REVIEW: Enzymes of Polyphosphate Metabolism in Yeast: Properties, Functions, Practical Significance
Tatiana V. Kulakovskaya1,a*, Nadezhda A. Andreeva1, Larisa A. Ledova1, Lubov P. Ryazanova1, Ludmila V. Trilisenko1, and Michail A. Eldarov2#
1Skryabin Institute of Biochemistry and Physiology of Microorganisms, Pushchino Research Center for Biology, Russian Academy of Sciences, 142290 Pushchino, Moscow Region, Russia2Institute of Bioengineering, Federal Scientific Center for Biotechnology, Russian Academy of Sciences, 117312 Moscow, Russia
* To whom correspondence should be addressed.
# Deceased.
Received April 19, 2020; Revised May 19, 2020; Accepted May 27, 2020
Inorganic polyphosphates (polyP) are the linear polymers of orthophosphoric acid varying in the number of phosphate residues linked by the energy-rich phosphoanhydride bonds. PolyP is an essential component in living cells. Knowledge of polyP metabolizing enzymes in eukaryotes is necessary for understanding molecular mechanisms of polyP metabolism in humans and development of new approaches for treating bone and cardiovascular diseases associated with impaired mineral phosphorus metabolism. Yeast cells represent a rational experimental model for this research due to availability of the methods for studying phosphorus metabolism and construction of knockout mutants and strains overexpressing target proteins. Multicomponent system of polyP metabolism in Saccharomyces cerevisiae cells is presented in this review discussing properties, functioning, and practical significance of the enzymes involved in the synthesis and degradation of this important metabolite.
KEY WORDS: polyphosphate, polyphosphatase, VTC, overexpression, enzyme properties, stress, Saccharomyces cerevisiaeDOI: 10.1134/S0006297921140078
Abbreviations: polyP, inorganic polyphosphates; VTC, vacuolar transporter chaperone complex.
Devoted to I. S. Kulaev, the pioneer of polyphosphate biochemistry, on
his 90th anniversary.
INTRODUCTION
At first glance, inorganic polyphosphates (polyP), linear polymers of orthophosphoric acid, occupy a rather modest and standalone place among the biologically significant polymers. It was not until fairly recently that they were considered only as microbial reserves of phosphorus, “molecular mineral resources” present in the cells of some species of microorganisms. Academician A. N. Belozersky, one of the founders of molecular biology in Russia, became interested in these energy-rich molecules as early as in the middle of the past century. He believed that in the early stages of life on the Earth polyP were formed abiogenically and preceded ATP, the major energy carrier in living organisms. Igor Stepanovich Kulaev was a disciple of A. N. Belozersky, and their pioneer works in the field of polyP biochemistry proved involvement of these phosphorus compounds in the central metabolism of microorganisms [1-3].
Later on, I. S. Kulaev, corresponding member of the Russian Academy of Sciences, became one of the leading scientists in the field of polyphosphate biochemistry, and his monograph [4, 5] was a basic manual on the properties and functions of these polymers. His idea that high-molecular weight polyphosphates are regulators of the metabolic processes is now commonly accepted. There is considerable evidence for the fact that they play a regulatory role in the control of gene expression, stress adaptation, membrane transport, and maintenance of cell motility [6, 7]. The ability of polyP to participate in quite different cellular processes is determined by their physicochemical properties.
These macroergic negatively charged polymers are able to bind either directly or indirectly to proteins, polyhydroxybutyrate, and polysaccharides in the presence of Ca2+, Mg2+, and K+ cations, thus changing their biological activity.
At present, the concept on involvement of polyP and enzymes of polyphosphate metabolism in various processes regulating vital activities of eukaryotes has been established. The data about the key role of polyP in bone tissue growth and development are quite convincing [8, 9]. It was established that calcium and polyP accumulate in osteoclasts as specific granules, which are released via exocytosis into the extracellular space in the places of bone growth or repair. Here the granules are destroyed and alkaline phosphatase hydrolyzes polyphosphates to orthophosphate. The structured bone apatite is formed from the released orthophosphate and calcium with involvement of the specific proteins. The literature addressing this problem includes more than two hundred references over the past ten years. The polyphosphate-containing granules have been found in platelets [10]. The polyP released into blood during platelet destruction participate in the coagulation cascade, binding to factor XII and activating it; then these polymers and calcium ions become part of a thrombus, increasing its stability [11, 12]. The polyPs of blood are participants of the inflammatory response [12]. Involvement of polyP in the signal transduction by nerve cells has been also shown [13, 14]. They transmit signals between astrocytes by activating purinergic receptors. PolyPs are involved in formation of the fibrils of amyloidogenic proteins (in the non-cytotoxic form) and prevent formation of amyloids disrupting the cells [15]. PolyP is a component of the specific calcium channel in mitochondrial membranes that regulates calcium level and stress response in these organelles [16, 17].
Thus, the knowledge on polyP metabolism in eukaryotes is necessary to understand the nature of disorders of human phosphorus metabolism and to develop new methods for treating cardiovascular and bone diseases, and disorders of the nervous system. New materials for treating bone diseases and injuries already being developed that include nanoparticles and bone implants containing polyP to stimulate apatite formation [18-20], as well as inhibitors of thrombus formation – polyP antagonists [21].
The Saccharomyces cerevisiae yeast cells, classical model of eukaryotic organisms, are similar to mammalian cells in terms of phosphorus metabolism. Under the conditions of glucose-induced catabolite repression in yeasts, there is biomineralization of mitochondria due to accumulation of inorganic polyphosphates [22]. It seems that the analogous process underlies formation of the phosphorus-calcium granules containing polyphosphates and calcium in platelets and osteoblasts. The polyphosphatase PPX1 protein in yeast was shown to be orthologous to the protein encoded by the human prune gene [23].
The S. cerevisiae cells have a multicomponent system of polyP metabolism, including the vacuolar transporter chaperone (VTC) complex consisting of five proteins responsible for polyP biosynthesis, as well as polyphosphatases Ppx1, Ppn1, Ddp1, and Ppn2 involved in polyP degradation. The aim of this review is to present comparative analysis of the data on physicochemical properties and functions of these enzymes.
VTC COMPLEX – POLYPHOSPHATE SYNTHASE OF YEAST
Polyphosphate kinases are responsible for polyP synthesis in the cells of most bacteria. These enzymes catalyze transfer of a phosphate residue from ATP to the growing polyP chain and the reverse reaction [6]. The polyphosphate kinase-encoding genes were found only in a few eukaryotic species, and their presence is explained by horizontal transfer from bacteria [24-26]. Transfer of the terminal phosphate residue from ATP to polyP in the yeast vacuolar membrane was discovered years ago [27]; however, the enzyme catalyzing this reaction could not be identified for a long time. Then it was observed that the VTC4 knockout mutants of S. cerevisiae [28] and Ustilago maydis [29] contained very little polyP in comparison with the parent strains. Vtc4 protein is part of the yeast VTC complex, which also includes Vtc1, Vtc2, and Vtc3 proteins [30, 31]. This complex is localized in the vacuolar membrane; it performs chaperone function with respect to V-ATPase of this membrane and participates in the fusion of the vacuolar membrane with other membrane structures [30, 31].
Evidence for the presence of the polyphosphate synthase activity in Vtc4 and its detailed characterization are given in the work by Hothorn et al. [32]. X-ray crystallography has shown that fragment of this protein, Vtc4p189-480, contains a long electron-dense domain responsible for the synthesis of polyP from ATP [32]. This fragment is able to catalyze polyP synthesis from ATP in solution in the presence of Mn2+. The enzyme can also use other nucleoside triphosphates and dATP and is moderately specific to divalent cations: Mn2+ > Zn2+ > Co2+ > Mg2+ > Fe2+ > Ni2+. The primers for polyP synthase reaction can be both orthophosphate and pyrophosphate (the latter is more effective). The catalytic domain faces cell cytoplasm, which suggests transport of polyP across the vacuolar membrane. Point mutations in the Vtc1-encoding gene reduce the level of polyP in the cell [32]. It has been suggested that this protein participates in the polyP transport across the vacuolar membrane [32].
