biological significance of this non-oxidative function of animal OADH could explain a high content of the 70-kDa isoform in the heart (Fig. 2a), where the rate of the complex-catalyzed overall reaction of oxidative decarboxylation of 2-oxoadipate (reaction 1) at its low concentration which is saturating for OADH, but not OGDH, is close to zero [3]. Under these conditions, the overall reaction of 2-oxoadipate oxidative decarboxylation is measurable only with the liver preparations of the enzyme [3], where both OADH isoforms are detected (Fig. 2a).
Fig. 5. Non-oxidative transformation of dicarboxylic 2-oxo acids, potentially catalyzed by OADH. The thiamine diphosphate ylide formed in the OADH active site, reacts with 2-oxoglutarate or 2-oxoadipate, catalyzing their decarboxylation. The reaction intermediate, stabilized by the resonance transition between its carbanion and enamine forms, is shown in square brackets. In the absence of acceptor substrates, the intermediate decomposes with the formation of semialdehyde of the dicarboxylic acid. In the presence of compounds with the activated carbon atom, such as glyoxylate, the 2-oxo acid dehydrogenases may catalyze the carboligase reaction. The reaction with glyoxylate producing 2-hydroxy-3-oxo acid, is shown.
Thus, the 70-kDa OADH isoform might be necessary for the non-oxidative functions of OADH (Fig. 5), which does not require the formation of the multienzyme complex essential for oxidative decarboxylation. At the same time, the presence of 130-kDa isoform coincides with the efficient catalytic transformation of 2-oxoadipate in the overall reaction of oxidative decarboxylation (reaction 1).
Characterization of human OADH produced in eukaryotic and bacterial systems. No low-molecular-weight OADH band is observed upon the human DHTKD1 expression in the Pichia pastoris eukaryotic system. The molecular weight of the produced protein is close to 100 kDa, corresponding to the weight calculated from the enzyme amino acid sequence (Fig. 6a). Similar result is obtained upon the human DHTKD1 expression in E. coli cells (Fig. 6b).
Fig. 6. Human OADH produced by the DHTKD1 expression in P. pastoris (a) or E. coli (b). I) Purification of human OADH produced in P. pastoris by affinity chromatography: 1) yeast lysate, 2-9) OADH fractions eluted from a Ni-NTA column by a gradient of increasing imidazole concentration from 30 mM (fraction 2) to 250 mM (fraction 9). The gel was stained with Coomassie Blue [31]. II) Immunoblot of P. pastoris lysates before and after induction of OADH expression. b) Immunoblot of purified OADH (Pur.) and lysate of E. coli BL21(DE3) cells expressing human DHTKD1.
The production of only one protein band with a molecular weight of 100 kDa upon the human DHTKD1 expression in bacteria or yeast (Fig. 6) indicates that the recombinant OADH is different from the enzyme isoforms existing in animal tissues (Figs. 2 and 6). This difference limits significance of the published functional studies on the full-length recombinant OADH produced by genetic engineering approaches, for understanding the biological functions and regulation mechanisms of OADH.
In conclusion, we have compared structural properties of mammalian OADH in tissues or produced in the yeast or bacterial expression systems. Immunostaining reveals that OADH in rat tissues is significantly modified, compared to the enzyme produced upon the human DHTKD1 expression in bacteria or yeast. In the tissues, OADH is represented by two isoforms with molecular weights of 130 and 70 kDa, while the weight of OADH produced in bacteria or yeast is close to the expected one (100 kDa). The characterized properties of OADH isoforms in mammalian tissues testify to specific regulation of the DHTKD1 expression and/or posttranslational modifications of the encoded protein. The functional significance of such regulation may be associated with the non-oxidative reactions catalyzed by mammalian OADH.
Acknowledgements. The authors are thankful to Dr. A. V. Graf and Dr. M. V. Maslova (Lomonosov Moscow State University) for providing rat tissue samples.
Funding. This work is supported by the Russian Foundation for Basic Research (project no. 18-54-7812) and the Council of National Research, Italy (grant SAC.AD002.020.017).
