REVIEW: Cold Shock Domain Proteins: Structure and Interaction with Nucleic Acids
K. S. Budkina1, N. E. Zlobin2, S. V. Kononova1, L. P. Ovchinnikov1,a*, and A. V. Babakov2
1Institute of Protein Research, Russian Academy of Sciences, 142290 Pushchino, Moscow Region, Russia2All-Russian Research Institute of Agricultural Biotechnology, Russian Academy of Sciences, 127550 Moscow, Russia
* To whom correspondence should be addressed.
Received September 9, 2019; Revised September 16, 2019; Accepted September 20, 2019
This review summarizes the features of cold shock domain (CSD) proteins in the context of their interactions with nucleic acids and describes similarities and differences in the structure of cold shock proteins of prokaryotes and CSD proteins of eukaryotes with special emphasis on the functions related to the RNA/DNA-binding ability of these proteins. The mechanisms and specificity of their interaction with nucleic acids in relation to the growing complexity of protein domain structure are described, as well as various complexes of the mammalian Y-box binding protein 1 (YB-1) with nucleic acids (filaments, globules, toroids). The role of particular amino acid residues in the binding of nitrogenous bases and the sugar-phosphate backbone of nucleic acids is emphasized. The data on the nucleic acid sequences recognized by the Y-box binding proteins are systematized. Post-translational modifications of YB-1, especially its phosphorylation, affect the recognition of specific sequences in the promoter regions of various groups of genes by YB-1 protein. The data on the interaction of Lin28 protein with let-7 miRNAs are summarized. The features of the domain structure of plant CSD proteins and their effect on the interaction with nucleic acids are discussed.
KEY WORDS: nucleic acid-binding proteins, cold shock domain, melting, annealing, domain structureDOI: 10.1134/S0006297920140011
Abbreviations: a.a., amino acid residue; A/P domain, alanine/proline-rich domain; CRS, cytoplasmic retention site; CSD, cold shock domain; CSP, cold shock protein; CTD, C-terminal domain; dsDNA, double-stranded DNA; (m)RNP, (messenger) ribonucleoprotein; NLS, nuclear localization signal; ssDNA, single-stranded DNA; ssRNA, single-stranded RNA; UTR, untranslated region; YB-1, Y-box binding protein 1.
Almost 2% of proteins encoded in the human genome have the ability to
bind both DNA and RNA. These proteins play a key role in the modulation
of gene expression, cell survival, and homeostasis [1]. Among them, there are proteins with the cold shock
domain (CSD). CSD proteins have been detected in organisms from
different taxa, both prokaryotic and eukaryotic.
The CSD structure is represented by five antiparallel β-strands forming a compact β-barrel. Strands β2 and β3 contain the RNA-binding motifs RNP1 and RNP2 [2] that are typical of all CSDs, as well as RRM domains of RNA-interacting proteins [3]. The side chains of aromatic amino acids in RNP1 and RNP2 ensure CSD binding to nucleic acids through the stacking interactions and are crucial for the RNA-binding and RNA-melting activities of CSD proteins [4]. In addition to aromatic residues, the surface of a protein molecule with RNP1 and RNP2 contains numerous basic amino acids. The overall positive charge typical of this CSD region provides non-specific electrostatic interaction with negatively charged molecules of nucleic acids, while aromatic amino acid side chains stabilize this binding through the hydrophobic and stacking interactions [4]. Analysis of eukaryotic CSD protein sequences by Kleene [5] revealed two additional RNA-binding motifs, one of which almost coincides with the β1 strand, while the other is located in the middle of the β3β4 loop.
In prokaryotic cold shock proteins (CSPs), which have a common spatial structure, aromatic amino acid residues are located in the same plane, thus forming a hydrophobic cluster of unusually large size on the protein surface [6, 7].
COLD SHOCK PROTEINS OF PROKARYOTES
General characteristics. The name cold shock domain originates from the discovery of this structure in the studies on the adaptation of bacteria to low temperatures.
Adaptation of prokaryotic organisms to a decreasing ambient temperature has been best studied in the mesophilic bacterium Escherichia coli. The optimal temperature for E. coli growth is 37°C; cultivation at temperatures below 20°C represents a low-temperature stress for this microorganism. The most obvious consequence of a sudden temperature downshift (cold shock) is cessation of growth and division of E. coli cells for a period of 2-4 h or more [8]. It was found that cold shock almost completely suppresses the synthesis of most proteins, which is the main cause for the cessation of bacterial growth [9, 10]. However, some proteins showed higher expression levels than before the onset of unfavorable temperatures [8-12]. When the temperature was decreased from 37 to 15°C, one of these proteins, called CspA (cold shock protein A), accumulated in amounts as high as 13% of the total pool of cellular proteins synthesized under the cold shock conditions [13]. Structurally, this protein is a β-barrel consisting of five antiparallel β-strands [14].
CspA-like proteins have been detected in bacteria from different ecological groups, such as psychrophiles, psychrotrophs, mesophiles, thermophiles, and hyperthermophiles [15]. Some of these proteins were found to be involved in the bacterial adaptation to the low-temperature stress and, therefore, named cold shock proteins (CSPs). The structural property of CSPs is the presence of the cold shock domains (CSDs).
In prokaryotic genomes, CSPs are usually encoded by a family of homologous genes [16]. In E. coli, the CSP family includes nine members named CspA to CspI. It was found that expression of CspA, CspB, CspG, and CspI (especially, the first three proteins) is induced at low ambient temperatures [10, 17], while CspC and CspE are expressed at 37°C [18, 19].
As demonstrated by deletion analysis, the functions of CSPs in E. coli overlap, at least, in the adaptation to low temperatures [20]. For example, strains carrying simultaneous deletions of two or three CspA homologs (ΔcspA∆cspB, ΔcspAΔcspG, ΔcspBΔcspG, ΔcspAΔcspI, ΔcspAΔcspBΔcspG) retained the ability to grow at low temperatures (15°C). Overexpression of the CspE gene was observed in the ΔcspAΔcspBΔcspG strain upon the temperature decrease. These facts indicate that members of the E. coli CSP family are functionally interchangeable and can compensate for the absence of expression of one or several homologs. Only the deletion of as many as four genes (ΔcspAΔcspBΔcspGΔcspE) was sufficient to generate an E. coli strain incapable of growing at low temperatures. The sensitivity of this strain to the temperature decrease could be compensated through the overexpression of any E. coli CSP, except CspD [20].
All bacterial CSPs share a common structure; they are typically small in size (67-73 a.a.) and lack any other sequences except the CSD. NMR and X-ray studies have determined the 3D structures of some bacterial CSPs, including CspA from E. coli, CspB from Bacillus subtilis, CspB from Bacillus caldolyticus, and CspB from Thermotoga maritima [6, 14, 21, 22]. Despite significantly different amino acid sequences, the spatial structures of these proteins are very similar. The studies of the tertiary structure of E. coli CspA demonstrated that the surface of this protein is composed of the β2 and β3 strands and carries a compact hydrophobic cluster containing aromatic amino acid residues Phe18, Phe20, Phe31, His33, and Phe34. The residues Phe18 and Phe20 belong to the RNP1 motif, while Phe31, His33, and Phe34 belong to RNP2 [23].
Functions of prokaryotic CSPs. All known functions of prokaryotic CSPs are associated with their ability to bind nucleic acids.
Interactions of bacterial CSPs with nucleic acids have been extensively studied in vitro. It was found that E. coli CspA has a low affinity for RNA and almost no specificity for its nucleotide sequence [24]. Other members of the E. coli CspA family can also bind RNA and ssDNA, but with slightly higher specificity. For example, CspB has the highest affinity for the UUUUU motif, while CspC preferentially binds the AGGGAGGGA sequence. CspE selectively interacts with AU-rich sequences [25]. CspB protein from B. subtilis exhibits an increased affinity for T-rich sequences; however, its specificity to nucleic acids is rather low [26]. Various prokaryotic CSPs not only bind nucleic acids, but also destabilize their secondary structure, i.e., demonstrate the so-called melting activity [27, 28]. A special case of the RNA-melting activity of CSPs is transcription antitermination. Through binding to the terminator sequences in RNAs, CSPs destabilize their secondary structure, thereby inhibiting termination of transcription [27, 29]. Such activity has been shown, for example, for CspE, which maintains high expression levels of the promoter-distant genes in the metY-rpsO operon [29]. It has been demonstrated that the expression of particular genes involved in the cold adaptation in bacteria in vivo is activated by the antitermination mechanism [29].
Long single-stranded mRNAs tend to form a variety of secondary structures that can interfere with the ribosome movement or hide the Shine–Dalgarno sequence, thereby adversely affecting the process of translation. This phenomenon becomes more pronounced with a decrease in the ambient temperature [30]. The RNA-melting activity of CSPs contributes to the destabilization of the RNA secondary structure, eliminates its adverse effects on translation, and maintains the operation of the protein synthesis machinery [24, 31-33]. It should be noted that, due to their low affinity for mRNAs, CSPs hardly make significant obstacles to the ribosome movement during translation [32].
The involvement of CSPs in the destabilization of the mRNA secondary structure was demonstrated in E. coli [12]. Deletion of the CspA gene caused a 30-40% decrease in the total translation efficiency at low temperatures. Additional deletion of genes encoding other E. coli CSPs (CspB, CspE, CspG) led to an almost complete loss of translational activity. The need for CSPs during cold acclimation can result from their ability to destabilize the mRNA secondary structure. Thus, it was found that in a mutant lacking four CSPs (ΔcspAΔcspBΔcspEΔcspG), mRNA molecules were highly structured at low temperatures. Interestingly, the proliferation rate of the mutant with five deleted CSPs (ΔcspAΔcspBΔcspCΔcspEΔcspG) was lower than that of the wild-type E. coli cells even at 37°C [12].
In addition to the general increase of translation efficiency, CSPs can regulate the stability of mRNAs by affecting their degradation [31]. Thus, E. coli CspC and CspE stabilize the mRNAs for RpoS (general regulator of stress response) and UspA (universal protein of stress response) [34]. CspE interacts with mRNA poly(A)-sequences and impedes mRNA degradation in the case of its treatment with polynucleotide phosphorylase (PNPase) and RNase E [35].
Therefore, prokaryotic CSPs regulate the synthesis of cell proteins via various mechanisms both at low temperatures and under conditions optimal for the growth of bacterial cells.
EUKARYOTIC CSD PROTEINS
General characteristics. Proteins containing CSDs have been also found in the multicellular organisms from different kingdoms of life. Unlike prokaryotic CSPs, which are almost always composed of the CSD only, eukaryotic proteins contain other domains as well. For this reason, they are often called CSD proteins to emphasize their multidomain nature. Eukaryotic proteins composed exclusively of CSDs are rare; they may have either one CSD copy (Clah8 from Cladosporium herbarum and zfY1 from Danio rerio) or several CSDs connected via the linker sequences (UNR/CSDEI from vertebrates) [36-38].
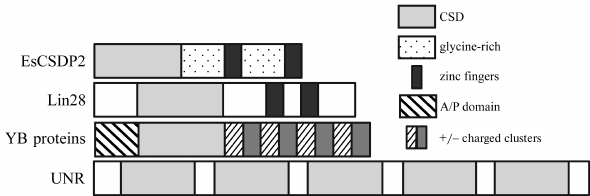
Additional domains present in the CSD proteins vary. In this review, we will examine proteins with glycine-rich sequences, CCHC zinc finger domains, and extended domains consisting of alternating clusters of positively and negatively charged amino acid residues, as well as their various combinations (Fig. 1). The presence of additional disordered domains in CSD proteins expands their multifunctionality [39] and allows them to act as organizers of various functional complexes, including those in the non-membranous structures, such as processing bodies (P-bodies) and stress granules [40].
Fig. 1. Domain structure of CSD proteins with and without additional domains.
The spatial structure of the eukaryotic CSDs is virtually identical to that of the prokaryotic CSPs and includes five β-strands forming the β-barrel. The main difference between the eukaryotic and prokaryotic proteins is that the former have a longer linker sequence connecting β3 and β4 strands that also has different amino acid composition [41].
YB PROTEINS
A large family of vertebrate CSD proteins is called Y-box binding proteins (YB proteins). It includes three subfamilies encoded by the YBX1, YBX2, and YBX3 genes, respectively [42]. The YBX3 transcript undergoes alternative splicing resulting in two mRNAs coding for the YB-3 protein long and short isoforms [5].
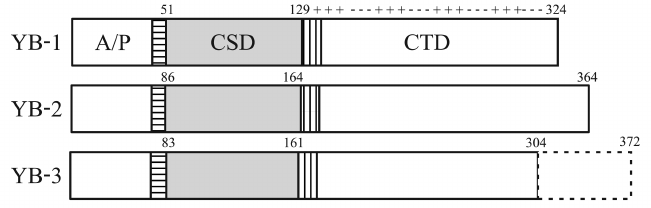
All YB proteins are basic (pI = 9.5-10.7) and composed of the structured CSD and two disordered domains: the N-terminal domain rich in Ala and Pro residues (A/P domain) and the C-terminal domain (CTD) containing four Arg-rich clusters that alternate with four clusters of negatively charged amino acid residues. The CSDs of all YB proteins are virtually identical. Besides, YB proteins have two highly homologous (~90%) CSD-flanking linkers: 9-a.a. N-terminal linker (NC9) and 13-a.a. C-terminal linker (CC13) [5] (Fig. 2).
Fig. 2. Domain structure of YB proteins. NC9 is horizontally shaded; CC13 is vertically shaded; pluses and minuses denote clusters of positively and negatively charged amino acid residues, respectively.