There are two forms of VTC complex present in the yeast cells: Vtc4/Vtc3/Vtc1 and Vtc4/Vtc2/Vtc1 [33]. The former is typical of the vacuolar membrane and the latter is typical of the endoplasmic reticulum membrane and nuclear envelope but is found in the vacuolar membrane under conditions of phosphate deficit [33]. It is considered that Vtc2 and Vtc3, which do not exhibit any activity in vitro, perform a regulatory function. These proteins, together with Vtc4, contain the SPX domains bound to inositol phosphates, the signaling molecules whose concentration varies depending on the phosphate availability [34]. Amino acid substitutions in this domain, which impair binding to inositol phosphates in vitro, lead to the decrease in polyP synthesis in yeast [34]. The activator of polyP synthesis is 5-PP-InsP5 [35]. The search of SPX domains in other proteins resulted in identification of one more component of the VTC complex: the Vtc5 subunit [36]. This protein physically interacts with VTC, and deletion in the respective gene leads to the reduction of the level of polyP synthesis, while its overexpression leads to its increase. Vtc5, like other protein components of VTC, is a transmembrane protein, with its C-terminus exposed to the vacuolar lumen and N-terminus exposed to the cytoplasm [36]. Let us note that the VTC4 mutants are characterized by low but reliably detected polyP level [28]. This fact implies presence of another enzyme systems capable of polyP synthesis. Among the potential candidates dolichyl diphosphate–polyphosphate phosphotransferase (EC 2.7.4.20) detected in the membrane fraction of S. cerevisiae cells [37] and 1,3-diphosphoglycerate polyphosphate phosphotransferase (EC 2.7.4.17) detected in the cell-free extract of Neurospora crassa should be mentioned [38]. Genes encoding the proteins responsible for these enzyme reactions have not been identified yet.
Thus, the yeast VTC complex plays the key role in polyP synthesis, has multiple-localizations in the cell, and is regulated by inositol phosphate 5-PP-InsP5.
YEAST POLYPHOSPHATASES
Structural peculiarities of yeast polyphosphatases. PolyP hydrolysis in yeast is catalyzed by the enzymes possessing exopolyphosphatase and/or endopolyphosphatase activity. Exopolyphosphatase (polyphosphate phosphohydrolase, EC 3.6.1.11) cleaves orthophosphate from the polyP chain end:
PolyPn + H2O→PolyPn-1 + Pi
Endopolyphosphatase (polyphosphate depolymerase, EC 3.6.1.10.) cleaves long polyP chains into shorter ones:
PolyPn + H2O→polyPm (m < n)
In S. cerevisiae, the genes of four polyphosphatases have been identified; the enzymes have been purified and characterized in detail [39-45]. PolyP degradation also seems to involve alkaline phosphatase Pho8 localized in vacuoles, because the PHO8 gene knockout mutant contains more polyP than the parent strain [46].
Yeast polyphosphatases belong to the different protein families (Table 1). The PPX1 gene has no introns; posttranslational modifications of this protein have not been reported. Polyphosphatase Ppx1 belongs to the DHH phosphoesterase family, which also includes inorganic pyrophosphatases of the family 2 in Gram-positive bacteria, the prune protein in insects and mammals, and the single-stranded DNA-specific exonuclease RecJ [47]. The X-ray crystallography data of Ppx1 demonstrated high structural similarity of the active site to that of other proteins belonging to the family 2 of inorganic pyrophosphatases [47]. A large long channel is present in the structure of this molecule, which is formed by the positively charged amino acids comprising the polyP-binding site and the site of hydrolysis. This structural peculiarity is believed to be responsible for the processive nature of hydrolysis and for the absence of activity with pyrophosphate [47]. The close orthologs of Ppx1 are the prune proteins of higher eukaryotes that demonstrate 25-30% structural similarity to Ppx1 of S. cerevisiae [23]. The human prune (h-prune) hydrolyzes short-chained polyP, though the best substrates are tripolyphosphate, tetrapolyphosphate, and nucleoside 5′ tetraphosphates [23]. The product of the PPN1 gene needs specific proteolytic cleavage for maturation and activation [41]. The Ppn1 protein contains several tentative glycosylation and ubiquitination sites with N-glycosylation required for proteolysis [48]. When purified, Ppn1 often forms large aggregates with a molecular mass of more than 800 kDa, and polyP is needed for homotetramer stabilization [49].
Table 1. Polyphosphatases of
Saccharomyces cerevisiae
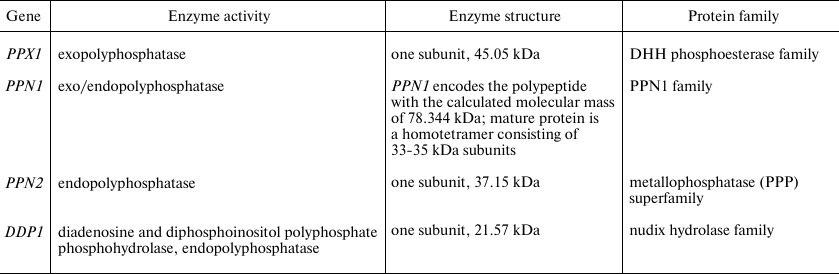
Note. The data are from the SDG database
(https://www.yeastgenome.org/)
The sphingomyelin- and calcineurin-like phosphoesterases are the Ppn1 orthologs in higher eukaryotes; however, the degree of similarity is no more than 15-20% [50].
In humans, there are three enzymes hydrolyzing inositol pyrophosphate and diadenosine hexaphosphate, DIPP1, DIPP2, and DIPP3a/b, with sequences similar to the yeast enzyme Ddp1 [43]. Like Ddp1, they can cleave long-chained polyP into shorter fragments though with the lower activity compared to the yeast enzyme [43]. The closest ortholog of Ppn2 is the diadenosine tetraphosphate hydrolase of Shigella flexnery 2a [44]. No prokaryotic orthologs have been identified for other polyphosphatases. Significant structural differences between the yeast polyphosphatases suggest their different evolutionary origins.
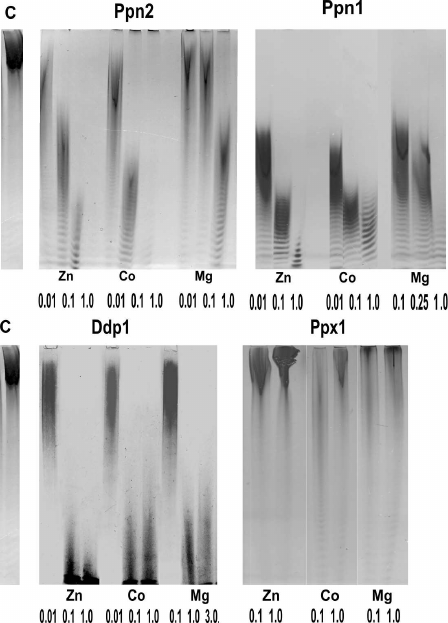
Substrate specificity, kinetics, inhibitors, and activators. The four known polyphosphatases of S. cerevisiae have different mechanisms of polyP hydrolysis: Ppx1 is an exopolyphosphatase, Ppn1 exhibits both exo- and endopolyphosphatase activities depending on divalent cations, Ppn2 and Ddp1 are mainly endopolyphosphatases. Figure 1 shows electrophoregrams of the polyP treated with the purified preparations of these polyphosphatases [45]. The treatment with Ppx1 did not lead to the change in the polyP chain length, which indicated absence of endopolyphosphatase activity and was in agreement with the processive mechanism of the substrate hydrolysis. The polyP chain length decreased after the treatment with other three enzymes, which indicated the presence of endopolyphosphatase activity. Exopolyphosphatase activities of the purified enzyme preparations are vary significantly depending on the method of purification and analytical conditions. The maximum exopolyphosphatase activities with polyP with the average chain length of about 200 phosphate residues were 300 U/mg protein for Ppx1 [51] and 900 U/mg protein for Ppn1 [52]. The detected exopolyphosphatase activities of Ddp1 and Ppn2 were lower by three orders of magnitude (0.05 and 0.1 U/mg protein, respectively) [45] and probably had no significant effect on the polyP metabolism. The pH optima for the yeast polyphosphatases are close to 7.0 [39-45].