Ethics declarations. The authors declare no conflict of interest in financial or any other sphere. All applicable international, national, and/or institutional guidelines for the care and use of animals were followed.
REFERENCES
1.Bunik, V. I., and Degtyarev, D. (2008)
Structure-function relationships in the 2-oxo acid dehydrogenase
family: substrate-specific signatures and functional predictions for
the 2-oxoglutarate dehydrogenase-like proteins, Proteins,
71, 874-890, doi: 10.1002/prot.21766.
2.Tsepkova, P. M., Artiukhov, A. V., Boyko, A. I.,
Aleshin, V. A., Mkrtchyan, G. V., Zvyagintseva, M. A., Ryabov, S. I.,
Ksenofontov, A. L., Baratova, L. A., Graf, A. V., and Bunik, V. I.
(2017) Thiamine induces long-term changes in amino acid profiles and
activities of 2-oxoglutarate and 2-oxoadipate dehydrogenases in rat
brain, Biochemistry (Moscow), 82, 723-736,
doi: 10.1134/S0006297917060098.
3.Artiukhov, A. V., Grabarska, A., Gumbarewicz, E.,
Aleshin, V. A., Kahne, T., Obata, T., Kazantsev, A. V., Lukashev, N.
V., Stepulak, A., Fernie, A. R., and Bunik, V. I. (2020) Synthetic
analogues of 2-oxo acids discriminate metabolic contribution of the
2-oxoglutarate and 2-oxoadipate dehydrogenases in mammalian cells and
tissues, Sci. Rep., 10, 1886,
doi: 10.1038/s41598-020-58701-4.
4.Nemeria, N. S., Gerfen, G., Yang, L., Zhang, X.,
and Jordan, F. (2018) Evidence for functional and regulatory cross-talk
between the tricarboxylic acid cycle 2-oxoglutarate dehydrogenase
complex and 2-oxoadipate dehydrogenase on the l-lysine, l-hydroxylysine
and l-tryptophan degradation pathways from studies in vitro,
Biochim. Biophys. Acta Bioenerg., 1859, 932-939,
doi: 10.1016/j.bbabio.2018.05.001.
5.Nemeria, N. S., Gerfen, G., Nareddy, P. R., Yang,
L., Zhang, X., Szostak, M., and Jordan, F. (2018) The mitochondrial
2-oxoadipate and 2-oxoglutarate dehydrogenase complexes share their E2
and E3 components for their function and both generate reactive oxygen
species, Free Radic. Biol. Med., 115, 136-145,
doi: 10.1016/j.freeradbiomed.2017.11.018.
6.Danhauser, K., Sauer, S. W., Haack, T. B., Wieland,
T., Staufner, C., Graf, E., Zschocke, J., Strom, T. M., Traub, T.,
Okun, J. G., Meitinger, T., Hoffmann, G. F., Prokisch, H., and Kolker,
S. (2012) DHTKD1 mutations cause 2-aminoadipic and 2-oxoadipic
aciduria, Am. J. Hum. Genet., 91, 1082-1087,
doi: 10.1016/j.ajhg.2012.10.006.
7.Stiles, A. R., Venturoni, L., Mucci, G.,
Elbalalesy, N., Woontner, M., Goodman, S., and Abdenur, J. E. (2016)
New cases of DHTKD1 mutations in patients with 2-ketoadipic aciduria,
JIMD Rep., 25, 15-19,
doi: 10.1007/8904_2015_462.
8.Amsterdam, A., Nissen, R. M., Sun, Z., Swindell, E.
C., Farrington, S., and Hopkins, N. (2004) Identification of 315 genes
essential for early zebrafish development, Proc. Natl. Acad. Sci.
USA, 101, 12792-12797,
doi: 10.1073/pnas.0403929101.
9.Yap, Z. Y., Strucinska, K., Matsuzaki, S., Lee, S.,
Si, Y., Humphries, K., Tarnopolsky, M. A., and Yoon, W. H. (2020) A
biallelic pathogenic variant in the OGDH gene results in a
neurological disorder with features of a mitochondrial disease, J.