Members of the YB protein family are interchangeable, although only partially. Each of them has its own expression profile in ontogenesis. YB-1 is expressed during almost the entire ontogenesis, especially at its early stages, and then gradually disappears with aging from the organs at different times. In old mice, YB-1 can be detected only in the liver [43]. YB-2 is present in large amounts in the gametes (oocytes, eggs, and sperm) of the clawed frog Xenopus laevis but completely disappears at the gastrula stage [44]. YB-3 is expressed in mammalian embryos; after birth, its amount decreases in all tissues, except heart, skeletal muscles, blood vessels, and testicles [45, 46]. It was recently reported that YB-3 is also detected in some brain regions in adult mammals (in glial cells), while YB-1 is expressed in neurons [47].
The YBX1 knockout disturbs normal mouse development as early as on the embryonic day 13.5 (E13.5). Usually the animals die either before birth or immediately after it. Simultaneous knockout of the YBX1 and YBX3 genes results in the animal’s death on an embryonic day 8.5-11.5 (E8.5-11.5) [48, 49].
General characteristics of YB-1. Human YB-1 is the best studied member of the YB protein family. The progress in its characterization is mostly associated with the involvement of this protein in oncogenesis [50-52]. Originally, YB-1 was discovered as the major protein of mRNPs (messenger ribonucleoproteins) [53-55] and named p50 in accordance with its electrophoretic mobility. Later, a protein interacting with the promoter regions of the major histocompatibility complex class II (MHCII) genes [56] and the enhancer region of the epidermal growth factor receptor gene [57] was discovered and sequenced. Since both target DNA regions contained the Y-box sequence (5′-CTGATTGGC/TC/TAA-3′), this protein was named Y-box binding protein 1 (YB-1). The sequencing of p50 demonstrated its identity to YB-1 [58].
Despite the fact that the electrophoretic mobility of YB-1 corresponds to a protein with a molecular mass of 50 kDa, its molecular mass calculated from the amino acid sequence is 36 kDa. Hence, the protein exhibits abnormal mobility during electrophoresis in the presence of sodium dodecyl sulfate [42].
YB-1 isolated from the rabbit reticulocyte mRNPs can form multimers with the sedimentation coefficient of ~18S and molecular mass up to 800 kDa [58]. According to the atomic force microscopy and electron microscopy on a substrate, multimeric YB-1 represents homogenous flattened granules with a diameter of 30-40 nm and height of 8-10 nm [41]. It is assumed that YB-1 protein forms multimers through the CTD when positively charged clusters of one protein molecule interact with negatively charged clusters of another molecule and vice versa [59]. It is possible that YB-1 CSD is also involved in the multimer formation via dimerization [60]. The YB-1 homodimer is formed by the interaction of Asp105 of one molecule and Asp105′ of the other molecule and stabilized by the proximity of Phe66 and Phe66′, as well as by the hydrogen bond between Tyr99 and Glu107′ [60].
Under certain conditions, YB-1 and its fragments can form reversible amyloid fibrils. CSD is responsible for the fibril formation, while the N-terminal domain stimulates the process. It was found that the first half of the CTD blocks the fibril formation, whereas its second half removes the blocking effect, but only in solutions with a high ionic strength [61].
YB-1 in the cytoplasm. YB-1 localized mostly to the cytoplasm, where it associates with translated and untranslated mRNAs. YB-1 was also detected in the complexes with miRNAs [62], tRNA fragments [63], and long non-coding RNAs [64]. In the cytoplasm, YB-1 participates in the global and specific regulation of mRNA translation at the initiation stage. YB-1 can either stimulate translation (at a comparatively low YB-1/mRNA ratio) or inhibit it (at a high YB-1/mRNA ratio) [65]. YB-1 also protects mRNAs against degradation and significantly prolongs their lifetime (up to 100 times) [66]. Free YB-1 and YB-1 in the unsaturated complexes with mRNAs can interact with actin and microfilaments to ensure localization of translationally active mRNAs to actin microfilaments [67]. Through binding to tubulin, YB-1 stimulates microtubule formation and can contribute to the localization and transport of translationally inactive mRNAs along the microtubules [68].
YB-1 interaction with the mitotic spindle microtubules facilitates their assembly and stabilization [69]. Excessive binding of YB-1 to the centrosomes can disturb their correct doubling in mitosis, promote the appearance of additional centrosomes, interfere with the correct chromosome separation, and cause partial aneuploidy, which, in turn, can lead to malignant cell transformation [70]. The role of YB-1 in stress and transport granules is less studied, and the obtained data are often contradictory. According to some reports, YB-1 is an obligatory component of stress granules that stimulates their formation; YB-1 knockout disturbs the process of stress granule assembly [71]. According to the other studies, increased YB-1 concentration interferes with the formation of stress granules [72].
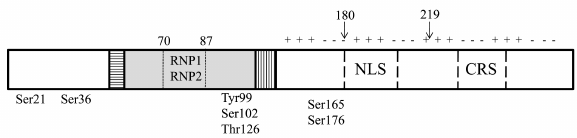
YB-1 in the nucleus. YB-1 translocation from the cytoplasm to the nucleus occurs during the G1/S cell cycle transition and stimulates expression of cyclin genes [73, 74]. Its nuclear localization is determined by growth factors and cytokines [42]. The process of translocation is stimulated by UV irradiation and xenobiotics causing DNA damage [42]. It was recently shown that nuclear translocation of YB-1 is under the circadian clock control [75]. Subcellular distribution of YB-1 is regulated by the nuclear localization signal (NLS) and cytoplasmic retention signal (CRS) (Fig. 3). Usually, CRS masks the NLS, and YB-1 is retained in the cytoplasm. Specific cleavage of CRS by the 20S proteasome leads to the translocation of C-truncated YB-1 (Fig. 3) to the nucleus [76]. Another mechanism of the full-length YB-1 transport to the nucleus is associated with its phosphorylation [77, 78]. In the nucleus, YB-1 co-localizes with RNA polymerase I and can be detected in Cajal bodies [79]. An increased concentration of YB-1 can contribute to the dissolution of nucleoli [80]. When in the nucleus, YB-1 participates in the DNA replication (including viral DNA), DNA repair, transcription of numerous genes, and alternative splicing of mRNA precursors [42, 65]. It has recently been shown that the activity of YB-1 can be inhibited by its interaction with circular RNAs in the nucleus [81].
Fig. 3. YB-1 amino acid residues undergoing phosphorylation resulting in the changes in the YB-1 activity. Arrows, sites of YB-1 truncation: 1-180, YB-1 fragment used for NMR analysis; 219, site of YB-1 cleavage by the 20S proteasome.
YB-1 interaction with nucleic acids. The ability of YB-1 to bind both RNA and DNA indicates low protein specificity for the type of nucleic acid. Nevertheless, YB-1 shows a higher affinity for RNA vs. ssDNA, which according to the molecular modeling, can be explained by additional H-bonding occurring due to the presence of 2′OH group in ribose. Molecular dynamics analysis also showed that YB-1 binding to the nucleic acid strand is orientation-dependent; its affinity is higher when the binding occurs in the 5′→3′ direction [82].
YB-1 exhibits almost identical affinity for rRNA and mRNA. The dissociation constant (KD) of the YB-1 complex with RNA is ~4 nM [83]. When interacting with RNA, YB-1 alters the secondary structure of the latter. Addition of YB-1 to the globin mRNA at room temperature causes melting of ~60% of its secondary structure. YB-1 can stimulate annealing of complementary DNA and RNA sequences, as well as the exchange of complementary strands of nucleic acids, leading to the formation of very long perfect duplexes [58, 84].
YB-1 shows a much higher affinity for ssDNA than for dsDNA. YB-1 binding to the CT-regions in one of the DNA strands results in the emergence of nuclease-sensitive regions and formation of the H-structure in the other strand. YB-1 is capable of binding to and breaking the double helix in dsDNA fragments with blunt ends, duplexes with protruding 5′- and 3′-ends, DNA molecules with unpaired bases and apurinic sites, and cisplatin-treated DNA [84-86]. The 3′→5′ exonuclease activity of YB-1 [87] and its elevated affinity for damaged DNA regions [85] confirm its involvement in the DNA repair in the nucleus.
As mentioned above, YB-1 binding to nucleic acids has a dual nature: it can either stabilize or destabilize the nucleic acid secondary structure [88]. Its interaction with dsDNA results in DNA melting and impedes recruitment of dsDNA-binding factors responsible for its transcription [89]. The ability of YB-1 to melt the secondary structure was confirmed in experiments on microchips, where dsDNA was used as a substrate. When interacting with the duplexes, YB-1 predominantly binds to one strand and destabilizes the duplex [90]. It was suggested that the CSD region responsible for the dsDNA binding is the loop connecting β3 and β4 strands, since its replacement with a shorter loop of prokaryotic YB-1 (which also differs in the amino acid composition) resulted in the loss of this activity [91].
Structures formed by YB-1 in the complexes with nucleic acids. As shown in in vitro experiments, the following structures can be formed depending on the YB-1/mRNA ratio: non-compact translated mRNPs or beads-on-a-string [41]. In the case of translated mRNAs (at a relatively low YB-1/mRNA ratio), YB-1 is present on the mRNA as a monomer (one YB-1 molecule per ~80 nt of mRNA) and binds to it through the CSD and CTD. In the case of untranslated, protein-saturated mRNAs, YB-1 forms multimers (beads) consisting of approximately 20 molecules, that have the molecular mass of ~700 kDa, diameter of 20 nm, and height of 7 nm (each bead per 600-700 nt of mRNA, i.e., one YB-1 molecule per 30-35 nt of mRNA) [41]. The formation of beads on the mRNA probably occurs by the same mechanism as the formation of multimers from free YB-1 molecules. Studying the role of YB-1 domains in the protein multimerization showed that isolated CTD forms structures smaller in size than those formed by the full-length protein, while CSD is uncapable of forming the beads [92]. This is consistent with the previously proposed model of saturated YB-1 complexes with untranslated mRNAs, according to which the beads of multimeric YB-1 are formed through the interaction of CTDs [41]. This model is also supported by the results of super-resolution microscopy and fluorescence correlation spectroscopy showing that in living cells, mRNAs are associated, inter alia, with YB-1 and form structures similar to the beads-on-a-string [93].
Recently, the complex of YB-1 CSD with the specific sequence CAUC within the UCAUCU hexamer was successfully crystallized despite a relatively low protein affinity for CAUC (KD = 1.26 μM), and the structure of this complex was determined by X-ray analysis with a 1.7-Å resolution [60]. These data allowed to identify amino acid residues involved in the formation of ππ-stacking pairs with the nitrogenous bases of CAUC: C1·His87, A2·Phe85, U3·Phe74, and C4·Trp65. Replacement of these amino acid residues resulted in the complete loss of YB-1 ability to bind the hexamer. An important role in the specific recognition of CAUC belongs to the H-bonds between C1 and Thr89, A2 and Lys118, and also U3 and both Asp83 and Lys64. Replacement of one of these residues leads to a minor decrease (2- or 3-fold) in the YB-1 affinity for the hexamer. Interestingly, Asn70, which is absent from other CSD-containing proteins (e.g., Lin28, bacterial CSPs), is involved in the CSD binding to the sugar-phosphate backbone [60].
Earlier, complexes of the C-truncated YB-1 (1-180) (Fig. 3) with poly(C), poly(T), and poly(U) of 5, 10, 20, or 30 nt were studied by NMR [94]. Analysis of the NMR spectra identified conserved amino acid residues Trp65, Phe74, and Phe85 involved in the interaction with oligonucleotides. The use of truncated YB-1(1-180) with a single cluster of positively/negatively charged amino acids in its CTD yielded identification of additional phosphate-interacting amino acid residues beyond the CSD, namely, Gly135, Ser136, and Lys137 [94]. These residues significantly contribute to the YB-1(1-180) affinity for RNA, as compared to the CSD.
Truncated YB-1(1-180) in a complex with mRNA is able to form still another (third) structure – linear nucleoprotein filament [94]. Small-angle X-ray scattering and molecular dynamics were used to show that YB-1(1-180) molecules form a single layer along the RNA/ssDNA strand, with 6 nt per protein molecule. Introduction of additional charged clusters in the CTD results in the formation of protein multimers and alters mRNA folding in mRNPs [94]. Binding to protein partners or posttranslational modifications in the CTD, which contains multiple phosphorylation sites [95], can neutralize the strong positive charge of the CTD and initiate formation of linear nucleoprotein filaments in the cell. Despite the tight packing of YB-1(1-180) in a complex with RNA, the protein shows no inhibitory activity, but on the contrary, stimulates mRNA translation [94].
The binding of dimeric YB-1 to RNA (5′-CAUCCAACAAGA-3′) and ssDNA (5′-TTGGCCAATCAG-3′) was studied by calorimetry [96]. The contribution of the A/P domain was neglectable, while the CTD sequence (a.a. 130-219) appeared to be most important for the thermodynamically favorable protein binding to RNA/ssDNA. This confirms the key role of CSD in YB-1 dimerization and its specific binding to RNA.
YB-1 preferentially binds to the supercoiled DNA at the intersection of helices, in particular, in the presence of a competing relaxed strand. Its CTD localizes mostly to the interface between the two intersecting DNA helices, so that the YB-1 molecules can interact with each other. Therefore, the multimers are formed at the sites of increased DNA condensation. Addition of YB-1 protein to the linearized DNA stimulates formation of complexes with a characteristic toroid shape [92].
Specific interactions of YB-1 with nucleic acids. The participation of YB-1 in the transcription of genes (including viral genes) occurs by different mechanisms described in a number of papers [42, 59]. By binding to DNA, YB-1 regulates the activity of many genes, whose protein products are involved in apoptosis, embryogenesis, immune response, multiple drug resistance, stress response, and tumorigenesis [42]. This multifunctionality indicates that YB-1 interacts with specific nucleic acid sequences.
YB-1 binds mostly to the single-stranded nucleic acid regions. Experiments on the competition between homopolyribonucleotides for YB-1 binding have shown that the YB-1 affinity for them decreases in the following order: poly(G) > poly(U) > poly(A) > poly(C) [83].