Fig. 1. Endopolyphosphatase activities of polyphosphatases of S. cerevisiae with polyP188 depending on the concentration of divalent cations (the numbers indicate cation concentrations in mM). Electrophoregrams of polyP188 after treatment with the preparations of purified polyphosphatases from superproducing strains are presented [45]; C – control, polyP188 was incubated without the addition of enzyme preparations.
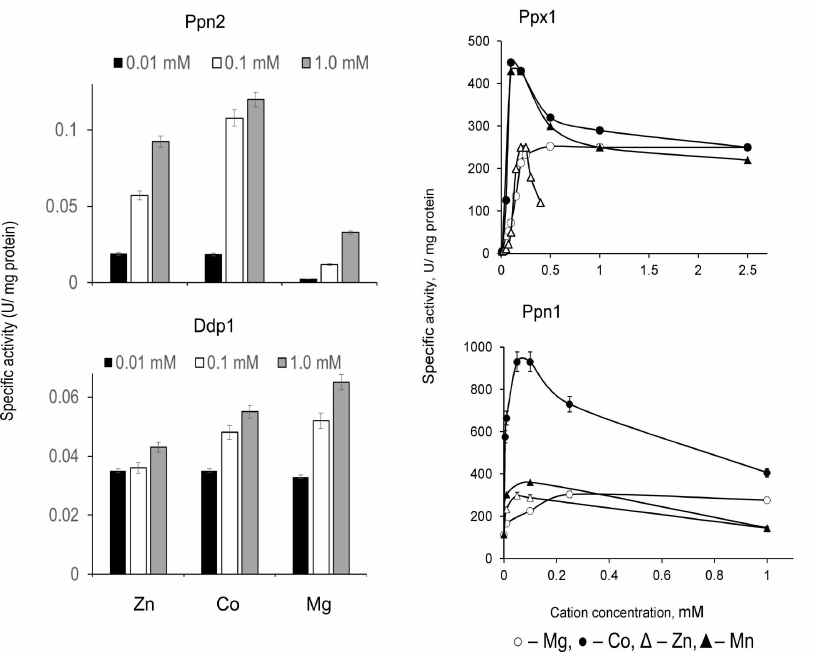
Presence of divalent metal cations is required for activity manifestation of all four polyphosphatases, but these enzymes are inactive in the presence of Ca2+ [39-45]. The dependencies of endo- and exopolyphosphatase activities on the type and concentration of metal ions are specific for each of these enzymes (Figs. 1 and 2).
Monovalent cations stimulate exopolyphosphatase activities of Ppx1 and Ppn1 with NH4+ ions being more effective than K+ and Na+ [39, 42]; NH4+ also stimulates endopolyphosphatase activities of Ppn1 and Ppn2 [45].
Fig. 2. Exopolyphosphatase activities of polyphosphatases from S. cerevisiae with polyP188 depending on concentration of divalent cations [45, 50].
Endopolyphosphatase activities of Ddp1 [43], Ppn1, and Ppn2 [52, 53] were examined using polyP with different degrees of polymerization and average chain length from 15 to 208 phosphate residues. Since the method for activity determination is based on monitoring the decrease in polyP chain length using electrophoresis, it is difficult to carry out quantitative assessment of the level of activity depending on the polyP chain length. With regards to the exopolyphosphatase activity, Ppn1 exhibits higher activitiy with the long-chain polyP and Ppx1 – with tripolyphosphate, respectively (Table 2). Yeast polyphosphatases also hydrolyze some organic compounds with phosphodiester bond, including second messengers. It is worth reminding that Ddp1 hydrolyzes diadenosine polyphosphates and inositol pyrophosphates, which are most likely the principal substrates of this enzyme in vivo [43]. Ppx1 cleaves terminal phosphate from adenosine 5′ tetraphosphate and guanosine 5′ tetraphosphate [54, 55] but cannot hydrolyze inositol pyrophosphate [43]. Ppx1 and Ppn1 demonstrate activities with different nucleoside phosphates. The ratios of these activities under optimal conditions for each enzyme are presented in Table 2. Ppx1 demonstrates only minor phosphodiesterase activity with cAMP [45] similar to the one exhibited by the orthologous protein h-prune [23], while Ppn1 has no such activity. Ppx1 and Ppn1 are inactive with p-nitrophenylphosphate, glucose-6-phosphate and glycerol-3-phosphate; the activity with pyrophosphates either is not exhibited or, under certain conditions (in the presence of Co2+), is no more than 7-10% of the activity with polyP188 [39, 42, 45].
Table 2. Specific activities of
exopolyphosphatases (% of the activity with polyP208) and
apparent Km in the hydrolysis of different substrates by
Ppx1 and Ppn1 enzymes of S. cerevisiae
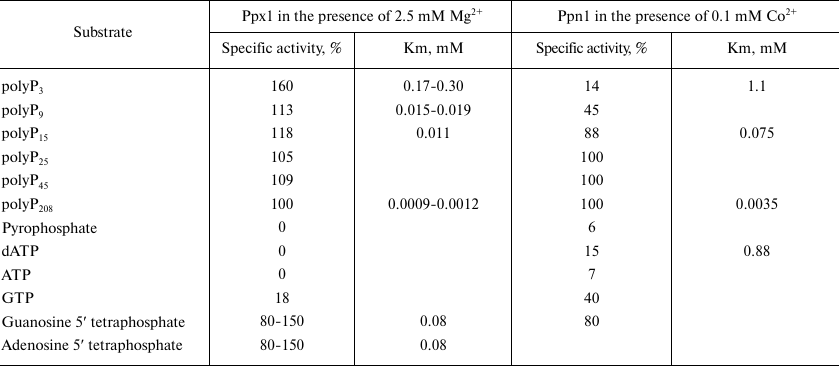
Note. The data are summarized from the works [39,
42, 45, 49, 54, 55].
The new type of posttranslational modification of proteins leading to modification of their properties and activity has been discovered recently that involves covalent attachment of polyP to lysine residues, the so-called lysine-polyphosphorylation [56, 57]. It has been shown that all four polyphosphatases are able to hydrolyze these polyP [56], and Ppn1 is the most active one [57]. Contributions of the individual polyphosphatases to the hydrolysis of polyP in the lysine-polyphosphorylated proteins in a cell depend on localization of these enzymes and target proteins [57].
The kinetics of polyP hydrolysis by Ppx1 is described by the Michaelis–Menten equation only under special conditions, namely, constant concentration of magnesium ions [39, 51], while the kinetics of polyP hydrolysis by Ppn1 is not described by this equation [42]. The values of apparent Km presented in Table 2, which are used for comparative analysis of the substrate affinity to these polyphosphatases, correspond to the substrate concentrations providing the reaction rate equal to half-maximum rate of the reaction [39, 32, 45, 49]. The substrate affinity is higher for the long-chain polyP for both enzymes. The kinetic parameters of Ppx1 are considered in more detail in the works [58, 59] suggesting the presence of an additional magnesium ion binding site in the enzyme molecule. The presence of such center seems to account for the extraordinary stimulating effect of EDTA on the activity of Ppx1: in the presence of 1 mM of this chelating agent and 2.5 mM of magnesium ions, the activity increases 1.5-fold [39, 51]. No such effect has been observed for Ppn1 [49].
Heparin is the commonly known inhibitor of polyphosphatases [49, 51]. Ppn1 is most sensitive to this competitive inhibitor, while Ppn2 and Ddp1 are the least sensitive. The endopolyphosphatase activity of Ppn1 was completely inhibited by heparin at concentration of 0.01 mg/ml, while the endopolyphosphatase activity of Ppn2 was not affected even by the heparin concentration of 0.25 mg/ml [45]. As to exopolyphosphatase activities, I50 for heparin is 0.001 mg/ml and 0.05 mg/ml for Ppn1 and Ppx1, respectively [45]. The exopolyphosphatase activities of Ppn2 and Ddp1 were inhibited by no more than 13-20% at the heparin concentration of 0.25 mg/ml [45]. In contrast to other polyphosphatases, Ddp1 is inhibited by 1 µM of fluoride [43].