Inherit. Metab. Dis., doi: 10.1002/jimd.12248.
10.Xu, W., Zhu, H., Gu, M., Luo, Q., Ding, J., Yao,
Y., Chen, F., and Wang, Z. (2013) DHTKD1 is essential for mitochondrial
biogenesis and function maintenance, FEBS Lett., 587,
3587-3592, doi: 10.1016/j.febslet.2013.08.047.
11.Plubell, D. L., Fenton, A. M., Wilmarth, P. A.,
Bergstrom, P., Zhao, Y., Minnier, J., Heinecke, J. W., Yang, X., and
Pamir, N. (2018) GM-CSF driven myeloid cells in adipose tissue link
weight gain and insulin resistance via formation of 2-aminoadipate,
Sci. Rep., 8, 11485,
doi: 10.1038/s41598-018-29250-8.
12.Timmons, J. A., Atherton, P. J., Larsson, O.,
Sood, S., Blokhin, I. O., Brogan, R. J., Volmar, C. H., Josse, A. R.,
Slentz, C., Wahlestedt, C., Phillips, S. M., Phillips, B. E.,
Gallagher, I. J., and Kraus, W. E. (2018) A coding and non-coding
transcriptomic perspective on the genomics of human metabolic disease,
Nucleic Acids Res., 46, 7772-7792,
doi: 10.1093/nar/gky570.
13.Lim, J., Liu, Z., Apontes, P., Feng, D., Pessin,
J. E., Sauve, A. A., Angeletti, R. H., and Chi, Y. (2014) Dual mode
action of mangiferin in mouse liver under high fat diet, PLoS
One, 9, e90137, doi: 10.1371/journal.pone.0090137.
14.Xu, W. Y., Zhu, H., Shen, Y., Wan, Y. H., Tu, X.
D., Wu, W. T., Tang, L., Zhang, H. X., Lu, S. Y., Jin, X. L., Fei, J.,
and Wang, Z. G. (2018) DHTKD1 deficiency causes
Charcot–Marie–Tooth disease in mice, Mol. Cell.
Biol., 38, doi: 10.1128/MCB.00085-18.
15.O’Neill, E., Chiara Goisis, R., Haverty,
R., and Harkin, A. (2019) L-alpha-aminoadipic acid restricts
dopaminergic neurodegeneration and motor deficits in an inflammatory
model of Parkinson’s disease in male rats, J. Neurosci.
Res., 97, 804-816, doi: 10.1002/jnr.24420.
16.Graf, A., Kabysheva, M. S., Klimuk, E. I.,
Trofimova, L., Dunaeva, T., Zundorf, G., Kahlert, S., Reiser, G.,
Storozhevykh, T. P., Pinelis, V. G., Sokolova, N., and Bunik, V. (2009)
Role of 2-oxoglutarate dehydrogenase in brain pathologies involving
glutamate neurotoxicity, J. Mol. Catal. B Enzym., 61,
80-87, doi: 10.1016/j.molcatb.2009.02.016.
17.Musayev, F. N., Di Salvo, M. L., Ko, T. P.,
Schirch, V., and Safo, M. K. (2003) Structure and properties of
recombinant human pyridoxine 5′-phosphate oxidase, Protein
Sci., 12, 1455-1463, doi: 10.1110/ps.0356203.
18.Wu, S., and Letchworth, G. J. (2004) High
efficiency transformation by electroporation of Pichia pastoris
pretreated with lithium acetate and dithiothreitol,
Biotechniques, 36, 152-154,
doi: 10.2144/04361DD02.
19.Aleshin, V. A., Mkrtchyan, G. V., Kaehne, T.,
Graf, A. V., Maslova, M. V., and Bunik, V. I. (2020) Diurnal regulation
of the function of the rat brain glutamate dehydrogenase by acetylation
and its dependence on thiamine administration, J. Neurochem.,
153, 80-102, doi: 10.1111/jnc.14951.