When interacting with ssDNA, YB-1 shows the highest preference for the G-rich regions. The longer the G-containing sequence, the higher the dissociation temperature of the DNA–protein complex [83, 90]. G-rich nucleotide sequences are capable of forming a structure called G-quadruplex. So far, the data on the specific recognition of G-quadruplexes by YB-1 are contradictory [97, 98].
The table presents some identified nucleotide sequences that specifically bind to YB-1 and can be involved in the selective regulation of gene expression.
Identified YB-1-interacting nucleotide sequences
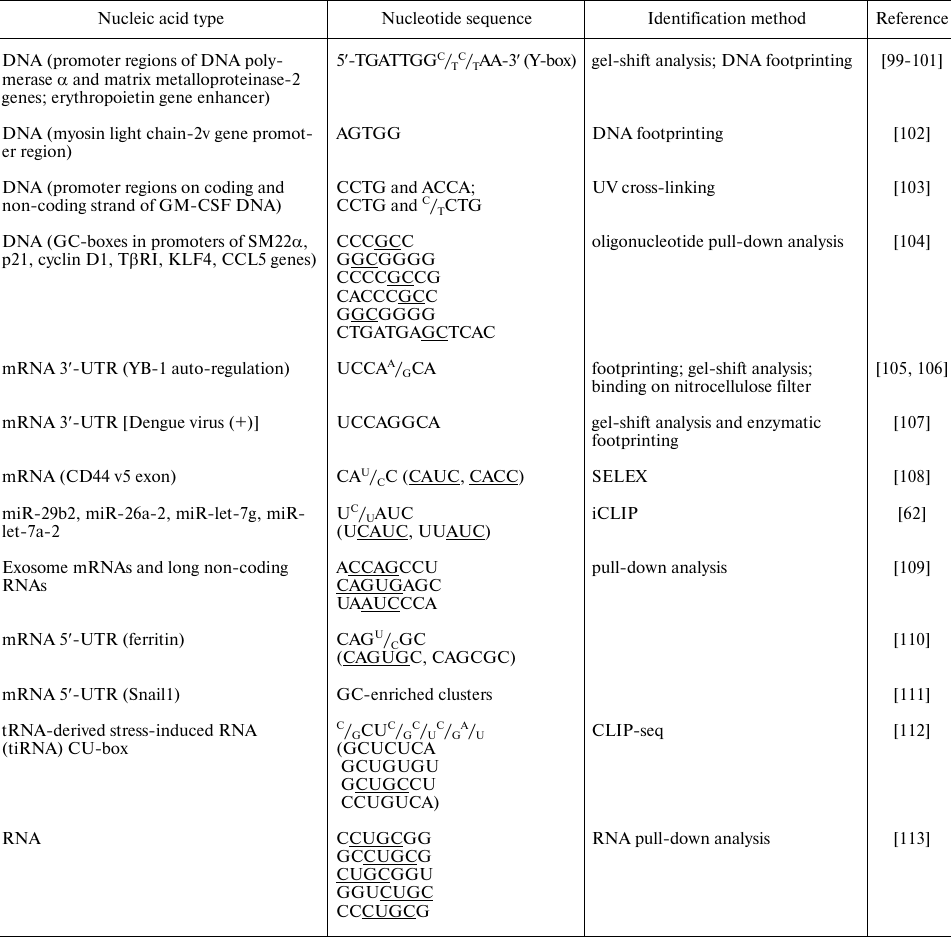
As follows from the table, YB-1 predominantly binds to the G- and C-rich sequences of DNA promoters, which is in agreement with the results of in vitro experiments. The same trend is observed for the YB-1-binding sites in RNA. YB-1 immunoprecipitation with RNA showed that the majority of binding sites are located in the 3′-untranslated regions (3′-UTRs) and exons, while the number of YB-1-binding sites in 5′-UTRs and intron sequences is minimal [112]. Analysis of the known binding sites identifies sequences rich in CANC, CU, and GC.
By binding to the promoter regions, YB-1 can inhibit or activate transcription of certain genes [100-102, 104, 114]. The process of translation is regulated through the recognition of specific sites in the 5′-UTRs [110, 111] and 3′-UTRs [106]. YB-1 binding to mRNA is cooperative [92], which can explain the fact that specific YB-1 binding to the mRNA 3′-UTRs leads to selective inhibition of protein synthesis initiation at the 5′-UTRs [105, 106]. By binding to the 3′-UTRs, YB-1 contributes to the regulation of mRNA localization, which is especially critical for neurons, in which protein synthesis takes place in axons [115]. Interestingly, YB-1 can be involved in the translation regulation not only via its direct binding to the translated mRNA but through other mechanisms as well. For example, YB-1 interacts with the 5′-fragments of angiogenin-induced tRNAAla and tRNACys and inhibits translation by displacing eIF4F from the mRNA cap structure [63]. Also, YB-1 binds to miRNAs and regulates their processing at the post-transcriptional level [62]; besides, YB-1 is required for sorting miRNAs into exosomes [116].
Specific recognition of modified nucleic acids by YB-1. In addition to the sequences described above, YB-1 can recognize modified nucleotides in mRNAs. The first studied interaction of this type was YB-1 binding through its CSD to the mRNA 5′-cap including guanine methylated at position 7 (m7G). This interaction allows YB-1 to regulate the stability and translational activity of mRNAs [117, 118].
Another modified residue recognized by YB-1 is 8-oxoguanine, which is usually a product of oxidative stress. YB-1 interaction with mRNAs containing this residue blocks their translation, thereby preventing errors in protein synthesis [119].
Recently, YB-1 has been characterized as a protein recognizing 5-methylcytosine (m5C)-containing mRNAs. It was shown by the isothermal titration calorimetry that CSD has a higher affinity for the 5′-UCAU(m5C)U-3′ oligonucleotide than for the non-methylated 5′-UCAUCU-3′. X-ray structure analysis of the YB-1–RNA complex identified Trp65 as the major residue responsible for the m5C recognition [120]. There are also data indicating that YB-1 is involved in the stabilization of maternal m5C mRNA at the early stages of embryonic development in Danio rerio [121]. Yet, the role of YB-1 binding to m5C remains unclear.
The role of post-translational modifications of YB-1 in its binding to nucleic acids. The role of YB-1 covalent modifications in nucleic acid binding is still poorly studied. These studies mostly used eukaryotic cells expressing YB-1 from a plasmid (Fig. 3).
Different stimuli activate appropriate kinases, thus triggering phosphorylation of YB-1 that, in turn, causes its nuclear translocation. It is believed that phosphorylation of particular amino acid residues plays a crucial role in the recognition of different DNA sequences through increasing protein affinity toward them. The resulting specific binding can entail activation or repression of transcription of various groups of genes [95, 122-125]. YB-1 represses VEGF gene transcription by binding to the promoter single-stranded hypoxia response region (HRR). This binding is enhanced in vitro by phosphorylation of Ser21 and Ser36 in the YB-1 A/P domain by GSK3β and ERK2. Therefore, modification of these amino acid residues can promote the inhibitory effect of YB-1 on the VEGF mRNA transcription in cells [122].
The IL-1β-triggered phosphorylation of YB-1 at Ser165 causes tumor growth via activation of the NF-κB signaling pathway [95]. As shown recently NF-κB signaling can be also activated by the IL-1β-induced YB-1 modification at Ser176. Hence, expression of different groups of genes controlled by NF-κB depends on which YB-1 residue (Ser165 or Ser176) is phosphorylated [125]. In addition, YB-1 phosphorylation at Ser165 results in the YB-1 nuclear translocation [95]. This suggests that differently modified YB-1 (at Ser165 or Ser176) not only activates NF-κB but also, together with this factor, recognizes different DNA motifs and regulates transcription of the corresponding group of genes.
Phosphorylation of YB-1 at Ser102 leads to the translation activation by the cap-dependent mechanism. Modified YB-1 loses its affinity for the cap, thereby promoting rapid assembly of the eIF4F complex on mRNA and facilitating translation initiation [123]. Under certain conditions, YB-1 phosphorylation at Ser102 leads to its translocation to the nucleus. It was found that nuclear localization of YB-1 stimulates cell growth [77]. Hence, it can be assumed that YB-1 modification at Ser102 affects gene expression at both transcriptional and translational levels.
Phosphorylation of YB-1 at Tyr99 results in its translocation to the nucleus and repression of the Col1a1 promoter, which exerts a positive effect in renal fibrosis [124]. Interestingly, Tyr99 residue is also involved in YB-1 dimerization [60], suggesting that its phosphorylation can affect protein oligomerization.
An interesting mechanism of CCL5 transcription regulation by the modified YB-1 during monocytes differentiation into macrophages was described in [126]. At the early stages of differentiation, YB-1 phosphorylated at Ser102 binds to the CCL5 promoter and activates gene expression. At the later stages, YB-1 is dephosphorylated by the serine/threonine phosphatase calcineurin. The YB-1 affinity for the promoter decreases and CCL5 synthesis ceases [126].
Other YB-1 modifications can affect its transcriptional activity, as it has been reported for YB-1 O-glycosylation at Thr126 [127]. YB-1 modifications might also affect DNA repair. YB-1, either free or in a complex with damaged DNA, can interact with the PARP1 polymerase. At a certain protein ratio (YB-1/PARP1 ˂ 10/1), auto-modification of PARP1 and poly(ADP-ribosyl)ation of YB-1 occur. This is accompanied by a decrease in the YB-1 affinity for DNA, allowing other proteins to repair the damage [128, 129].
Due to its multifunctional nature, YB-1 can be involved in the development of diseases. It is now considered as a prognostic marker in some types of cancer, a novel anti-cancer therapeutic target [50, 52], and even a potential therapeutic agent against Alzheimer’s disease [130].
LIN28
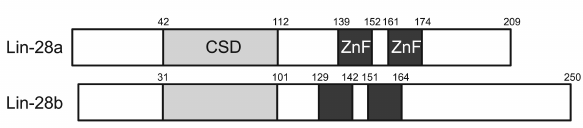
Lin28 is another well-studied RNA-binding CSD protein. It was detected in both invertebrates and vertebrates, and its structure is typical for CSD proteins. In Lin28, the CSD is located in the N-terminus, while the C-terminus contains two zinc fingers of the CCНC type. Some forms of this protein have additional sequences of several tens amino acids located upstream of the CSD and downstream of the zinc fingers [131]. There are two isoforms of human Lin28: Lin28A (209 a.a.) and Lin28B (250 a.a.) (Fig. 4). Their CSDs are 85.5% identical. According to the results reported in [132], Lin28A localizes to the cytoplasm and Lin28B is present in the nucleus. However, recent reports demonstrated that Lin28B is mostly a cytoplasmic protein [133, 134].
Fig. 4. Domain structure of two Lin28 isoforms. ZnF, zinc finger.
The study of Lin28 functioning at different developmental stages of various tissues and organs characterized this protein as a cell division activator and inhibitor of differentiation [131]. Low expression of Lin28A and Lin28B results in a lower rate of growth and development of the organism [135]. Lin28 is directly involved in the reprogramming of somatic cells to stem cells [136, 137]. High expression levels of Lin28 were observed in the least differentiated and most aggressive tumors [138, 139]. Lin28 acts as an oncoprotein and stimulates tumor development and metastasis in various human cancers [140]. In addition to the above functions, Lin28 participates in metabolism regulation, including glycolysis and related processes [141, 142].
Originally, Lin28 together with the let-7 miRNAs were discovered in the nematode Caenorhabditis elegans as a heterochronic gene that controls developmental timing [143, 144]. Recent studies have provided evidence for the Lin28B/let-7 involvement in the regulation of kidney development timing in mice [145]. In mammals, the let-7 family includes 12 members that target oncogenes and numerous pluripotency-maintaining genes [146]. Special attention has been focused on Lin28/let-7 due to the fact that Lin28 is one of the four factors required for the reprogramming of differentiated human cells into induced pluripotent stem cells [136].
Many studies have shown that Lin28 blocks maturation of let-7 [131]. Lin28-induced inhibition of the mature let-7 formation can occur by three mechanisms. One is the inhibition of pri-let-7 processing in the nucleus by the nuclease Drosha [147, 148]. Another is the suppression of the pre-let-7 processing by the nuclease Dicer [149]. Lastly, Lin28 promotes the polyuridylation of pre-let-7 at its 3′-end by recruiting terminal uridyl transferase TUT4 to the Lin28/pre-let-7 complex [150, 151]. Next, uridylated pre-let-7 is degraded by the 3′→5′ exonuclease Dis3I2 [152]. There is a reason to believe that the main role in the formation of the Lin28/pre-let-7/TUT4 ternary complex belongs to the Lin28 CTD containing two zinc finger domains (ZKDs) [133]. Interestingly, polyuridylation of the ternary complex requires the presence of a double-stranded fragment of pre-let-7 that should contain at least 15 nucleotide pairs.
Both CSD and ZKDs of Lin28 are involved in the interaction with pre-let-7 [153, 154]. X-ray structure analysis of Lin28 complexes with the precursors of pre-let-7d, pre-let-7-f1, and pre-let-7g showed that Lin28 binds to pre-let-7 through two distinct single-stranded regions in the terminal loop of let-7 [153]. ZKDs bind to the GGAG motif, while CSD binds to GNGAY, where Y is pyrimidine and N is any base. NMR spectroscopy results reported in the same study showed that the linker connecting CSD and ZKDs is a flexible structure that presumably allows Lin28 to interact with all members of the let-7 family, except let-7a-3 (ortholog of murine let-7c-2) lacking the GGAG sequence [155]. The study of the affinity of the full-length Lin28 and its two domains toward different members of the pre-let-7 family showed that the full-length Lin28 exhibits the highest affinity for pre-let-7, while CSD shows the intermediate affinity, and ZKD shows the lowest affinity for it [133, 153, 154]. The above results suggest that Lin28 binding to pre-let-7 is a two-step process [154]. Firstly, CSD binds to pre-let-7, thereby altering the conformation of the latter and making its GGAG motif accessible for ZKD. Next, ZKD binding to GGAG secures the Lin28/pre-let-7 conformation that disrupts the interaction between Dicer and pre-let-7.