ADP was shown to have an activating effect on the endopolyphosphatase activities of Ppn1 [52] and Ppn2 [53]. Arginine (100 mM) activated the endopolypohsphatase activity of Ppn1 [53].
Thus, total set of the properties such as mechanism of polyP hydrolysis, ability to hydrolyze low-molecular substrates of different structure, stimulating effect of divalent cations and sensitivity to heparin suggests that the four polyphosphatases of S. cerevisiae cells are considerably different indicating specific set of cellular functions for each of them.
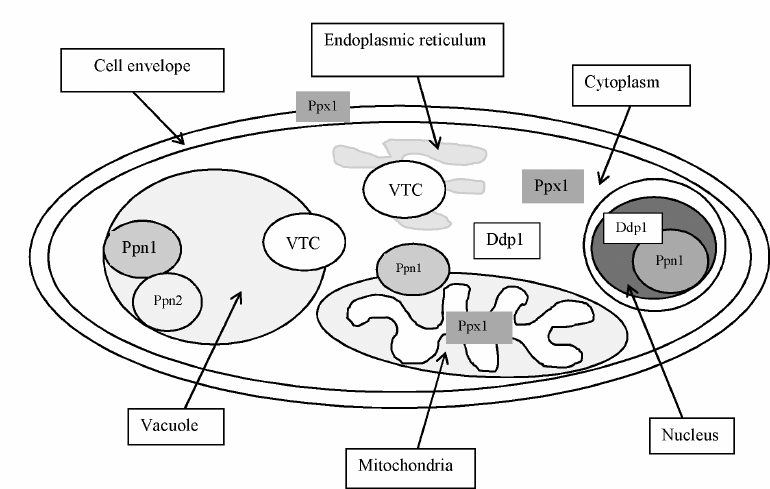
Localization of polyphosphatases. Multiple localization of polyP in yeast cells was presumed already in the first monograph by I. S. Kulaev [4, 5]. This assumption was confirmed by the analysis of polyP content and chain length in the purified fractions of vacuoles, nuclei, mitochondria, and cytoplasm demonstrating that these subcellular compartments contained their own pools of polyP different from the polyP of other compartments in the chain length as well as the effects of cultivation conditions and of knockout mutations in the PPX1 and PPN1 genes [60]. It is obvious that the polyP-containing organelles and compartments must be equipped with the enzymes for their metabolism. In S. cerevisiae, the enzymes with exopolyphosphatase activities from the cell envelope [39], the cytoplasm [49], vacuoles [61], mitochondria [62, 63] and nuclei [64] were purified and characterized, while the enzymes with endopolyphosphatase activities were purified from the cytoplasm [43, 65] and vacuoles [44]. Analysis of the effects of knockout mutations in the PPX1, PPN1 [60], and PPN2 genes [44] on polyphosphatase activities in subcellular fractions allowed identification of the genes responsible for the polyphosphatase activities in organelles and compartments. For example, the exopolyphosphatase activities in the nuclear, vacuolar, and mitochondrial membrane fractions of the Δppx1 mutant did not change compared to the parent strain, the cell envelope extract did not contain polyphosphatase, while the cytoplasm and the mitochondrial matrix contained high-molecular weight enzyme aggregates instead of the 45-kDa enzyme, similar in properties to the PPN1 gene product [60, 66-69]. In the Δppn1 mutant, the polyphosphatase activity was absent in the mitochondrial membrane and dramatically decreased in the vacuoles and nuclei [60]. These data, together with the comparative analysis of the properties of polyphosphatases purified from the separate subcellular fractions, demonstrate that Ppx1 is localized in the cytoplasm, cell envelope, and mitochondrial matrix, and Ppn1 is localized in the vacuoles, nuclei, and mitochondrial membranes. Ddp1 is localized mainly in the cytoplasm [43] and, in lower amounts, in the nuclei [56]. Ppn2 and Ppn1 localized in the vacuoles are able to form a complex [44].
The polyphosphatase localization changes not only as a result of the Δppx1 knockout mutation but also under some specific cultivation conditions. Transfer of the phosphate-starved S. cerevisiae cells to a complete medium leads to the decrease in the activity of Ppx1 in the cytoplasm and mitochondrial matrix and activity of Ppn1 appears [68, 69]. The causes of such interrelationship between Ppx1 and Ppn1 have not been elucidated; this phenomenon probably involves signaling molecules that are hydrolyzed by both enzymes, e.g., nucleoside tetraphosphates.
Figure 3 shows the scheme of localization of the known enzymes of polyP metabolism in the wild type S. cerevisiae cells growing on a phosphate-rich medium.
Fig. 3. Localization of the enzymes of polyphosphate metabolism in yeast cells.
Effects of knockout mutations in polyphosphatase genes on polyP metabolism and other cellular functions. The first work devoted to identification and functions of polyphosphatase Ppn1 showed that the PPN1 gene mutations leading to the absence of this polyphosphatase in S. cerevisiae cells caused an increase in the level of polyP level and its chain length [41]. The more detailed analysis of the content of different polyP fractions in the Δppn1 mutant showed a twofold increase in the content of the shortest chain fraction, polyP1, in the cells at stationary growth phase, while the content of other fractions varied only slightly [70]. The analysis of polyP content and chain length in the isolated subcellular fractions showed that the polyP level increased in the mitochondria and vacuoles of the Δppn1 mutant and the polyP chain length increased in the mitochondria, vacuoles and cytoplasm of this mutant strain [60].
It was also reported that the polyP levels in the Δppn1, Δppn2, and Δppn1Δppn2 mutants did not vary compared to the parent strain in cells at the logarithmic and stationary growth phases [44]. These mutations led to the increase in the polyP chain length, and this effect was more pronounced in the stationary phase for the Δppn1 mutant and in the logarithmic phase for the Δppn2 mutant [44]. The increased polyP chain length in the Δppn1 mutants was shown by a number of researchers who used different genetic constructs to obtain these strains [43, 44, 57].
The Δppx1 knockout mutant demonstrated an insignificant (~15%) increase in the content of the shortest chain fraction, polyP1, while the content of other polyP fractions was unchanged [70]. The changes in the polyP chain length were not observed in the isolated subcellular fractions of this mutant [60]. The polyP content in the cells of the Δddp1 knockout mutant even decreased (by ~20%) compared to the parent strain, though the content of inositol pyrophosphate increased more than 2-fold and the polyP chain length did not change [43].
Analysis of the effects of different combinations of knockout mutations in the genes of four yeast polyphosphatases on the chain length of polyP extracted from the cells by phenol and lithium salts demonstrated that the knockout mutation in the PPN1 gene provided the largest contribution to the increase in the polyP chain length; even more noticeable increase in the polyP chain length was observed in the double mutant Δppn1Δppn2, and the mutations Δppx1 and Δddp1, both in combination and separately, had no effect on the polyP chain length [57].
The study of peculiarities of polyP content and chain length in the cells with knock-out polyphosphatase genes allows concluding that polyphosphatases Ppn1 and Ppn2 are key contributors to the regulation of polyP content and chain length in the S. cerevisiae cells.
Now let us consider what other cellular functions are changed in the case of knockout mutations in the polyphosphatase genes. Note that the Δppn1Δppn2Δppx1Δddp1 mutant is viable [57] and it is not a simple problem to find out the conditions when mutants in the polyphosphatase genes exhibit growth defects. Such conditions were found for the mutants not expressing the Ppn1 polyphosphatase. The reduced survival of the Δppn1 mutant was observed in the stationary growth phase [41]. This reduction was due to the fact that the Δppn1 mutant strain was unable to utilize ethanol and other oxidizable substrates [67]. In the mitochondria of the parent strain, the content of acid-soluble polyP decreased from ~1 µmole P per 1 mg protein in the logarithmic growth phase to 0.3 µmole P per 1 mg protein in the stationary growth phase. In the Δppn1 mutants, the polyP level increased from 0.3 µmole per 1 mg protein in the logarithmic growth phase to 0.72 µmole P per 1 mg protein in the stationary growth phase. The polyP chain length decreased in the mitochondria of strains with preserved function of the PPN1 gene upon transition to the stationary growth phase but increased in the strains with mutations in this gene. During the growth phase when the functionally active mitochondria should be formed, the promitochondria of the Δppn1 mutant contained polyP with an average chain length of 80-130 phosphate residues but not the short-chain polyP (~15 phosphate residues) typical of the control strains [67]. Thus, the Ppn1 polyphosphatase (the membrane fraction of mitochondria being one of the sites of its localization) plays a key role in the development of mitochondria during transition from the state of catabolite repression to the growth on oxidizable substrates.