20.Altschul, S. F., Gish, W., Miller, W., Myers, E.
W., and Lipman, D. J. (1990) Basic local alignment search tool, J.
Mol. Biol., 215, 403-410,
doi: 10.1016/S0022-2836(05)80360-2.
21.Kumar, S., Stecher, G., Li, M., Knyaz, C., and
Tamura, K. (2018) MEGA X: molecular evolutionary genetics analysis
across computing platforms, Mol. Biol. Evol., 35,
1547-1549, doi: 10.1093/molbev/msy096.
22.Roy, A., Kucukural, A., and Zhang, Y. (2010)
I-TASSER: a unified platform for automated protein structure and
function prediction, Nat. Protoc., 5, 725-738,
doi: 10.1038/nprot.2010.5.
23.Payne, S. H., and Loomis, W. F. (2006) Retention
and loss of amino acid biosynthetic pathways based on analysis of
whole-genome sequences, Eukaryot. Cell, 5, 272-276,
doi: 10.1128/EC.5.2.272-276.2006.
24.Bunik, V. (2017) Vitamin-Dependent Multienzyme
Complexes of 2-Oxo Acid Dehydrogenases: Structure, Function, Regulation
and Medical Implications, Hauppaughe, NY, Nova Science Publishers
Inc.
25.Bunik, V., Shoubnikova, A., Bisswanger, H., and
Follmann, H. (1997) Characterization of thioredoxins by sodium dodecyl
sulfate-slab gel electrophoresis and high performance capillary
electrophoresis, Electrophoresis, 18, 762-766,
doi: 10.1002/elps.1150180517.
26.Bonacci, T., Audebert, S., Camoin, L., Baudelet,
E., Bidaut, G., Garcia, M., Witzel, II, Perkins, N. D., Borg, J. P.,
Iovanna, J. L., and Soubeyran, P. (2014) Identification of new
mechanisms of cellular response to chemotherapy by tracking changes in
post-translational modifications by ubiquitin and ubiquitin-like
proteins, J. Proteome Res., 13, 2478-2494,
doi: 10.1021/pr401258d.
27.Akimov, V., Barrio-Hernandez, I., Hansen, S. V.
F., Hallenborg, P., Pedersen, A. K., Bekker-Jensen, D. B., Puglia, M.,
Christensen, S. D. K., Vanselow, J. T., Nielsen, M. M., Kratchmarova,
I., Kelstrup, C. D., Olsen, J. V., and Blagoev, B. (2018) UbiSite
approach for comprehensive mapping of lysine and N-terminal
ubiquitination sites, Nat. Struct. Mol. Biol., 25,
631-640, doi: 10.1038/s41594-018-0084-y.
28.Hornbeck, P. V., Zhang, B., Murray, B.,
Kornhauser, J. M., Latham, V., and Skrzypek, E. (2015) PhosphoSitePlus,
2014: mutations, PTMs and recalibrations, Nucleic Acids Res.,
43, D512-520, doi: 10.1093/nar/gku1267.
29.Udeshi, N. D., Mertins, P., Svinkina, T., and
Carr, S. A. (2013) Large-scale identification of ubiquitination sites
by mass spectrometry, Nat. Protoc., 8, 1950-1960,
doi: 10.1038/nprot. 2013.120.
30.McCartney, R. G., Rice, J. E., Sanderson, S. J.,
Bunik, V., Lindsay, H., and Lindsay, J. G. (1998) Subunit interactions
in the mammalian alpha-ketoglutarate dehydrogenase complex. Evidence
for direct association of the alpha-ketoglutarate dehydrogenase and
dihydrolipoamide dehydrogenase components, J. Biol. Chem.,
273, 24158-24164.
31.Arndt, C., Koristka, S., Feldmann, A., Bartsch,
H., and Bachmann, M. (2012) Coomassie-Brilliant Blue staining of
polyacrylamide gels, Methods Mol. Biol., 869, 465-469,
doi: 10.1007/978-1-61779-821-4_40.