More accurate identification of the pre-let-7 motif that binds to the CSD was performed using bioinformatics techniques [156-158] specifically developed for analyzing the results of HITS-Clip and Clip-sec studies that included Lin28–RNA cross-linking and immunoprecipitation, followed by large-scale sequencing. The developed computer techniques were supplementary to the databases formed based on the results of Lin28–RNA immunoprecipitation from the embryonic stem cell lysates [159] and K652 and HepG2 cell lines [160]. This analysis yielded the (U)GAU motif [161] located in the region of the previously predicted GNGAY motif [153].
Only half of let-7 family members contain both GAU and GGAG-like motifs (CSD+ subfamily). These include pre-let-7b, pre-let-7d, pre-let-7f-1, pre-let-7g, and mir-98, plus pre-let-7i that has GAC, a variant of GAU. The rest, except let-7a-3, have only the ZKD-binding motif GGAG (CSD– subfamily). The study [161] also reported that Lin28 shows a higher affinity for the pre-let-7 sequences from the CSD+ subfamily and that members of this subfamily are much more susceptible to polyuridylation and subsequent degradation in vivo.
In addition to pri- and pre-let-7 processing, Lin28 controls the level of brain-specific miRNA-9 via a polyuridylation-independent mechanism [162]. Interestingly, in contrast to pre-let-7, Lin28 interaction with pre-miRNA-9 is independent of GGAG [163]. During neural differentiation in vitro, constitutive expression of Lin28A can either increase or decrease the levels of miRNAs. This effect of Lin28 overexpression on miRNAs mediated by a decrease in the let-7 content can result from the redistribution of Argonaute-associated miRNAs within their families [164]. In turn, this redistribution can entail significant changes in translation.
There are also numerous data indicating the existence in mammalian cells of pathways other than Lin28/let-7 by which Lin28 exerts its influence on a wide variety of intracellular processes. First of all, Lin28 interacts with a large set of mRNAs (about several thousand), which has been demonstrated by cross-linking and immunoprecipitation with subsequent large-scale sequencing [159, 165-167] and by analysis of RNA directly in the immunoprecipitation product [168]. It was shown that Lin28 binds mostly to the coding mRNA regions or to the non-coding 3′-UTRs. Analysis of the Lin28-binding sites in RNA identified two motifs, AAGNNG and AAGNG, supposedly recognized by Lin28 and usually located in the terminal loop of the RNA hairpin structure [159]. Studies of the Lin28-targeted RNAs indicated probable auto-regulation of Lin28 and its involvement in the regulation of splicing [165]. Among the most frequent RNA targets of Lin28 in HEK293 cells are its own mRNA and mRNAs encoding other RNA-binding proteins and cell cycle regulators [167].
Studies of embryonic stem cells and cancer cells by overexpression or RNA interference also indicated the existence of Lin28-mediated pathways, other than let-7, in the regulation of cellular events [131, 140, 169]. However, their exact mechanisms remain unknown.
Large-scale studies of the proteome and phosphoproteome of human embryonic stem cells during their differentiation [170, 171] identified four phosphorylated amino acid residues in Lin28: Ser3/Ser5, Ser120, Ser180, and Ser200 (the latter is within the MAPK recognition sequence) [172]. Using various MAPK inhibitors, it was confirmed that Ser200 is phosphorylated by MAPK/ERK and that its phosphorylation increases Lin28 stability. Although Lin28 phosphorylation does not affect the amount of let-7, phosphorylated Lin28 shows higher efficiency in the reprogramming of differentiated cells to the pluripotent ones.
Previously, Lin28 had been considered exclusively as an RNA-binding protein. However, recent studies [173] have shown that in mouse embryonic stem cells, Lin28 regulates transcription through the epigenetic DNA modification. At the initial stage, Lin28 binds to specific DNA sites (“CAGnACC”-nn-“GGACAG”) in the promoter regions located in the immediate vicinity of the transcription initiation sites. In turn, this facilitates recruitment and binding of 5-methylcytosine dioxygenase (Tet1) that converts 5-methylcytosine to 5-hydroxymethylcytosine. The knockdown of Lin28 or Tet1 results in the disturbed regulation of DNA methylation and altered gene expression. These observations offer a completely new view on the mechanisms of Lin28 participation in the regulation of various processes in mammalian cells.
CSD PROTEINS IN PLANTS
Genes encoding CSD proteins have been detected in plants of different systematic groups, such as lower plants, monocotyledonous, dicotyledonous, and arboreous plants [174]. The genomes of all examined species contained at least two genes for the CSD proteins, as in rice (Oryza sativa), maize (Zea mays), sorghum (Sorghum bicolor), and grapes (Vitis vinifera). The largest number of CSD protein genes (seven) was found in the soybean (Glycine max) [175].
All plant CSD proteins are structurally very similar [175] and mostly close to Lin28, with CSD located in the N-terminal domain and 2 to 7 zinc fingers of the CCHC type in the CTD [175]. In lower plants, proteins of this family can contain several CSDs [176], which is untypical of higher plants. Unlike Lin28, plant CSD proteins have an increased number of glycine and arginine residues in their CTDs. Their CCHC-type zinc fingers consist of 14 a.a. (CNNCNNNNHNNNNC; residues shown in bold coordinate zinc ions). The amino acid sequences of zinc fingers differ from one another; the most variable are the 2nd, 3rd, 5th, and 6th residues that vary both within the zinc fingers of the same protein and between different proteins. The reason for the variability of the zinc finger primary structure still remains unclear. It was suggested in [177] that in plants, the diversity of partner proteins for each of the CSD proteins depends on the number of zinc fingers and their amino acid sequence.
The three-dimensional structure of plant CSD proteins has not yet been determined.
The functions of CSD proteins in plants are characterized much less than in animals. It was found that expression of plant CSD proteins is activated by a decrease in the ambient temperature [174, 176, 178-181]. Cold acclimation of soft winter wheat results in a significant accumulation of CSD proteins in the apical meristem tissues, whose preservation determines general frost resistance of the plant; however, no such effect was observed for the spring wheat [182]. Using the red osier dogwood shrub (Cornus sericea), it was shown by Western blotting that the highest accumulation of CSD proteins occurs during the periods of its maximal cold hardiness [176].
At the optimal temperatures, expression of genes encoding various CSD proteins is observed mainly in young plants, as well as in meristematic and generative tissues, as it was shown for Arabidopsis thaliana, wheat, and rice [178-180, 182-184].
The study of A. thaliana with artificially reduced or enhanced expression of CSD protein genes confirmed the involvement of these proteins in cold resistance and plant development. Arabidopsis thaliana AtCSDP3-null lines displayed a higher sensitivity to freezing temperatures than the wild-type plants, both with and without cold acclimation. Overexpression of AtCSDP3 increased the tolerance of A. thaliana plants to freezing [180]. Overexpression of AtCSDP4 caused a number of abnormalities in the plant development; specifically, decreased the length of siliqua, reduced the viability of ovules, and induced early browning and shrunken seed formation starting at the late heart embryo stage [185]. The mutant lines with up- or downregulated AtCSDP2 expression exhibited various abnormalities in the apical domination, flowering timing, and generative sphere development [178].
The molecular mechanisms through which CSD proteins contribute to the stress tolerance and plant development have not yet been elucidated. By analogy with animal and bacterial CSD proteins, the functions of plant proteins are believed to be mediated by their interactions with nucleic acids. Various plant CSD proteins have been demonstrated to localize to the cytoplasm, which suggests their involvement in events related to mRNA formation and functioning [178, 180, 186-188]. Interaction of plant CSD proteins with DNA and RNA was shown in vitro using synthetic oligonucleotides. Similar to their homologs from animals and bacteria, these proteins are capable of melting nucleic acid secondary structures [186, 188].
Interaction between plant CSD proteins and nucleic acids in vitro was studied in detail using EsCSDP1, EsCSDP2, and EsCSDP3 from Eutrema salsugineum [177, 189, 190]. These proteins were able to melt the secondary structure of DNA and RNA molecules with different nucleotide sequences and spatial structures. Experiments with DNA oligonucleotides showed that melting of the secondary structure required protein binding to a single-stranded region adjacent to the secondary structure from the 3′-direction; for effective melting, the single-stranded region should be at least 7-8-nt-long. Proteins with a larger number of zinc fingers in their CTD displayed higher DNA- and RNA-melting activity [191].
Although CSD proteins display a higher affinity for ssRNA, their binding to RNA, in contrast to DNA, does not require the presence of single-stranded regions [177]. Similar to the DNA binding, the major role in the non-specific RNA binding belongs to the zinc fingers of the CTD; the higher the number of zinc fingers, the higher the affinity for RNA. In comparison with the affinity of Lin28 or its domains for pre-let-7 (Lin28 > CSD > ZKD), the non-specific EsCSDP1 binding to both RNA and DNA shows a different order in the affinity: EsCSDP1 > ZKD > CSD [177, 189]. In addition, EsCSDP binding to RNA in vitro requires the presence of G in the binding site sequence both in ssRNA and dsRNA.
As in the case of Lin28 [159, 165-168], the product of A. thaliana AtCSDP1 immunoprecipitation [192] contained several thousand different mRNAs, which indicates a rather non-specific interaction of this protein with RNA in vivo. Most of these mRNAs encoded proteins involved in the energy-consuming cell processes, such as ribosome biogenesis. The nucleotide composition of their 5′-UTRs was characterized by an increased content of GC pairs, which potentially hinders translation of these mRNAs under conditions unfavorable for the plant [193, 194]. It was found that these 5′-UTRs are prone to the formation of secondary structures; hence, their efficient translation requires interaction with the RNA-melting protein. It is believed that AtCSDP1, by analogy with prokaryotic CSPs, exhibits this activity towards certain mRNAs and facilitates their interaction with the 43S pre-initiation complex, and therefore, their translation [192]. Interestingly, AtCSDP1 was detected in the polysome fraction, which indicates its participation in translation; a temperature downshift promoted enrichment of polysome fraction with this protein.
In addition to the nonspecific interaction of plant CSD proteins with RNA, their specific interaction with certain target RNAs cannot be ruled out. This is evidenced, e.g., by the recently reported ability of the CSD protein PpCSP1 from the moss Physcomitrella patens to regulate the reprogramming of differentiated leaf cells into chloronema apical stem cells [195]. It should be noted that Lin28, which closely resembles PpCSP1 in the amino acid sequence and domain structure, is involved in the reprogramming of human fibroblasts to pluripotent stem cells [136].
A characteristic feature of CSD proteins is their upregulated expression in actively dividing cells. Prokaryotic CSPs are required for the cell growth under unfavorable conditions and during active cell proliferation. Eukaryotic CSD proteins are involved in the cell growth regulation and cell differentiation and dedifferentiation. In eukaryotes, these ontogenetic processes show intricate time dependence and are controlled by numerous cellular regulators.
The involvement of CSD proteins in the regulation of gene expression at different levels is related to their ability to bind DNA and RNA, with the specificity and tightness of the binding dictated by the growing complexity of the protein structure. Prokaryotic proteins composed of CSD only regulate gene expression through their RNA-melting activity. The presence of additional domains in the CSD eukaryotic proteins provides their more complex interactions with nucleic acids. In particular, under certain conditions, their DNA/RNA-melting activity may be replaced by the annealing activity. This dual activity allows CSD proteins to stimulate formation of more perfect duplexes and facilitates transition of RNA molecules to more energy-favorable conformations. The functioning of CSD proteins is controlled by various post-translational modifications resulting from the action of stress factors or cellular effectors. Hence, these proteins are frequently involved in various pathological processes in the host organism, including tumor formation.
The search for specific nucleotide sequences and identification of amino acid residues involved in the interactions with these sequences are necessary to understand the mechanisms of CSD protein functioning. Over the past year, several YB-1 complexes with different oligonucleotides have been generated to provide experimental identification of amino acid residues involved in the nucleic acid binding. Identification of functionally important amino acid residues will promote the development of new approaches to the treatment of cancer and neurodegenerative diseases.
Funding. This study was performed within the State Assignment no. 0574-2019-0001 and supported in part (section “YB proteins”) by the Russian Science Foundation (project 19-74-20129).
Acknowledgements. The authors thank E. V. Serebrova for the help in the manuscript preparation.
Conflict of interest. The authors declare no conflict of interest.
Compliance with ethical norms. This article does not contain any studies with human participants or animals performed by any of the authors.
REFERENCES
1.Hudson, W. H., and Ortlund, E. A. (2014) The
structure, function and evolution of proteins that bind DNA and RNA,
Nat. Rev. Mol. Cell Biol., 15, 749-760.
2.Horn, G., Hofweber, R., Kremer, W., and Kalbitzer,
H. R. (2007) Structure and function of bacterial cold shock proteins,
Cell. Mol. Life Sci., 64, 1457-1470.
3.Maris, C., Dominguez, C., and Allain, F. H. T.
(2005) The RNA recognition motif, a plastic RNA‐binding platform
to regulate post‐transcriptional gene expression, FEBS
J., 272, 2118-2131.
4.Schroder, K., Graumann, P., Schnuchel, A., Holak,
T. A., and Marahiel, M. A. (1995) Mutational analysis of the putative
nucleic acid‐binding surface of the cold‐shock domain,
CspB, revealed an essential role of aromatic and basic residues in
binding of single‐stranded DNA containing the Y‐box
motif, Mol. Microbiol., 16, 699-708.
5.Kleene, K. C. (2018) Y-box proteins combine
versatile cold shock domains and arginine-rich motifs (ARMs) for
pleiotropic functions in RNA biology, Biochem. J., 475,
2769-2784.