The polyP level in yeast cells regularly oscillates during the cell cycle. On entering the S-phase, the polyP content decreases and is restored to its initial level at the stage of mitosis. At the same time, in spite of the changing polyP level, concentration of the cytoplasmic phosphate remains constant (20 mM) [71]. What is the role of cyclic variations in the polyP level and what systems are responsible for the phosphate homeostasis during the cell cycle? The use of mutant stains with mutations in the genes involved in polyP biosynthesis and degradation proved to be very useful for answering these questions. The normal cell cycle progression was disturbed in the strains that had defects in polyP synthesis (Δvtc4) and mobilization (Δppn1Δppx1) [71]. The time of DNA replication increased 2-fold in both mutants cultivated on a phosphate-deficient medium [71]. This effect, as well as the higher genome instability observed in the mutants, is obviously related to the decreased level of dNTP [71].
The findings allowed the authors to validate the key role of polyP as a source of phosphate for the synthesis of dNTP under conditions of phosphate limitation [71]. Due to the multiple functions of polyP, one can expect existence of other effects of the polyphosphatase gene knockout mutants on the ability of cells to overcome unfavorable environmental conditions.
Overexpression of polyphosphatase genes. Examination of the physiological features of knockout mutants is a classical approach to determine functions of the specific proteins. In the case of yeast polyphosphatases, this approach happens to be insufficient due to pleiotropic action of such mutations and sometimes absence of the significant changes in the polyP level. Another approach is to construct the strains overexpressing specific polyphosphatases. It should be noted that cloning of the yeast polyphosphatase genes in bacterial cells is not always appropriate; in particular, presence of the specific proteinase is required to obtain the mature active form of Ppn1 [41]. Construction of the S. cerevisiae strains overproducing polyphosphatases solves two problems: facilitating production of pure polyphosphatase preparations from the cell-free extracts with significantly higher activity than in the case of the wild strain, and possibility to study effects of such overexpression on the polyP metabolism and other cell functions. It should be noted that the wild-type yeast strains proved to be inefficient for such construction, because the resultant strains were characterized by reduced viability, at least in case of the PPN1 gene. The PPN1 mutant was found to be a more suitable recipient; the resultant strains overexpressing all four polyphosphatases separately showed no difference in viability from the initial strain. The polyphosphatase genes were expressed in the yeast cells under control of a strong constitutive promoter of the S. cerevisiae glyceraldehyde-3-phosphate dehydrogenase gene (TDH3) in composition of the specialized vector PMB1 [53, 72-74]. It is a low-copy, segregationally stable vector, but it provides substantial increase in the level of production of the studied polyphosphatases in the transformants compared to the wild type strains. Viability of such strains was actually the same as in wild strains, allowing us to carry out comparative analysis of physiology of all strains under the same conditions and to obtain all four polyphosphatases with a high degree of purity from the cell-free extract using standard chromatographic procedures [45]. The transformed strains showed a significant increase in polyphosphatase activities in the cell-free extract; in the case of exopolyphosphatases, this increase was assessed quantitatively: specific activity increased 10- to 20-fold depending on the cultivation conditions [72, 73]. Table 3 shows how overexpression of polyphosphatases influenced cellular level of the relatively low-molecular weight (with an average chain length of about 15 phosphate residues) acid-soluble polyP and high molecular weight (25 to 200 phosphate residues) acid-insoluble polyP. In the strains overexpressing Ppx1 and Ddp1, there are only slight changes in the polyP level, which is in agreement with the fact that mutations in the respective genes had no substantial effect on the content of polyP. The maximum changes were observed in the overproducers of Ppn1 and Ppn2. These changes are different: the Ppn1 overexpression most and foremost leads to the decrease in the content of short-chain polyP, while the Ppn2 overexpression leads to its increase, and only the level of long-chain polyP decreases. It should be noted that in both cases the overexpressed enzymes appear in the cytoplasm in considerable amounts in the soluble form [53, 72-74]; in the wild type strains, they are localized mainly in vacuoles or other organelles in the case of Ppn1. As a result, the low molecular weight polyP of the cytoplasm can be exposed to Ppn1 as an exopolyphosphatase and to Ppn2 as an endopolyphosphatase. The increased levels of Ppx1 and Ddp1 in the cytoplasm did not produce any significant effect on these low molecular weight polyPs [73, 74]. It should be taken into account that these two enzymes are localized mostly in the cytoplasm in the wild strains and, hence, the polyP of this compartment is somehow protected from them.
Table 3. Content of polyP in the cells of
S. cerevisiae strains overproducing polyphosphatases (% of the
polyP content in the parent strain cells)
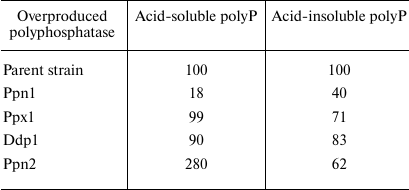
Note. The cells were grown on a selective YNB medium till the stationary
growth phase. The results are summarized from the works [53, 72-74].
Overexpression of polyphosphatases Ppn1 and Ppn2 had a marked effect on the physiology of S. cerevisiae cells. The Ppn1-overexpressing strain showed enhanced expression of the large groups of genes associated with the response to external stimuli, organization of the cytoplasmic membrane, and redox reactions [75]. As a result, resistance of this strain to oxidative and manganese stresses also increased [75]. The Ppn2-overproducing strain proved to be more resistant to alkali and peroxide exposure [53]. We suggest that the short-chain polyP can play a role of signaling molecules and participate in the maintenance of pH homeostasis in the cytoplasm; therefore, considerable decrease or increase in their level influences the adaptive potential of yeast cells. The obtained data show that the polyphosphatase-overproducing strains are a good model for studying the role of particular polyP fractions in the adaptation of yeast cells to different types of stresses.
PRACTICAL SIGNIFICANCE OF POLYPHOSPHATASES
Quantitative analysis of polyP in human blood and other biological samples is a pressing challenge crucial for monitoring the state of patients with cardiovascular diseases, especially with the enhanced risk of thrombosis, some types of blood clotting disorders, osteoporosis, and osteochondrosis. The necessity of controlling polyP levels in food products, natural waters and soils is dictated by the fact that these polymers are used as water deionizers, fertilizers, and permitted food additives in candies, frozen fish, seafood, meat and sausage products, and bakery products. It is well known that polyP can be absorbed in human intestinal tract [76]. In the developed countries, people get more inorganic phosphorous than before; however, the problem of its effect on health has so far been studied insufficiently.
Nowadays, the most sensitive and highly specific methods of polyP detection are based either on measuring fluorescence of the polyP complexes with DAPI or monitoring of enzymatic activity of the specific enzymes [77]. Yeast polyphosphates are more active than bacterial polyphosphatases by two orders of magnitude [78], which is an advantage, when they are used as analytical reagents. There are two enzymes suitable for polyP detection, Ppx1 and Ppn1, with the following properties: they are highly specific to polyP and have long shelf-life stored in a freezer in a medium containing Triton X-100 (according to our observations, for more than two years without loss of activity). These two enzymes have different substrate specificities: Ppx1 is more active with short-chain polyP, including tripolyphosphate, and Ppn1 is more active with long-chain polyP, which gives additional opportunities to estimate prevalence of the polymers with different chain lengths in the test samples. The superproducing strains and purification methods that we have developed provide a rather high yield of the enzymes: 1 g of yeast biomass can yield the amount of enzyme sufficient to conduct 200 tests [79]. The assay procedure is relatively simple: a sample is incubated at pH 7 with addition of magnesium or cobalt ions and an aliquot of the enzyme; next the increase in the phosphate level is determined by the standard colorimetric technique with nanomolar sensitivity [79]. Applications of Ppx1 for assaying polyP in biological objects has been described in detail in the literature [79-82]. With respect to endopolyphosphatases, they are of interest for producing polyP with the specified average chain length from commercial high-molecular weight polyphosphates. It is possible to obtain products with the chain length of about 60 phosphate residues by varying treatment conditions [52]. This is precisely the length of polyP present in mammals and, hence, required for the study of the effects of polyP on different biological processes [10, 11].