6.Kremer, W., Schuler, B., Harrieder, S., Geyer, M.,
Gronwald, W., Welker, C., Jaenicke, R., and Kalbitzer, H. R. (2001)
Solution NMR structure of the cold‐shock protein from the
hyperthermophilic bacterium Thermotoga maritima, Eur. J.
Biochem., 268, 2527-2539.
7.Schnuchel, A., Wiltscheck, R., Czisch, M., Herrler,
M., Willimsky, G., Graumann, P., Marahiel, M. A., and Holak, T. A.
(1993) Structure in solution of the major cold-shock protein from
Bacillus subtilis, Nature, 364, 169-171.
8.Jones, P. G., VanBogelen, R. A., and Neidhardt, F.
C. (1987) Induction of proteins in response to low temperature in
Escherichia coli, J. Bacteriol., 169,
2092-2095.
9.Etchegaray, J. P., Jones, P. G., and Inouye, M.
(1996) Differential thermoregulation of two highly homologous
cold‐shock genes, CspA and CspB, of Escherichia coli,
Genes Cells, 1, 171-178.
10.Etchegaray, J. P., and Inouye, M. (1999) CspA,
CspB, and CspG, major cold shock proteins of Escherichia coli,
are induced at low temperature under conditions that completely block
protein synthesis, J. Bacteriol., 181, 1827-1830.
11.Gualerzi, C. O., Giuliodori, A. M., and Pon, C.
L. (2003) Transcriptional and post-transcriptional control of
cold-shock genes, J. Mol. Biol., 331, 527-539.
12.Zhang, Y., Burkhardt, D. H., Rouskin, S., Li, G.
W., Weissman, J. S., and Gross, C. A. (2018) A stress response that
monitors and regulates mRNA structure is central to cold shock
adaptation, Mol. Cell, 70, 274-286.
13.Goldstein, J., Pollitt, N. S., and Inouye, M.
(1990) Major cold shock protein of Escherichia coli,
PNAS, 87, 283-287.
14.Schindelin, H., Jiang, W., Inouye, M., and
Heinemann, U. (1994) Crystal structure of CspA, the major cold shock
protein of Escherichia coli, PNAS, 91,
5119-5123.
15.Graumann, P. L., and Marahiel, M. A. (1998) A
superfamily of proteins that contain the cold-shock domain, Trends
Biochem. Sci., 23, 286-290.
16.Phadtare, S., Alsina, J., and Inouye, M. (1999)
Cold-shock response and cold-shock proteins, Curr. Opin.
Microbiol., 2, 175-180.
17.Wang, N., Yamanaka, K., and Inouye, M. (1999)
CspI, the ninth member of the CspA family of Escherichia coli,
is induced upon cold shock, J. Bacteriol., 181,
1603-1609.
18.Bae, W., Phadtare, S., Severinov, K., and Inouye,
M. (1999) Characterization of Escherichia coli CspE,
whose product negatively regulates transcription of CspA, the
gene for the major cold shock protein, Mol. Microbiol.,
31, 1429-1441.
19.Yamanaka, K., Mitani, T., Ogura, T., Niki, H.,
and Hiraga, S. (1994) Cloning, sequencing, and characterization of
multicopy suppressors of a mukB mutation in Escherichia coli,
Mol. Microbiol., 13, 301-312.
20.Xia, B., Ke, H., and Inouye, M. (2001)
Acquirement of cold sensitivity by quadruple deletion of the cspA
family and its suppression by PNPase S1 domain in Escherichia
coli, Mol. Microbiol., 40, 179-188.
21.Mueller, U., Perl, D., Schmid, F. X., and
Heinemann, U. (2000) Thermal stability and atomic-resolution crystal
structure of the Bacillus caldolyticus cold shock protein, J.
Mol. Biol., 297, 975-988.
22.Schindelin, H., Marahiel, M. A., and Heinemann,
U. (1993) Universal nucleic acid-binding domain revealed by crystal
structure of the B. subtilis major cold-shock protein,
Nature, 364, 164-168.
23.Newkirk, K., Feng, W., Jiang, W., Tejero, R.,
Emerson, S. D., Inouye, M., and Montelione, G. T. (1994) Solution NMR
structure of the major cold shock protein (CspA) from Escherichia
coli: identification of a binding epitope for DNA, PNAS,
91, 5114-5118.
24.Jiang, W., Hou, Y., and Inouye, M. (1997) CspA,
the major cold-shock protein of Escherichia coli, is an RNA
chaperone, J. Biol. Chem., 272, 196-202.
25.Phadtare, S., and Inouye, M. (1999)
Sequence‐selective interactions with RNA by CspB, CspC and CspE,
members of the CspA family of Escherichia coli, Mol.
Microbiol., 33, 1004-1014.
26.Lopez, M. M., Yutani, K., and Makhatadze, G. I.
(2001) Interactions of the cold shock protein CspB from Bacillus
subtilis with single-stranded DNA. Importance of the T base content
and position within the template, J. Biol. Chem., 276,
15511-15518.
27.Phadtare, S., Inouye, M., and Severinov, K.
(2002) The nucleic acid melting activity of Escherichia coli
CspE is critical for transcription antitermination and cold acclimation
of cells, J. Biol. Chem., 277, 7239-7245.
28.Rennella, E., Sara, T., Juen, M., Wunderlich, C.,
Imbert, L., Solyom, Z., Favier, A., Ayala, I., Weinhaupl, K., Shanda,
P., Konrat, R., Kreutz., K., and Brutscher, B. (2017) RNA binding and
chaperone activity of the E. coli cold-shock protein CspA,
Nucleic Acids Res., 45, 4255-4268.
29.Bae, W., Xia, B., Inouye, M., and Severinov, K.
(2000) Escherichia coli CspA-family RNA chaperones are
transcription antiterminators, PNAS, 97, 7784-7789.
30.Ermolenko, D. N., and Makhatadze, G. I. (2002)
Bacterial cold-shock proteins, Cell. Mol. Life Sci., 59,
1902-1913.
31.Barria, C., Malecki, M., and Arraiano, C. M.
(2013) Bacterial adaptation to cold, Microbiology, 159,
2437-2443.
32.Rudan, M., Schneider, D., Warnecke, T., and
Krisko, A. (2015) RNA chaperones buffer deleterious mutations in E.
coli, Elife, 4, 1-16.
33.Yamanaka, K. (1999) Cold shock response in
Escherichia coli, J. Mol. Microbiol. Biotechnol.,
1, 193-202.
34.Phadtare, S., and Inouye, M. (2001) Role of CspC
and CspE in regulation of expression of RpoS and UspA, the stress
response proteins in Escherichia coli, J. Bacteriol.,
183, 1205-1214.
35.Feng, Y., Huang, H., Liao, J., and Cohen, S. N.
(2001) Escherichia coli poly(A)-binding proteins that interact
with components of degradosomes or impede RNA decay mediated by
polynucleotide phosphorylase and RNase E, J. Biol. Chem.,
276, 31651-31656.
36.Chang, B. E., Lin, C. Y., and Kuo, C. M. (1999)
Molecular cloning of a cold-shock domain protein, zfY1, in zebrafish
embryo, BBA Protein Struct. Mol. Enzymol., 1433,
343-349.
37.Falsone, F. S., Weichel, M., Crameri, R.,
Breitenbach, M., and Kungl, A. J. (2002) Unfolding and double-stranded
DNA binding of the cold shock protein homologue Clah8 from
Cladosporium herbarum, J. Biol. Chem., 277,
16512-16516.
38.Ferrer, N., Garcia-Espana, A., Jeffers, M., and
Pellicer, A. (1999) The unr gene: evolutionary considerations
and nucleic acid-binding properties of its long isoform product, DNA
Cell Biol., 18, 209-218.
39.Varadi, M., Zsolyomi, F., Guharoy, M., Tompa, P.,
and Levy, Y. K. (2015) Functional advantages of conserved intrinsic
disorder in RNA-binding proteins, PLoS One, 10,
e0139731.
40.Kedersha, N., and Anderson, P. (2017) Mammalian
stress granules and processing bodies, Methods Enzymol.,
431, 61-81.
41.Skabkin, M. A., Kiselyova, O. I., Chernov, K. G.,
Sorokin, A. V., Dubrovin, E. V., Yaminsky, I. V., Vasiliev, V. D., and
Ovchinnikov, L. P. (2004) Structural organization of mRNA complexes
with major core mRNP protein YB-1, Nucleic Acids Res.,
32, 5621-5635.
42.Eliseeva, I. A., Kim, E. R., Guryanov, S. G.,
Ovchinnikov, L. P., and Lyabin, D. N. (2011) Y-box-binding protein 1
(YB-1) and its functions, Biochemistry (Moscow), 76,
1402-1433.
43.Miwa, A., Higuchi, T., and Kobayashi, S. (2006)
Expression and polysome association of YB-1 in various tissues at
different stages in the lifespan of mice, Biochim. Biophys. Acta
Gen. Subj., 1760, 1675-1681.
44.Murray, M. T., Schiller, D. L., and Franke, W. W.
(1992) Sequence analysis of cytoplasmic mRNA-binding proteins of
Xenopus oocytes identifies a family of RNA-binding proteins,
PNAS, 89, 11-15.
45.Berghella, L., De Angelis, L., De Buysscher, T.,
Mortazavi, A., Biressi, S., Forcales, S., Sirabella, D., Cossu, G., and
Wold, B. (2008) A highly conserved molecular switch binds MSY-3 to
regulate myogenin repression in postnatal muscle, Genes Dev.,
22, 2125-2138.
46.Lima, B., Forrester, M., Hess, D., and Stamler,
J. (2014) S-Nitrosylation in cardiovascular signaling, Circ.
Res., 106, 633-646.
47.Bernstein, H., Lindquist, J., Keilhoff, G.,
Dobrowolny, H., Brandt, S., Steiner, J., Bogerts, B., and Mertens, P.
(2014) Differential distribution of Y-box-binding protein 1 and cold
shock domain protein A in developing and adult human brain, Brain
Struct. Funct., 220, 2235-2245.
48.Lu, Z. H., Books, J. T., and Ley, T. J. (2005)
YB-1 is important for late-stage embryonic development, optimal
cellular stress responses, and the prevention of premature senescence,
Mol. Cell. Biol., 25, 4625-4637.
49.Lu, Z. H., Books, J. T., and Ley, T. J. (2006)
Cold shock domain family members YB-1 and MSY4 share essential
functions during murine embryogenesis, Mol. Cell. Biol.,
26, 8410-8417.
50.Lasham, A., Print, C. G., Woolley, A. G., Dunn,
S. E., and Braithwaite, A. W. (2013) YB-1: oncoprotein, prognostic
marker and therapeutic target? Biochem. J., 449,
11-23.
51.Prabhu, L., Hartley, A. V., Martin, M., Warsame,
F., Sun, E., and Lu, T. (2015) Role of post-translational modification
of the Y box binding protein 1 in human cancers, Genes Dis.,
2, 240-246.
52.Maurya, P., Mishra, A., Yadav, B., Singh, S.,
Kumar, P., Chaudhary, A., Srivastava, S., Murugesan, S., and Mani,
A. (2017) Role of Y box protein-1 in cancer: as potential
biomarker and novel therapeutic target, J. Cancer, 8,
1900-1907.
53.Morel, C., Kayibanda, B., and Scherrer, K. (1971)
Proteins associated with globin messenger RNA in avian erythroblasts:
isolation and comparison with the proteins bound to nuclear
messenger-like RNA, FEBS Lett., 18, 84-88.
54.Blobel, G. (1972) Protein tightly bound to globin
mRNA, Biochem. Biophys. Res. Commun., 47, 88-95.
55.Morel, C., Gander, E. S., Herzberg, M., Dubochet,
J., and Scherrer, K. (1973) The duck-globin
messenger–ribonucleoprotein complex resistance to high ionic
strength, particle gel electrophoresis, composition and visualisation
by dark-field electron microscopy, Eur. J. Biochem., 36,
455-464.
56.Didier, D. K., Schiffenbauer, J., Woulfe, S. L.,
and Zacheis, M. (1988) Characterization of the cDNA encoding a protein
binding to the major histocompatibility complex class II Y box,
PNAS, 85, 7322-7326.
57.Hiroshi, S., Toshio, M., Fumio, I., Kunio, Y.,
and Shunsuke, I. (1988) Two human genes isolated by a novel method
encode DNA-binding proteins containing a common region of homology,
Gene, 73, 499-507.
58.Evdokimova, V. M., Wei, C. L., Sitikov, A. S.,
Simonenko, P. N., Lazarev, O. A., Vasilenko, K. S., Ustinov, V. A.,
Hershey, J. W., and Ovchinnikov, L. P. (1995) The major protein
of messenger ribonucleoprotein particles in somatic cells is a member
of the Y-box binding transcription factor family, J. Biol.
Chem., 270, 3186-3192.
59.Skabkin, M. A., Lyabin, D. N., and Ovchinnikov,
L. P. (2006) Nonspecific and specific interaction of Y-box binding
protein 1 (YB-1) with mRNA and posttranscriptional regulation of
protein synthesis in animal cells, Mol. Biol., 40,
551-563.
60.Yang, X., Zhu, H., Mu, S., Wei, W., Yuan, X.,
Wang, M., Liu, Y., Hui, J., and Huang, Y. (2019) Crystal structure of a
Y-box binding protein 1 (YB-1)–RNA complex reveals key features
and residues interacting with RNA, J. Biol. Chem., 294,
10998-11010.
61.Guryanov, S. G., Filimonov, V. V., Timchenko, A.
A., Melnik, B. S., Kihara, H., Kutyshenko, V. P., Ovchinnikov, L. P.,
and Semisotnov, G. V. (2013) The major mRNP protein YB-1: structural
and association properties in solution, BBA Proteins Proteom.,
1834, 559-567.
62.Wu, S., Fu, X., Huang, J., Jia, T., Zong, F., Mu,
S., Zhu, H., Yan, Y., Qiu, S., Wu, Q., Yan, W., Peng, Y., Chen, J., and
Hui, J. (2015) Genome-wide analysis of YB-1–RNA
interactions reveals a novel role of YB-1 in miRNA processing in
glioblastoma multiforme, Nucleic Acids Res., 43,
8516-8528.
63.Ivanov, P., Emara, M., Villen, J., Gygi, S. P.,
and Anderson, P. (2011) Angiogenin-induced tRNA fragments
inhibit translation initiation, Mol. Cell, 43,
613-623.
64.Dimartino, D., Colantoni, A., Ballarino, M.,
Martone, J., Mariani, D., Danner, J., Bruckmann, A., Meister, G.,
Morlando, M., and Bozzoni, I. (2018) The long non-coding RNA lnc-31
interacts with Rock1 mRNA and mediates its YB-1-dependent translation,
Cell Rep., 23, 733-740.
65.Lyabin, D. N., Eliseeva, I. A., and Ovchinnikov,
L. P. (2014) YB-1 protein: functions and regulation, Wiley
Interdiscip. Rev. RNA, 5, 95-110.
66.Evdokimova, V. M., Kovrigina, E. A., Nashchekin,
D. V., Davydova, E. K., Hershey, J. W. B., and Ovchinnikov, L. P.
(1998) The major core protein of messenger ribonucleoprotein particles
(p50) promotes initiation of protein biosynthesis in vitro,
J. Biol. Chem., 273, 3574-3581.
67.Ruzanov, P. V., Evdokimova, V. M., Korneeva, N.
L., Hershey, J. W. B., and Ovchinnikov, L. P. (1999) Interaction
of the universal mRNA-binding protein, p50, with actin: a possible link
between mRNA and microfilaments, J. Cell Sci., 112,
3487-3496.
68.Chernov, K. G., Curmi, P. A., Hamon, L.,
Mechulam, A., Ovchinnikov, L. P., and Pastre, D. (2008) Atomic force
microscopy reveals binding of mRNA to microtubules mediated by two
major mRNP proteins YB-1 and PABP, FEBS Lett., 582,
2875-2881.
69.Jurchott, K., Royer, H., and Centrum, M. (2000)
Y-box factor YB-1 is associated with the centrosome during mitosis,
Gene Funct. Dis., 1, 57-59.
70.Davies, A. H., Barrett, I., Hu, K., Stratford, A.
L., Freeman, S., Berquin, I. M., Pelech, S., Hieter, P., Maxwell, C.,
and Dunn, S. E. (2011) YB-1 evokes susceptibility to cancer through
cytokinesis failure, mitotic dysfunction and HER2 amplification,
Oncogene, 30, 3649-3660.
71.Somasekharan, S. P., El-Naggar, A., Leprivier,
G., Cheng, H., Hajee, S., Grunewald, T., Zhang, F., Ng, T., Delattre,
O., Evdokimova, V., Wang, Y., Gleave, M., and Sorensen, P. H. (2015)
YB-1 regulates stress granule formation and tumor progression by
translationally activating G3BP1, J. Cell Biol., 208,
913-929.
72.Bounedjah, O., Desforges, B., Wu, T.,
Pioche-Durieu, C., Marco, S., Hamon, L., Curmi, P., Guerquin-Kern, J.,
Pietrement, O., and Pastre, D. (2014) Free mRNA in excess upon polysome
dissociation is a scaffold for protein multimerization to form stress,
Nucleic Acids Res., 42, 8678-8691.
73.Jurchott, K., Bergmann, S., Stein, U., Walther,
W., Janz, M., Manni, I., Piaggio, G., Fietze, E., Dietel, M., and
Royer, H. (2003) YB-1 as a cell cycle-regulated transcription factor
facilitating cyclin A and cyclin B1 gene expression, J. Biol.
Chem., 278, 27988-27996.
74.Harada, M., Kotake, Y., Ohhata, T., Kitagawa, K.,
Niida, H., Matsuura, S., Funai, K., Sugimura, H., Suda, T., and
Kitagawa, M. (2014) YB-1 promotes transcription of cyclin D1 in human
non-small-cell lung cancers, Genes Cells, 19,
504-516.
75.Pagano, C., Martino, O., Ruggiero, G., Guarino,
A., Mueller, N., Siauciunaite, R., Reischl, M., Foulkes, N., Vallone,
D., and Calabro, V. (2017) The tumor-associated YB-1 protein: new
player in the circadian control of cell proliferation,
Oncotarget, 8, 6193-6205.
76.Sorokin, A. V., Selyutina, A. A., Skabkin, M. A.,
Guryanov, S. G., Nazimov, I. V., Richard, C., Th'Ng, J., Yau, J.,
Sorensen, P., Ovchinnikov, L. P., and Evdokimova, V. (2005)
Proteasome-mediated cleavage of the Y-box-binding protein 1 is linked
to DNA-damage stress response, EMBO J., 24,
3602-3612.
77.Sutherland, B., Kucab, J., Wu, J., Lee, C.,
Cheang, M., Yorida, E., Turbin, D., Dedhar, S., Nelson, C., Pollak, M.,
Grimes, H., Miller, K., Badve, S., Huntsman, D., Chen, M., Pallen, C.,
and Dunn, S. (2005) Akt phosphorylates the Y-box binding protein 1 at
Ser102 located in the cold shock domain and affects the
anchorage-independent growth of breast cancer cells, Oncogene,
24, 4281-4292.
78.Basaki, Y., Hosoi, F., Oda, Y., Fotovati, A.,
Maruyama, Y., Oie, S., Ono, M., Izumi, H., Kohno, K., Sakai, K.,
Shimoyama, T., Nishio, K., and Kuwano, M. (2007) Akt-dependent nuclear
localization of Y-box-binding protein 1 in acquisition of malignant
characteristics by human ovarian cancer cells, Oncogene,
26, 2736-2746.
79.Bogolyubova, I. O., Lyabin, D. N., Bogolyubov, D.
S., and Ovchinnikov, L. P. (2014) Immunocytochemical study of YB-1
nuclear distribution in different cell types, Tissue Cell,
46, 457-461.
80.Gonda, K., Wudel, J., Nelson, D., Katoku-Kikyo,
N., Reed, P., Tamada, H., and Kikyo, N. (2006) Requirement of the
protein B23 for nucleolar disassembly induced by the FRGY2a family
proteins, J. Biol. Chem., 281, 8153-8160.
81.Fang, J., Hong, H., Xue, X., Zhu, X., Jiang, L.,
Qin, M., Liang, H., and Gao, L. (2019) A novel circular RNA,
circFAT1(e2), inhibits gastric cancer progression by targeting miR-548g
in the cytoplasm and interacting with YBX1 in the nucleus, Cancer
Lett., 442, 222-232.
82.Kljashtorny, V., Nikonov, S., Ovchinnikov, L.,
Lyabin, D., Volodar, N., Curmi, P., and Manivet, P. (2015) The cold
shock domain of YB-1 segregates RNA from DNA by non-bonded
interactions, PLoS One, 10, e0130318.
83.Minich, W. B., Maidebura, I. P., and Ovchinnikov,
L. P. (1993) Purification and characterization of the major
50‐kDa repressor protein from cytoplasmic mRNP of rabbit
reticulocytes, Eur. J. Biochem., 212, 633-638.
84.Skabkin, M., Evdokimova, V., Thomas, A., and
Ovchinnikov, L. (2001) The major messenger ribonucleoprotein particle
protein p50 (YB-1) promotes nucleic acid strand annealing, J. Biol.
Chem., 276, 44841-44847.
85.Hasegawa, S., Doetsch, P., Hamilton, K., Martin,
A., Okenquist, S., Lenz, J., and Boss, J. (1991) DNA binding properties
of YB-1 and dbpA: binding to double-stranded, single-stranded, and
abasic site containing DNAs, Nucleic Acids Res., 19,
4915-4920.
86.Gaudreault, I., Guay, D., and Lebel, M. (2004)
YB-1 promotes strand separation in vitro of duplex DNA
containing either mispaired bases or cisplatin modifications, exhibits
endonucleolytic activities and binds several DNA repair proteins,
Nucleic Acids Res., 32, 316-327.
87.Izumi, H., Imamura, T., Nagatani, G., Ise, T.,
Murakami, T., Uramoto, H., Torigoe, T., Ishiguchi, H., Yoshida, Y.,
Nomoto, M., Okamoto, T., Uchiumi, T., Kuwano, M., Funa, K., and Kohno,
K. (2001) Y box-binding protein-1 binds preferentially to
single-stranded nucleic acids and exhibits 3′→5′
exonuclease activity, Nucleic Acids Res., 29,
1200-1207.
88.Swamynathan, S. K., Nambiar, A., and Guntaka, R.
V. (1998) Role of single-stranded DNA regions and Y-box proteins in
transcriptional regulation of viral and cellular genes, FASEB
J., 12, 515-522.
89.MacDonald, G. H., Itoh-Lindstrom, Y., and Ting,
J. (1995) The transcriptional regulatory protein, YB-1, promotes
single-stranded regions in the DRA promoter, J. Biol. Chem.,
270, 3527-3533.
90.Zasedateleva, O. A., Krylov, A. S., Prokopenko,
D. V., Skabkin, M. A., Ovchinnikov, L. P., Kolchinsky, A., and
Mirzabekov, A. D. (2002) Specificity of mammalian Y-box binding
protein p50 in interaction with ss and ds DNA analyzed with generic
oligonucleotide microchip, J. Mol. Biol., 324, 73-87.
91.Wang, N., Yamanaka, K., and Inouye, M. (2000)
Acquisition of double-stranded DNA-binding ability in a hybrid protein
between Escherichia coli CspA and the cold shock domain of human
YB-1, Mol. Microbiol., 38, 526-534.
92.Kretov, D. A., Curmi, P. A., Hamon, L., Abrakhi,
S., Desforges, B., Ovchinnikov, L. P., and Pastre, D. (2015) mRNA and
DNA selection via protein multimerization: YB-1 as a case study,
Nucleic Acids Res., 43, 9457-9473.
93.Mateu-regue, A., Christiansen, J., Bagger, F.,
Hellriegel, C., and Nielsen, F. (2019) Single mRNP analysis by
super-resolution microscopy and fluorescence correlation spectroscopy
reveals that small mRNP granules represent mRNA singletons,
bioRxiv, 558098, 1-37.
94.Kretov, D., Clement, M., Lambert, G., Durand, D.,
Lyabin, D., Bollot, G., Bauvais, C., Samsonova, A., Budkina, K.,
Maroun, R., Hamon, L., Bouhss, A., Lescop, E., Toma, F., Curmi, P.,
Maucuer, A., Ovchinnikov, L., and Pastre, D. (2019) YB-1, an abundant
core mRNA-binding protein, has the capacity to form an RNA
nucleoprotein filament: a structural analysis, Nucleic Acids
Res., 47, 3127-3141.
95.Prabhu, L., Mundade, R., Wang, B., Wei, H.,
Hartley, A., Martin, M., McElyea, K., Temm, C., Sandusky, G., Liu, Y.,
and Lu, T. (2015) Critical role of phosphorylation of serine 165 of
YBX1 on the activation of NF-κB in colon cancer,
Oncotarget, 6, 29396-29412.
96.Tanabe, Y., Nagatoishi, S., and Tsumoto, K.
(2015) Molecular biosystems thermodynamic characterization of the
interaction between the human Y-box binding protein YB-1 and nucleic
acids, Mol. Biosyst., 11, 2441-2448.
97.Von Hacht, A., Seifert, O., Menger, M., Schutze,
T., Arora, A., Neubauer, P., Wagner, A., Weise, C., and Kurreck,
J. (2014) Identification and characterization of RNA
guanine-quadruplex binding proteins, Nucleic Acids Res.,
42, 6630-6644.
98.Ivanov, P., O'Day, E., Emara, M., Wagner, G.,
Lieberman, J., and Anderson, P. (2014) G-quadruplex structures
contribute to the neuroprotective effects of angiogenin-induced tRNA
fragments, PNAS, 111, 18201-18206.
99.Mertens, P., Harendza, S., Pollock, A., and
Lovett, D. (1997) Glomerular mesangial cell-specific transactivation of
matrix metalloproteinase 2 transcription is mediated by YB-1, J.
Biol. Chem., 272, 22905-22912.
100.En-Nia, A., Yilmaz, E., Klinge, U., Lovett, D.,
Stefanidis, I., and Mertens, P. (2005) Transcription factor YB-1
mediates DNA polymerase α gene expression, J. Biol. Chem.,
280, 7702-7711.
101.Rauen, T., Frye, B., Wang, J., Raffetseder, U.,
Alidousty, C., En-Nia, A., Floege, J., and Mertens, P. (2016) Cold
shock protein YB-1 is involved in hypoxia-dependent gene transcription,
Biochem. Biophys. Res. Commun., 478, 982-987.
102.Zou, Y., and Chien, K. R. (1995) EFIA/YB-1 is a
component of cardiac HF-1A binding activity and positively regulates
transcription of the myosin light-chain 2v gene, Mol. Cell.
Biol., 15, 2972-2982.
103.Coles, L. S., Diamond, P., Occhiodoro, F.,
Vadas, M. A., and Shannon, M. F. (2000) An ordered array of cold shock
domain repressor elements across tumor necrosis factor-responsive
elements of the granulocyte-macrophage colony-stimulating factor
promoter, J. Biol. Chem., 275, 14482-14493.
104.Shi, J., Zheng, B., Li, Y., Sun, Y., Han, A.,
Zhang, X., Lv, X., Chen, S., and Wen, J. (2013) Novel insight into
Y-box binding protein 1 in the regulation of vascular smooth muscle
cell proliferation through targeting GC box-dependent genes, FEBS
Lett., 587, 1326-1332.