CONCLUSIONS
Yeasts possess a multicomponent system of polyP metabolism, including the proteins of VTC complex and four polyphosphatases with different properties, cellular localizations, and functions that belong to different protein families. Ppn1 and Ppn2 play the key role in the polyP metabolism in the wild type yeast cells. The role of Ddp1 is mainly to control the level of second messengers, inositol pyrophosphate and adenosine polyphosphates; the functions of Ppx1 require further studies. All polyphosphatases can to a certain extent hydrolyze the second messengers with ester bond, while Ppx1 and Ppn1 are able to cleave polyP from the lysine-polyphosphorylated proteins thereby regulating their activity. These properties of polyphosphatases indicate that they involve polyP in a wide range of regulatory processes in yeast cells, including regulation of the cell cycle and adaptation to stresses. Further studies of the functional role of these enzymes are important also due to the presence of structurally similar proteins in mammalian cells.
Yeast exopolyphosphatases could serve as promising analytical reagents for highly specific and sensitive analysis of polyP and for obtaining polyP of the specified length for the purposes of research and drug development.
Ethics declarations. The authors declare no conflict of interest in financial or any other sphere. This article does not contain any studies involving animals or human participants performed by any of the authors.
REFERENCES
1.Belozersky, A. N., and Kulaev, I. S. (1957)
Polyphosphates and their significance for the processes of the
development of Aspergillusniger, Biokhimiya, 22,
29-39.
2.Kulaev, I. S., and Belozersky, A. N. (1962)
Condensed inorganic phosphates in the metabolism of living organisms.
Part I, Izv. Akad. Nauk SSSR, 3, 354-368.
3.Kulaev, I. S., and Belozersky, A. N. (1962)
Condensed inorganic phosphates in the metabolism of living organisms.
Part II, Izv. Akad. Nauk SSSR, 3, 502-521.
4.Kulaev, I. S. (1975) The Biochemistry of
Inorganic Polyphosphates, MGU, Moscow, p. 246.
5.Kulaev, I. S. (1979) The Biochemistry of
Inorganic Polyphosphates, Chichester, John Willey & Sons, p.
255.
6.Rao, N. N., Gómez-García, M. R., and
Kornberg, A. (2009) Inorganic polyphosphate: Essential for growth and
survival, Ann. Rev. Biochem., 78, 605-647.
7.Clotet, J. (2017) Polyphosphate: popping up from
oblivion, Curr. Genet., 63, 15-18.
8.Omelon, S., Georgiou, J., Henneman, Z. J., Wise, L.
M., Sukhu, B., et al. (2009) Control of vertebrate skeletal
mineralization by polyphosphates, PLoS One, 4, e5634.
9.Müller, W. E., Wang, X., and Schröder, H.
C. (2017) New target sites for treatment of osteoporosis, Progr.
Mol. Subcell. Biol., 55, 187-219.
10.Ruiz, F. A., Lea, C. R., Oldfield, E., and
Docampo, R. (2004) Human platelet dense granules contain polyphosphate
and are similar to acidocalcisomes of bacteria and unicellular
eukaryotes, J. Biol. Chem., 279, 44250-44257.
11.Smith, S. A., and Morrissey, J. H. (2008)
Polyphosphate as a general procoagulant agent, J. Thromb.
Haemost., 6, 1750-1756.
12.Baker, C. J., Smith, S. A., and Morrissey, J. H.
(2019) Polyphosphate in thrombosis, hemostasis, and inflammation,
Res. Pract. Thromb. Haemost., 3, 18-25.
13.Holmstrom, K. M., Marina, N., Baev, A. Y., Wood,
N. W., Gourine, A. V., and Abramov, A. Y. (2013) Signaling properties
of inorganic polyphosphate in the mammalian brain, Nat. Commun.,
4, 1362, doi: 10.1038/ncomms2364.
14.Angelova, P. R., Iversen, K. Z., Teschemacher, A.
G., Kasparov, S., Gourine, A. V., and Abramov, A. Y. (2018) Signal
transduction in astrocytes: localization and release of inorganic
polyphosphate, Glia, 66, 2126-2136.
15.Lempart, J., and Jakob, U. (2019) Role of
polyphosphate in amyloidogenic processes, Cold Spring Harb.
Perspect. Biol., 11, a034041, doi:
10.1101/cshperspect.a034041.
16.Seidlmayer, L. K., Juettner, V. V., Kettlewell,
S., Pavlov, E. V., Blatter, L. A., and Dedkova, E. N. (2015) Distinct
mPTP activation mechanisms in ischaemia-reperfusion: contributions of
Ca2+, ROS, pH, and inorganic polyphosphate, Cardiovasc.
Res., 106, 237-248.
17.Baev, A. Y., Negoda, A., and Abramov, A. Y.
(2017) Modulation of mitochondrial ion transport by inorganic
polyphosphate – essential role in mitochondrial permeability
transition pore, J. Bioenerg. Biomembr., 49, 49-55.
18.Pilliar, R. M., Kandel, R. A., Grynpas, M. D.,
Theodoropoulos, J., Hu, Y., et al. (2017) Calcium polyphosphate
particulates for bone void filler applications, J. Biomed. Mater.
Res. B Appl. Biomater., 105, 874-884.
19.Müller, W. E. G., Wang, S., Tolba, E.,
Neufurth, M., Ackermann, M., et al. (2018) Transformation of amorphous
polyphosphate nanoparticles into coacervate complexes: an approach for
the encapsulation of mesenchymal stem cells, Small, 14,
e1801170, doi: 10.1002/smll.201801170.
20.Wang, Y., Li, M., Li, P., Teng, H., Fan, D., et
al. (2019) Progress and applications of polyphosphate in bone and
cartilage regeneration, Biomed. Res. Int., 27, 5141204,
doi: 10.1155/2019/5141204.
21.Kalathottukaren, M. T., Abraham, L., Kapopara,
P., Lai, B. F., Shenoi, R. A., et al. (2017) Alteration of blood
clotting and lung damage by protamine are avoided using the heparin and
polyphosphate inhibitor UHRA, Blood, 129, 1368-1379.
22.Pestov, N. A., Kulakovskaya, T. V., and Kulaev,
I. S. (2004) Inorganic polyphosphate in mitochondria of
Saccharomyces cerevisiae at phosphate limitation and phosphate
excess, FEMS Yeast Res., 4, 643-648.
23.Tammenkoski, M., Koivula, K., Cusanelli, E.,
Zollo, M., Steegborn, C., et al. (2008) Human metastasis regulator
protein H-prune is a short-chain exopolyphosphatase,
Biochemistry, 47, 9707-9713.
24.McGrath, J. W., Kulakova, A. N., Kulakov, L. A.,
and Quinn, J. P. (2005) In vitro detection and characterization
of a polyphosphate synthesising activity in the yeast Candida
humicola G-1, Res. Microbiol., 156, 485-491.
25.Hooley, P., Whitenhead, M. P., and Brown, M. R.
(2008) Eucaryote polyphosphate kinases: is the “Kornberg”
complex ubiquitous? Trends Biochem. Sci., 33,
577-582.
26.Whitehead, M. P., Hooley, P. W., and Brown, M. R.
(2013) Horizontal transfer of bacterial polyphosphate kinases to
eukaryotes: implications for the ice age and land colonization, BMC
Res. Notes, 6, 221, doi: 10.1186/1756-0500-6-221.
27.Shabalin, Yu. A., Vagabov, V. M., Tsiomenko, A.
B., Zemlyanukhina, O. A., and Kulaev, I. S. (1977) The study of
polyphosphate kinase activity in yeast vacuoles, Biokhimiya,
42, 1642-1648.
28.Boyce, K. J., Kretschmer, M., and Kronstad, J. W.
(2006) The vtc4 gene influences polyphosphate storage,
morphogenesis, and virulence in the maize pathogen Ustilago
maydis, Eucaryot. Cell, 5, 1399-1409.
29.Ogawa, N., DeRisi, J., and Brown, P. O. (2000)
New components of a system for phosphate accumulation and polyphosphate
metabolism in Saccharomyces cerevisiae revealed by genomic
expression analysis, Mol. Biol. Cell, 11, 4309-4321.
30.Müller, O., Bayer, M. J., Peters, C.,
Andersen, J. S., Mann, M., and Mayer, A. (2002) The Vtc proteins in
vacuole fusion: coupling NSF activity to V(0) trans-complex
formation, EMBO J., 21, 259-269.
31.Müller, O., Neumann, H., Bayer, M. J., and
Mayer, A. (2003) Role of Vtc proteins in V-ATPase stability and
membrane trafficking, J. Cell Sci., 116, 1107-1115.
32.Hothorn, M., Neumann, H., Lenherr, E. D., Wehner,
M., Rybin, V., et al. (2009) Catalytic core of a membrane-associated
eucaryotic polyphosphate polymerase, Science, 324,
513-516.
33.Gerasimaitė, R., and Mayer, A. (2016)
Enzymes of yeast polyphosphate metabolism: structure, enzymology and
biological roles, Biochem. Soc. Trans., 44, 234-239.
34.Wild, R., Gerasimaite, R., Jung, J. Y.,
Truffault, V., Pavlovic, I., Schmidt, A., et al. (2016) Control of
eukaryotic phosphate homeostasis by inositol polyphosphate sensor
domains, Science, 352, 986-990.
35.Gerasimaitė, R., Pavlovic, I., Capolicchio,
S., Hofer, A., Schmidt, A., et al. (2017) Inositol pyrophosphate
specificity of the SPX-dependent polyphosphate polymerase VTC, ACS
Chem. Biol., 12, 648-653.
36.Desfougères, Y., Gerasimaitė, R. U.,
Jessen, H. J., and Mayer, A. (2016) Vtc5, a novel subunit of the
vacuolar transporter chaperone complex, regulates polyphosphate
synthesis and phosphate homeostasis in yeast, J. Biol. Chem.,
291, 22262-22275.
37.Shabalin, Yu. A., and Kulaev, I. S., (1989)
Solubilization and properties of yeast dolichyl
diphosphate:polyphosphate phosphotransferase, Biokhimiya,
54, 68-75.
38.Kulaev, I. S., Shimona, O., and Bobyk, M. A.
(1968) On the biosynthesis of inorganic polyphosphates in
Neurosporacrassa, Biokhimiya, 33, 419-443.
39.Andreeva, N. A., and Okorokov, L. A. (1993)
Purification and characterization of highly active and stable
polyphosphatase from Saccharomyces cerevisiae cell envelope,
Yeast, 9, 127-139.
40.Wurst, H., Shiba, T., and Kornberg, A. (1995) The
gene for a major exopolyphosphatase of Saccharomyces cerevisiae,
J. Bacteriol., 177, 898-906.
41.Sethuraman, A., Rao, N. N., and Kornberg, A.
(2001) The endopolyphosphatase gene: essential in Saccharomyces
cerevisiae, Proc. Natl. Acad. Sci. USA, 98,
8542-8547.
42.Andreeva, N. A., Kulakovskaya, T. V., and Kulaev,
I. S. (2006) High molecular mass exopolyphosphatase from the cytosol of
the yeast Saccharomyces cerevisiae is encoded by the PPN1
gene, Biochemistry (Moscow), 71, 975-977, doi:
10.1134/S0006297906090045.
43.Lonetti, A., Szijgyarto, Z., Bosch, D., Loss, O.,
Azevedo, C., and Saiardi, A. (2011) Identification of an evolutionarily
conserved family of inorganic polyphosphate endopolyphosphatases, J.
Biol. Chem., 286, 31966-31974.
44.Gerasimaitė, R., and Mayer, A. (2017) Ppn2,
a novel Zn2+-dependent polyphosphatase in the
acidocalcisome-like yeast vacuole, J. Cell Sci., 130,
1625-1636.
45.Andreeva, N., Ledova, L., Ryazanova, L.,
Tomashevsky, A., Kulakovskaya, T., and Eldarov, M. (2019) Ppn2
endopolyphosphatase overexpressed in Saccharomyces cerevisiae:
comparison with Ppn1, Ppx1, and Ddp1 polyphosphatases,
Biochimie, 163, 101-107.
46.Kizawa, K., Aono, T., and Ohtomo, R. (2017)
PHO8 gene coding alkaline phosphatase of Saccharomyces
cerevisiae is involved in polyphosphate metabolism, J. Gen.
Appl. Microbiol., 62, 297-302.
47.Ugochukwu, E., Lovering, A. L., Mather, O. C.,
Young, T. W., and White, S. A. (2007) The crystal structure of the
cytosolic exopolyphosphatase from Saccharomyces cerevisiae
reveals the basis for substrate specificity, J. Mol. Biol.,
371, 1007-1021.
48.Shi, X., and Kornberg, A. (2005)
Endopolyphosphatase in Saccharomyces cerevisiae undergoes
post-translational activations to produce short-chain polyphosphates,
FEBS Lett., 579, 2014-2018.
49.Andreeva, N. A., Kulakovskaya, T. V., and Kulaev,
I. S. (2004) Purification and properties of exopolyphosphatase from the
cytosol of Saccharomyces cerevisiae not encoded by the PPX1
gene, Biochemistry (Moscow), 69, 387-393.
50.Andreeva, N., Lichko, L., Trilisenko, L.,
Kulakovskiy, I. V., and Kulakovskaya, T. (2016) Yeast polyphosphatases
PPX1 and PPN1: properties, functions, and localization, in Inorganic
Polyphosphates in Eukaryotic Cells (Kulakovskaya, T., Pavlov, E.,
and Dedkova, E. N., eds.) Springer, Cham, pp. 15-33.
51.Andreeva, N. A., Kulakovskaya, T. V., and Kulaev,
I. S. (1996) Purification and characterization of polyphosphatase from
Saccharomyces cerevisiae cytosol, Biochemistry (Moscow),
61, 1213-1220.
52.Andreeva, N., Trilisenko, L., Eldarov, M., and
Kulakovskaya, T. (2015) Polyphosphatase PPN1 of Saccharomyces
cerevisiae: switching of exopolyphosphatase and endopolyphosphatase
activities, PLoS One, 10, e0119594, doi:
10.1371/journal.pone.0119594.
53.Ryazanova, L. P., Ledova, L. A., Andreeva, N. A.,
Zvonarev, A. N., Eldarov, M. A., and Kulakovskaya, T. V. (2020)
Inorganic polyphosphates and peculiarities of physiology of the yeast
Saccharomyces cerevisiae under overexpression of the PPN2 gene,
Biokhimiya, 85, 598-606, doi:
10.31857/S0320972520040120.
54.Kulakovskaya, T. V., Andreeva, N. A., and Kulaev,
I. S. (1997) Adenosine-5′-tetraphosphate and
guanosine-5′-tetraphosphate – new substrates of the cytosol
exopolyphosphatase of Saccharomyces cerevisiae, Biochemistry
(Moscow), 62, 1180-1184.
55.Guranowski, A., Starzynska, E., Barnes, L. D.,
Robinson, A. K., and Liu, S. (1998) Adenosine 5′-tetraphosphate
phosphohydrolase activity is an inherent property of soluble
exopolyphosphatase from Saccharomyces cerevisiae, Biochim.
Biophys. Acta, 1380, 232-238.
56.Azevedo, C., Livermore, T., and Saiardi, A.