105.Skabkina, O. V., Lyabin, D. N., Skabkin, M. A.,
and Ovchinnikov, L. P. (2005) YB-1 autoregulates translation of its own
mRNA at or prior to the step of 40S ribosomal subunit joining, Mol.
Cell. Biol., 25, 3317-3323.
106.Lyabin, D. N., Eliseeva, I. A., Skabkina, O.
V., and Ovchinnikov, L. P. (2011) Interplay between Y-box-binding
protein 1 (YB-1) and poly(A) binding protein (PABP) in specific
regulation of YB-1 mRNA translation, RNA Biol., 8,
883-892.
107.Paranjape, S. M., and Harris, E. (2007) Y
box-binding protein-1 binds to the dengue virus 3′-untranslated
region and mediates antiviral effects, J. Biol. Chem.,
282, 30497-30508.
108.Wei, W. J., Mu, S. R., Heiner, M., Fu, X., Cao,
L. J., Gong, X. F., Bindereif, A., and Hui, J. (2012) YB-1 binds to
CAUC motifs and stimulates exon inclusion by enhancing the recruitment
of U2AF to weak polypyrimidine tracts, Nucleic Acids Res.,
40, 8622-8636.
109.Yanshina, D., Kossinova, O., Gopanenko, A.,
Krasheninina, O., Malygin, A., Venyaminova, A., and Karpova, G. (2017)
Structural features of the interaction of the 3′-untranslated
region of mRNA containing exosomal RNA-specific motifs with YB-1, a
potential mediator of mRNA sorting, Biochimie, 144,
134-143.
110.Ashizuka, M., Fukuda, T., Nakamura, T.,
Shirasuna, K., Iwai, K., Izumi, H., Kohno, K., Kuwano, M., and Uchiumi,
T. (2002) Novel translational control through an iron-responsive
element by interaction of multifunctional protein YB-1 and IRP2,
Mol. Cell. Biol., 22, 6375-6383.
111.Evdokimova, V., Tognon, C., Ng, T., Ruzanov,
P., Melnyk, N., Fink, D., Sorokin, A., Ovchinnikov, L. P., Davicioni,
E., Triche, T. J., and Sorensen, P. H. B. (2009) Translational
activation of Snail1 and other developmentally regulated transcription
factors by YB-1 promotes an epithelial-mesenchymal transition,
Cancer Cell, 15, 402-405.
112.Goodarzi, H., Liu, X., Nguyen, H., Zhang, S.,
Fish, L., and Tavazoie, S. (2015) Endogenous tRNA-derived fragments
suppress breast cancer progression via YBX1 displacement, Cell,
161, 790-802.
113.Ray, D., Kazan, H., Chan, E. T., Castillo, L.
P., Chaudhry, S., Talukder, S., Blencowe, B. J., Morris, Q., and
Hughes, T. R. (2009) Rapid and systematic analysis of the RNA
recognition specificities of RNA-binding proteins, Nat.
Biotechnol., 27, 667-670.
114.Matsumoto, K. I., Abiko, S., and Ariga, H.
(2005) Transcription regulatory complex including YB-1 controls
expression of mouse matrix metalloproteinase-2 gene in NIH3T3 cells,
Biol. Pharm. Bull., 28, 1500-1504.
115.Gomes, C., Merianda, T., Lee, S., Yoo, S., and
Twiss, J. (2014) Molecular determinants of the axonal mRNA
transcriptome, Dev. Neurobiol., 74, 218-232.
116.Shurtleff, M., Temoche-Diaz, M., Karfilis, K.,
Ri, S., and Schekman, R. (2016) Y-box protein 1 is required to sort
microRNAs into exosomes in cells and in a cell-free reaction,
Elife, 5, 1-23.
117.Evdokimova, V., Ruzanov, P., Imataka, H.,
Raught, B., Svitkin, Y., Ovchinnikov, L. P., and Sonenberg, N. (2001)
The major mRNA-associated protein YB-1 is a potent 5ʹ
cap-dependent mRNA stabilizer, EMBO J., 20,
5491-5502.
118.Nekrasov, M., Ivshina, M., Chernov, K.,
Kovrigina, E., Evdokimova, V., Thomas, A., Hershey, J., and
Ovchinnikov, L. P. (2003) The mRNA-binding protein YB-1 (p50) prevents
association of the eukaryotic initiation factor eIF4G with mRNA and
inhibits protein synthesis at the initiation stage, J. Biol.
Chem., 278, 13936-13943.
119.Hayakawa, H., Uchiumi, T., Fukuda, T.,
Ashizuka, M., Kohno, K., Kuwano, M., and Sekiguchi, M. (2002) Binding
capacity of human YB-1 protein for RNA containing 8-oxoguanine,
Biochemistry, 41, 12739-12744.
120.Chen, X., Li, A., Sun, B., Yang, Y., Han, Y.,
Yuan, X., Chen, R., Wei, W., Liu, Y., Gao, C., Chen, Y., Zhang, M., Ma,
X., Liu, Z., Luo, J., Lyu, C., Wang, H., Ma, J., Zhao, Y., Zhou, F.,
Huang, Y., Xie, D., and Yang, Y. (2019) 5-Methylcytosine promotes
pathogenesis of bladder cancer through stabilizing mRNAs, Nat. Cell
Biol., 21, 978-990.
121.Yang, Y., Wang, L., Han, X., Yang, W., Zhang,
M., Ma, H., Sun, B., Li, A., Xia, J., Chen, J., Heng, J., Wu, B., Chen,
Y., Xu, J., Yang, X., Yao, H., Sun, J., Lyu, C., Wang, H., Huang, Y.,
Sun, Y., Zhao, Y., Meng, A., Ma, J., Liu, F., and Yang, Y. (2019) RNA
5-methylcytosine facilitates the maternal-to-zygotic transition by
preventing maternal mRNA decay, Mol. Cell, 75, 1-15.
122.Coles, L. S., Lambrusco, L., Burrows, J.,
Hunter, J., Diamond, P., Bert, A. G., Vadas, M. A., and Goodall, G. J.
(2005). Phosphorylation of cold shock domain/Y‐box proteins by
ERK2 and GSK3β and repression of the human VEGF promoter, FEBS
Lett., 579, 5372-5378.
123.Evdokimova, V., Ruzanov, P., Anglesio, M.,
Sorokin, A., Ovchinnikov, L., Buckley, J., Triche, T., Sonenberg, N.,
and Sorensen, P. (2006) Akt-mediated YB-1 phosphorylation activates
translation of silent mRNA species, Mol. Cell. Biol., 26,
277-292.
124.Wang, J., Gibbert, L., Djudjaj, S., Alidousty,
C., Rauen, T., Kunter, U., Rembiak, A., Enders, D., Jankowski, V.,
Braun, G., Floege, J., Ostendorf, T., and Raffetseder, U. (2016)
Therapeutic nuclear shuttling of YB-1 reduces renal damage and
fibrosis, Kidney Int., 90, 1226-1237.
125.Martin, M., Hua, L., Wang, B., Wei, H., Prabhu,
L., Hartley, A. V., Jiang, G., Liu, Y., and Lu, T. (2017) Novel serine
176 phosphorylation of YBX1 activates NF-κB in colon cancer,
J. Biol. Chem., 292, 3433-3444.
126.Alidousty, C., Rauen, T., Hanssen, L., Wang,
Q., Alampour-Rajabi, S., Mertens, P. R., Bernhagen, J., Floege, J.,
Ostendorf, T., and Raffetseder, U. (2014) Calcineurin-mediated YB-1
dephosphorylation regulates CCL5 expression during monocyte
differentiation, J. Biol. Chem., 289, 21401-21412.
127.Liu, Q., Tao, T., Liu, F., Ni, R., Lu, C., and
Shen, A. (2016) Hyper-O-GlcNAcylation of YB-1 affects Ser102
phosphorylation and promotes cell proliferation in hepatocellular
carcinoma, Exp. Cell Res., 349, 230-238.
128.Alemasova, E., Pestryakov, P., Sukhanova, M.,
Kretov, D., Moor, N., Curmi, P., Ovchinnikov, L., and Lavrik, O. (2015)
Poly(ADP-ribosyl)ation as a new posttranslational modification of YB-1,
Biochimie, 119, 36-44.
129.Alemasova, E. E., and Lavrik, O. I. (2019)
Poly(ADP-ribosyl)ation by PARP1: reaction mechanism and regulatory
proteins, Nucleic Acids Res., 47, 3811-3827.
130.Bobkova, N. V., Lyabin, D. N., Medvinskaya, N.
I., Samokhin, A. N., Nekrasov, P. V., Nesterova, I. V., Aleksandrova,
I. Y., Tatarnikova, O. G., Bobylev, A. G., Vikhlyantsev, I. M.,
Kukharsky, M. S., Ustyugov, A. A., Polyakov, D. N., Eliseeva, I. A.,
Kretov, D. A., Guryanov, S. G., and Ovchinnikov, L. P. (2015) The Y-box
binding protein 1 suppresses Alzheimer’s disease progression in
two animal models, PLoS One, 10, e0138867.
131.Tsialikas, J., and Romer-Seibert, J. (2015)
LIN28: roles and regulation in development and beyond,
Development, 142, 2397-2404.
132.Piskounova, E., Polytarchou, C., Thornton, J.
E., LaPierre, R. J., Pothoulakis, C., Hagan, J. P., Iliopoulos, D., and
Gregory, R. I. (2011) Lin28A and Lin28B inhibit let-7 microRNA
biogenesis by distinct mechanisms, Cell, 147,
1066-1079.
133.Wang, L., Nam, Y., Lee, A. K., Yu, C., Roth,
K., Chen, C., Ransey, E. M., and Sliz, P. (2017) LIN28 zinc knuckle
domain is required and sufficient to induce let-7 oligouridylation,
Cell Rep., 18, 2664-2675.
134.Hafner, M., Max, K. E., Bandaru, P., Morozov,
P., Gerstberger, S., Brown, M., Molina, H., and Tuschl, T. (2013)
Identification of mRNAs bound and regulated by human LIN28 proteins and
molecular requirements for RNA recognition, RNA, 19,
613-626.
135.Shinoda, G., Shyh‐Chang, N., Soysa, T.
Y. D., Zhu, H., Seligson, M. T., Shah, S. P., Abo-Sido, N., Yabuuchi,
A., Hagan, J. P., Gregory, R. I., Asara, J. M., Cantley, L. C., Moss,
E. G., and Daley, G. Q. (2013) Fetal deficiency of Lin28 programs
life‐long aberrations in growth and glucose metabolism, Stem
Cells, 31, 1563-1573.
136.Yu, J., Vodyanik, M. A., Smuga-Otto, K.,
Antosiewicz-Bourget, J., Frane, J. L., Tian, S., Nie, J., Jonsdottir,
G. A., Ruotti, V., Stewart, R., Slukvin, I. I., and Thomson, J. A.
(2007) Induced pluripotent stem cell lines derived from human somatic
cells, Science, 318, 1917-1920.
137.Zhang, J., Ratanasirintrawoot, S.,
Chandrasekaran, S., Wu, Z., Ficarro, S. B., Yu, C., Ross, C. A.,
Cacchiarelli, D., Xia, Q., Seligson, M., Shinoda, G., Xie, W., Cahan,
P., Wang, L., Ng, S.-C., Tintara, S., Trapnell, C., Onder, T., Loh,
Y.-H., Mikkelsen, T., Sliz, P., Teitell, M., Asara, J. M., Marto, J.
A., Li, H., Collins, J., and Daley, G. Q. (2016) LIN28 regulates stem
cell metabolism and conversion to primed pluripotency, Cell Stem
Cell, 19, 66-80.
138.Hamano, R., Miyata, H., Yamasaki, M., Sugimura,
K., Tanaka, K., Kurokawa, Y., Nakajima, K., Takiguchi, S., Fujiwara,
Y., Mori, M., and Doki, Y. (2012) High expression of Lin28 is
associated with tumour aggressiveness and poor prognosis of patients in
oesophagus cancer, Brit. J. Cancer, 106, 1415-1423.
139.Wang, T., Wang, G., Hao, D., Liu, X., Wang, D.,
Ning, N., and Li, X. (2015) Aberrant regulation of the LIN28A/LIN28B
and let-7 loop in human malignant tumors and its effects on the
hallmarks of cancer, Mol. Cancer, 14, 125.
140.Jiang, S., and Baltimore, D. (2016) RNA-binding
protein Lin28 in cancer and immunity, Cancer Lett., 375,
108-113.
141.Zhu, H., Shyh-Chang, N., Segre, A. V., Shinoda,
G., Shah, S. P., Einhorn, W. S., Takeuchi, A., Engreitz, J. M., Hagan,
J. P., Kharas, M. G., Urbach, A., Thornton, J. E., Triboulet, R.,
Gregory, R. I., DIAGRAM Consortium, MAGIC Investigators, Altshuler, D.,
and Daley, G. Q. (2011) The Lin28/let-7 axis regulates glucose
metabolism, Cell, 147, 81-94.
142.Docherty, C. K., Salt, I. P., and Mercer, J. R.
(2016) Lin28A induces energetic switching to glycolytic metabolism in
human embryonic kidney cells, Stem Cell Res. Ther., 7,
78.
143.Ambros, V., and Horvitz, H. R. (1984)
Heterochronic mutants of the nematode Caenorhabditis elegans,
Science, 226, 409-416.
144.Moss, E. G., Lee, R. C., and Ambros, V. (1997)
The cold shock domain protein LIN-28 controls developmental timing in
C. elegans and is regulated by the lin-4 RNA, Cell,
88, 637-646.
145.Yermalovich, A. V., Osborne, J. K., Sousa, P.,
Han, A., Kinney, M. A., Chen, M. J., Robinton, D. A., Montie, H.,
Pearson, D. S., Wilson, S. B., Combes, A. N., Little, M. H., and Daley,
G. Q. (2019) Lin28 and let-7 regulate the timing of cessation of murine
nephrogenesis, Nat. Commun., 10, 168.