(2015) Protein polyphosphorylation of lysine residues by inorganic
polyphosphate, Mol. Cell, 58, 71-82.
57.Azevedo, C., Desfougères, Y., Jiramongkol,
Y., Partington, H., Trakansuebkul, S., et al. (2020) Development of a
yeast model to study the contribution of vacuolar polyphosphate
metabolism to lysine polyphosphorylation, J. Biol. Chem.,
295, 1439-1451.
58.Kulakovskaya, T. V., Andreeva, N. A., Karpov, A.
V., Sidorov, I. A., and Kulaev, I. S. (1999) Hydrolysis of
tripolyphosphate by purified exopolyphosphatase of Saccharomyces
cerevisiae cytosol: kinetic model, Biochemistry (Moscow),
64, 990-993.
59.Tammenkoski, M., Moiseev, V. M., Lahti, M.,
Ugochukwu, E., Brondijk, T., et al. (2007) Kinetic and mutational
analyses of the major cytosolic exopolyphosphatase from
Saccharomyces cerevisiae, J. Biol. Chem., 282,
9302-9311.
60.Lichko, L., Kulakovskaya, T., Pestov, N., and
Kulaev, I. (2006) Inorganic polyphosphates and exopolyphosphatases in
cell compartments of the yeast Saccharomyces cerevisiae under
inactivation of PPX1 and PPN1 genes, Biosci. Rep., 26,
45-54.
61.Andreeva, N. A., Kulakovskaya, T. V., and Kulaev,
I. S. (1998) Purification and properties of exopolyphosphatase isolated
from Saccharomyces cerevisiae vacuoles, FEBS Lett.,
429, 194-196.
62.Lichko, L. P., Kulakovskaya, T. V., and Kulaev,
I. S. (1998) Membrane-bound and soluble polyphosphatases of
mitochondria of Saccharomycres cerevisiae: Identification and
comparative characterization, Biochim. Biophys. Acta,
1372, 153-162.
63.Lichko, L. P., Kulakovskaya, T. V., and Kulaev,
I. S. (2000) Purification and characterization of a soluble
polyphosphatase from mitochondria of Saccharomyces cerevisiae,
Biochemistry (Moscow), 65, 355-361.
64.Lichko, L. P., Kulakovskaya, T. V., and Kulaev,
I. S. (2004) Partial purification and characterization of nuclear
exopolyphosphatase from Saccharomyces cerevisiae strain with
inactivated PPX1 gene encoding a major yeast exopolyphosphatase,
Biochemistry (Moscow), 69, 270-274.
65.Lichko, L. P., Kulakovskaya, T. V., and Kulaev,
I. S. (2010) Properties of partially purified endopolyphosphatase of
the yeast Saccharomyces cerevisiae, Biochemistry
(Moscow), 75, 1404-1407, doi: 10.1134/S0006297910110131.
66.Lichko, L., Kulakovskaya, T., and Kulaev, I.
(2004) Inactivation of endopolyphosphatase gene PPN1 results in
inhibition of expression of exopolyphosphatase PPX1 and
high-molecular-mass exopolyphosphatase not encoded by PPX1 in
Saccharomyces cerevisiae, Biochim. Biophys. Acta,
1674, 98-102.
67.Pestov, N. A., Kulakovskaya, T. V., and Kulaev,
I. S. (2005) Effects of inactivation of the PPN1 gene on
exopolyphosphatases, inorganic polyphosphates and function of
mitochondria in the yeast Saccharomyces cerevisiae, FEMS
Yeast Res., 5, 823-828.
68.Kulakovskaya, T. V., Andreeva, N. A., Trilisenko,
L. V., Vagabov, V. M., and Kulaev, I. S. (2004) Two exopolyphosphatases
in Saccharomyces cerevisiae cytosol at different culture
conditions, Process Biochem., 39, 1625-1630.
69.Andreeva, N. A., Kulakovskaya, T. V.,
Kulakovskaya, E. V., and Kulaev, I. S. (2008) Polyphosphates and
exopolyphosphatases in cytosol and mitochondria of Saccharomyces
cerevisiae during growth on glucose or ethanol under phosphate
surplus, Biochemistry (Moscow), 73, 65-69, doi:
10.1134/S0006297908010094.
70.Kulakovskaya, T. V., Trilisenko, L. V., Lichko,
L. P., Vagabov, V. M., and Kulaev, I. S. (2006) Effect of inactivation
of exopolyphosphatase genes PPX1 and PPN1 on the level of
polyphosphates of different fractions in Saccharomyces
cerevisiae, Mikrobiologiya, 75, 35-39.
71.Bru, S., Martínez-Laínez, J. M.,
Hernández-Ortega, S., Quandt, E., Torres-Torronteras, J., et al.
(2016) Polyphosphate is involved in cell cycle progression and genomic
stability in Saccharomyces cerevisiae, Mol. Microbiol.,
101, 367-380.
72.Eldarov, M. A., Baranov, M. V., Dumina, M. V.,
Shgun, A. A., Andreeva, N. A., et al. (2013) Polyphosphates and
exopolyphosphatase activities in the yeast Saccharomyces
cerevisiae under overexpression of homologous and heterologous
PPN1 genes, Biochemistry (Moscow), 78, 946-953,
doi: 10.1134/S0006297913080129.
73.Lichko, L. P., Eldarov, M. A., Dumina, M. V., and
Kulakovskaya, T. V. (2014) PPX1 gene overexpression has no influence on
polyphosphates in Saccharomyces cerevisiae, Biochemistry
(Moscow), 79, 1211-1215, doi: 10.1134/S000629791411008X.
74.Trilisenko, L. V., Andreeva, N. A., Eldarov, M.
A., Dumina, M. V., Kulakovskaya, T. V. (2015) Polyphosphates and
polyphosphatase activity in the yeast Saccharomyces cerevisiae
during overexpression of the DDP1 gene, Biochemistry
(Moscow), 80, 1312-1317, doi: 10.1134/S0006297915100120.
75.Trilisenko, L., Zvonarev, A., Valiakhmetov, A.,
Penin, A. A., Eliseeva, I. A., et al. (2019) The reduced level of
inorganic polyphosphate mobilizes antioxidant and manganese-resistance
systems in Saccharomyces cerevisiae, Cells, 8,
461, doi: 10.3390/cells8050461.
76.Segawa, S., Fujiya, M., Konishi, H., Ueno, N.,
Kobayashi, N., et al. (2011) Probiotic-derived polyphosphate enhances
the epithelial barrier function and maintains intestinal homeostasis
through integrin-p38 MAPK pathway, PLoS One, 6, e23278,
doi: 10.1371/journal.pone.0023278.
77.Solesio, M. E., and Pavlov, E. V. (2016) Methods
of inorganic polyphosphate (PolyP) assay in higher eukaryotic cells, in
Inorganic Polyphosphates in Eukaryotic Cells (Kulakovskaya, T.,
Pavlov, E., and Dedkova, E. N., eds) Springer, Cham, pp. 81-89.
78.Akiyama, M., Crooke, E., and Kornberg, A. (1993)
An exopolyphosphatase of Escherichia coli. The enzyme and its
ppx gene in a polyphosphate operon, J. Biol. Chem.,
268, 633-639.
79.Lichko, L., and Kulakovskaya, T. (2015)
Polyphosphatase PPX1 of Saccharomyces cerevisiae as a tool for
polyphosphate assay, Adv. Enzyme Res., 3, 61624, doi:
10.4236/aer.2015.34013.
80.Ohtomo, R., Sekiguchi, Y., Kojima, T., and Saito,
M. (2008) Different chain length specificity among three polyphosphate
quantification methods, Anal. Biochem., 383, 210-216.
81.Zvonarev, A. N., Crowley, D. E., Ryazanova, L.
P., Lichko, L. P., Rusakova, T. G., et al. (2017) Cell wall canals
formed upon growth of Candida maltosa in the presence of
hexadecane are associated with polyphosphates, FEMS Yeast Res.,
17, fox026.
82.Christ, J. J., and Blank, L. M. (2018) Enzymatic
quantification and length determination of polyphosphate down to a
chain length of two, Anal. Biochem., 548, 82-90.