146.Worringer, K. A., Rand, T. A., Hayashi, Y.,
Sami, S., Takahashi, K., Tanabe, K., Narita, M., Srivastava, D., and
Yamanaka, S. (2014) The let-7/LIN-41 pathway regulates reprogramming to
human induced pluripotent stem cells by controlling expression of
prodifferentiation genes, Cell Stem Cell, 14, 40-52.
147.Newman, M. A., Thomson, J. M., and Hammond, S.
M. (2008) Lin-28 interaction with the Let-7 precursor loop mediates
regulated microRNA processing, RNA, 14, 1539-1549.
148.Wiswanathan, S. R., Daley, G. Q., and Gregory,
R. I. (2008) Selective blockade of microRNA processing by Lin28,
Science, 320, 97-100.
149.Rybak, A., Fuchs, H., Smirnova, L., Brandt, C.,
Pohl, E. E., Nitsch, R., and Wulczyn, F. G. (2008) A feedback loop
comprising lin-28 and let-7 controls pre-let-7 maturation during neural
stem-cell commitment, Nat. Cell Biol., 10, 987-993.
150.Heo, I., Joo, C., Cho, J., Ha, M., Han, J., and
Kim, V. N. (2008) Lin28 mediates the terminal uridylation of let-7
precursor microRNA, Mol. Cell, 32, 276-284.
151.Heo, I., Joo, C., Kim, Y. K., Ha, M., Yoon, M.
J., Cho, J., and Kim, V. N. (2009) TUT4 in concert with Lin28
suppresses microRNA biogenesis through pre-microRNA uridylation,
Cell, 138, 696-708.
152.Chang, H. M., Triboulet, R., Thornton, J. E.,
and Gregory, R. I. (2013) A role for the Perlman syndrome exonuclease
Dis3l2 in the Lin28–let-7 pathway, Nature, 497,
244-248.
153.Nam, Y., Chen, C., Gregory, R. I., Chou, J. J.,
and Sliz, P. (2011) Molecular basis for interaction of let-7 microRNAs
with Lin28, Cell, 147, 1080-1091.
154.Mayr, F., Schutz, A., Doge, N., and Heinemann,
U. (2012) The Lin28 cold-shock domain remodels pre-let-7 microRNA,
Nucleic Acids Res., 40, 7492-7506.
155.Triboulet, R., Pirouz, M., and Gregory, R. I.
(2015) A single let-7 microRNA bypasses LIN28-mediated repression,
Cell Rep., 13, 260-266.
156.Zhang, C., and Darnell, R. B. (2011) Mapping
in vivo protein–RNA interactions at single-nucleotide
resolution from HITS-CLIP data, Nat. Biotechnol., 29,
607-614.
157.Weyn-Vanhentenryck, S. M., Mele, A., Yan, Q.,
Sun, S., Farny, N., Zhang, Z., Xue, C., Herre, M., Silver, P. A.,
Zhang, M. Q., Krainer, A. R., Darnell, R. B., and Zhang, C. (2014)
HITS-CLIP and integrative modeling define the Rbfox splicing-regulatory
network linked to brain development and autism, Cell Rep.,
6, 1139-1152.
158.Shah, A., Qian, Y., Weyn-Vanhentenryck, S. M.,
and Zhang, C. (2016) CLIP Tool Kit (CTK): a flexible and robust
pipeline to analyze CLIP sequencing data, Bioinformatics,
33, 566-567.
159.Cho, J., Chang, H., Kwon, S. C., Kim, B., Kim,
Y., Choe, J., Ha, M., Kim, K. Y., and Kim, V. N. (2012) LIN28A is a
suppressor of ER-associated translation in embryonic stem cells,
Cell, 151, 765-777.
160.Van Nostrand, E. L., Pratt, G. A., Shishkin, A.
A., Gelboin-Burkhart, C., Fang, M. Y., Sundararaman, B., Blue, S. M.,
Nguyen, T. B., Surka, C., Elkins, K., Stanton, R., Rigo, F., Guttman,
M., and Yeo, G. W. (2016) Robust transcriptome-wide discovery of
RNA-binding protein binding sites with enhanced CLIP (eCLIP), Nat.
Methods, 13, 508-514.
161.Ustianenko, D., Chiu, H. S., Treiber, T.,
Weyn-Vanhentenryck, S. M., Treiber, N., Meister, G., Sumazin, P., and
Zhang, C. (2018) LIN28 selectively modulates a subclass of let-7
microRNAs, Mol. Cell, 71, 271-283.
162.Nowak, J. S., Choudhury, N. R., de Lima Alves,
F., Rappsilber, J., and Michlewski, G. (2014) Lin28a regulates neuronal
differentiation and controls miR-9 production, Nat. Commun.,
5, 3687.
163.Nowak, J. S., Hobor, F., Velasco, A. D. R.,
Choudhury, N. R., Heikel, G., Kerr, A., Ramos, A., and Michlewski, G.
(2017) Lin28a uses distinct mechanisms of binding to RNA and affects
miRNA levels positively and negatively, RNA, 23,
317-332.
164.Tan, F. E., Sathe, S., Wheeler, E. C.,
Nussbacher, J. K., Peter, S., and Yeo, G. W. (2019) A
transcriptome-wide translational program defined by LIN28B expression
level, Mol. Cell, 73, 304-313.
165.Wilbert, M. L., Huelga, S. C., Kapeli, K.,
Stark, T. J., Liang, T. Y., Chen, S. X., Yan, B. Y., Nathanson, J. L.,
Hutt, K. R., Lovci, M. T., Kazan, H., Vu, A. Q., Massirer, K. B.,
Morris, Q., Hoon, S., and Yeo, G. W. (2012) LIN28 binds messenger RNAs
at GGAGA motifs and regulates splicing factor abundance, Mol.
Cell, 48, 195-206.
166.Graf, R., Munschauer, M., Mastrobuoni, G.,
Mayr, F., Heinemann, U., Kempa, S., Rajewsky, N., and Landthaler, M.
(2013) Identification of LIN28B-bound mRNAs reveals features of target
recognition and regulation, RNA Biol., 10, 1146-1159.
167.Hafner, M., Max, K. E., Bandaru, P., Morozov,
P., Gerstberger, S., Brown, M., Molina, H., and Tuschl, T. (2013)
Identification of mRNAs bound and regulated by human LIN28 proteins and
molecular requirements for RNA recognition, RNA, 19,
613-626.
168.Peng, S., Chen, L. L., Lei, X. X., Yang, L.,
Lin, H., Carmichael, G. G., and Huang, Y. (2011) Genome‐wide
studies reveal that Lin28 enhances the translation of genes important
for growth and survival of human embryonic stem cells, Stem
Cells, 29, 496-504.
169.Shyh-Chang, N., and Daley, G. Q. (2013) Lin28:
primal regulator of growth and metabolism in stem cells, Cell Stem
Cell, 12, 395-406.
170.Rigbolt, K. T., Prokhorova, T. A., Akimov, V.,
Henningsen, J., Johansen, P. T., Kratchmarova, I., Kassem, M., Mann,
M., Olsen, J. V., and Blagoev, B. (2011) System-wide temporal
characterization of the proteome and phosphoproteome of human embryonic
stem cell differentiation, Sci. Signal., 4, rs3-rs3.
171.Van Hoof, D., Munoz, J., Braam, S. R., Pinkse,
M. W., Linding, R., Heck, A. J., Mummery, C. L., and Krijgsveld, J.
(2009) Phosphorylation dynamics during early differentiation of human
embryonic stem cells, Cell Stem Cell, 5, 214-226.
172.Tsanov, K. M., Pearson, D. S., Wu, Z., Han, A.,
Triboulet, R., Seligson, M. T., Powers, J. T., Osborne, J. K., Kane,
S., Gygi, S. P., Gregory, R. I., and Daley, G. Q. (2017) LIN28
phosphorylation by MAPK/ERK couples signaling to the
post-transcriptional control of pluripotency, Nat. Cell Biol.,
19, 60-67.
173.Zeng, Y., Yao, B., Shin, J., Lin, L., Kim, N.,
Song, Q., Liu, S., Su, Y., Guo, J. U., Huang, L., Wan, J., Wu, H.,
Qian, J., Cheng, X., Zhu, H., Ming, G.-L., Jin, P., and Song, H. (2016)
Lin28A binds active promoters and recruits Tet1 to regulate gene
expression, Mol. Cell, 61, 153-160.
174.Karlson, D., and Imai, R. (2003) Conservation
of the cold shock domain protein family in plants, Plant
Physiol., 131, 12-15.
175.Sasaki, K., and Imai, R. (2012) Pleiotropic
roles of cold shock domain proteins in plants, Front. Plant
Sci., 2, 116.
176.Karlson, D. (2009) Plant cold-shock domain
proteins: on the tip of an iceberg, in Plant Cold Hardiness: From
the Laboratory to the Field, CABI, Cambridge, pp. 43-54.
177.Taranov, V. V., Zlobin, N. E., Evlakov, K. I.,
Shamustakimova, A. O., and Babakov, A. V. (2018) Contribution of
Eutrema salsugineum cold shock domain structure to the
interaction with RNA, Biochemistry (Moscow), 83,
1369-1379.
178.Fusaro, A. F., Bocca, S. N., Ramos, R. L. B.,
Barroco, R. M., Magioli, C., Jorge, V. C., Coutinho, T. C.,
Rangel-Lima, C. M., De Rycke, R., Inze, D., Engler, G., and
Sachetto-Martins, G. (2007) AtGRP2, a cold-induced nucleo-cytoplasmic
RNA-binding protein, has a role in flower and seed development,
Planta, 225, 1339-1351.
179.Yang, Y., and Karlson, D. (2013) AtCSP1
regulates germination timing promoted by low temperature, FEBS
Lett., 587, 2186-2192.
180.Kim, M. H., Sasaki, K., and Imai, R. (2009)
Cold shock domain protein 3 regulates freezing tolerance in
Arabidopsis thaliana, J. Biol. Chem., 284,
23454-23460.
181.Taranov, V. V., Berdnikova, M. V., Babakov, A.
V., Nosov, A. V., and Galkin, A. V. (2010) Cold shock domain proteins
in the extremophyte Thellungiella salsuginea (salt cress): gene
structure and differential response to cold, Mol. Biol.
(Moscow), 44, 787-794.
182.Radkova, M., Vitamvas, P., Sasaki, K., and
Imai, R. (2014) Development- and cold-regulated accumulation of cold
shock domain proteins in wheat, Plant Physiol. Bioch.,
77, 44-48.
183.Chaikam, V., and Karlson, D. (2008) Functional
characterization of two cold shock domain proteins from Oryza
sativa, Plant Cell Environ., 31, 995-1006.
184.Nakaminami, K., Hill, K., Perry, S. E.,
Sentoku, N., Long, J. A., and Karlson, D. T. (2009) Arabidopsis
cold shock domain proteins: relationships to floral and silique
development, J. Exp. Bot., 60, 1047-1062.
185.Yang, Y., and Karlson, D. T. (2011)
Overexpression of AtCSP4 affects late stages of embryo development in
Arabidopsis, J. Exp. Bot., 62, 2079-2091.
186.Nakaminami, K., Karlson, D. T., and Imai, R.
(2006) Functional conservation of cold shock domains in bacteria and
higher plants, PNAS, 103, 10122-10127.
187.Park, S. J., Kwak, K. J., Oh, T. R., Kim, Y.
O., and Kang, H. (2009) Cold shock domain proteins affect seed
germination and growth of Arabidopsis thaliana under abiotic
stress conditions, Plant Cell Physiol., 50, 869-878.
188.Sasaki, K., Kim, M. H., and Imai, R. (2007)
Arabidopsis COLD SHOCK DOMAIN PROTEIN2 is an RNA chaperone that is
regulated by cold and developmental signals, Biochem. Biophys. Res.
Commun., 364, 633-638.
189.Zlobin, N., Evlakov, K., Alekseev, Y.,
Blagodatskikh, K., Babakov, A., and Taranov, V. (2016) High DNA melting
activity of extremophyte Eutrema salsugineum cold shock domain
proteins EsCSDP1 and EsCSDP3, Biochem. Biophys. Rep., 5,
502-508.
190.Zlobin, N., Evlakov, K., Tikhonova, O.,
Babakov, A., and Taranov, V. (2018) RNA melting and RNA chaperone
activities of plant cold shock domain proteins are not correlated,
RNA Biol., 15, 1040-1046.
191.Zlobin, N. E. (2019) The Interaction of
Proteins with the Cold Shock Domain of the Extremophyte Plant Eutrema
salsugineum with Nucleic Acids: PhD (in biology) dissertation [in
Russian], All-Russian Research Institute of Agricultural Biotechnology,
Moscow.
192.Juntawong, P., Sorenson, R., and
Bailey‐Serres, J. (2013) Cold shock protein 1 chaperones mRNAs
during translation in Arabidopsis thaliana, Plant J.,
74, 1016-1028.
193.Kawaguchi, R., and Bailey-Serres, J. (2005)
mRNA sequence features that contribute to translational regulation in
Arabidopsis, Nucleic Acids Res., 33, 955-965.
194.Puckette, M., Iyer, N. J., Tang, Y., Dai, X.
B., Zhao, P., and Mahalingam, R. (2012) Differential mRNA translation
in Medicago truncatula accessions with contrasting responses to
ozone-induced oxidative stress, Mol. Plant, 5,
187-204.
195.Li, C., Sako, Y., Imai, A., Nishiyama, T.,
Thompson, K., Kubo, M., Hiwatashi, Y., Kabeya, Y., Karlson, D., Wu, S.,
Ishikawa, M., Murata, T., Benfey, P., Sato, Y., Tamada, Y., and Hasebe,
M. (2017) A Lin28 homologue reprograms differentiated cells to stem
cells in the moss Physcomitrella patens, Nat. Commun.,
8, 14242.